Abstract
The efficient and timely degradation of proteins is crucial for many cellular processes and to maintain general proteostasis. The proteasome, a complex multisubunit protease, plays a critical role in protein degradation. Therefore, it is important to understand the assembly, regulation, and localization of proteasome complexes in the cell under different conditions. Fluorescent tags are often utilized to study proteasomes. A GFP-tag on the β5 subunit, one of the core particle (CP) subunits with catalytic activity, has been shown to be incorporated into proteasomes and commonly used by the field. We report here that a tag on this subunit results in aberrant phenotypes that are not observed when several other CP subunits are tagged. These phenotypes appear in combination with other proteasome mutations and include poor growth, and, more significantly, altered 26S proteasome localization. In strains defective for autophagy, β5-GFP tagged proteasomes, unlike other CP tags, localize to granules upon nitrogen starvation. These granules are reflective of previously described proteasome storage granules but display unique properties. This suggests proteasomes with a β5-GFP tag are specifically recognized and sequestered depending on physiological conditions. In all, our data indicate the intricacy of tagging proteasomes, and possibly, large complexes in general.
Similar content being viewed by others
Introduction
Proteasomes are the major protease involved in the selective degradation of proteins in eukaryotic cells. As many necessary cellular processes require the timely degradation of regulators, proteasomes are essential complexes1, 2. Proteasomes are composed of a regulatory particle (RP) that can be sub-divided into base and lid subcomplexes, and a core particle (CP). Assembly of the 66 subunit proteasome complex is a well-regulated process involving a number of proteasome-dedicated chaperones3. Mutations that impair proteasome assembly or function often cause severe growth defects or even lead to cell death under conditions of proteotoxic stress. For example, deletion of the proteasome assembly chaperones Rpn14 and Nas6, or truncation of the C-terminus of the proteasome subunit Rpt5, results in poor growth at high temperatures (37 °C) or in the presence of the arginine analog canavanine4,5. Similarly, in mammals, several mutations that alter proteasome function are responsible for a variety of pathologies6,7. However, not all proteasome mutations induce these phenotypes even if they compromise function. This is because there is some functional redundancy within the proteasome, and regulatory mechanisms exists that compensate for reduced proteasome activity by upregulating proteasome levels. For instance, when the proteasome assembly chaperones Hsm3 or Nas2 are deleted individually, cells typically grow at rates similar to wild type5,8,9. However, yeast harboring deletions of both chaperones are very sensitive to heat and other protein folding stresses. These phenotypes are further exacerbated when the ability to upregulate transcription of proteasome subunits, as a response to a loss in proteasome activity, is eliminated. In yeast, stabilization of the transcription factor Rpn4, which itself is a proteasome substrate, results in an upregulation of proteasome levels10,11,12. In all, cells require a certain threshold of proteasome activity that is lower under conditions of optimal growth and higher under conditions that induce proteotoxic stress. Consequently, mutations like the complete deletion of many subunits are lethal under all conditions. The threshold of proteasome activity under optimal conditions is never reached. However, other mutations, such as those discussed above, are tolerated under normal conditions where proteasome activity meets a minimal threshold. Under conditions that require higher proteasome activity, these mutants fail, thus, revealing phenotypes.
Proteasomes are primarily localized to the nuclei of dividing yeast13,14. This localization is routinely monitored using fluorescent tags on proteasome subunits. Indeed, the localization of proteasomes has been visualized with fluorescent tags on base, lid, and CP subunits15,16. A number of tags on proteasome subunits have also been used to purify these complexes in a variety of organisms17,18. Here, we report that tagging the active site subunit β5 (Pre2) resulted in tag specific phenotypes and poor cell growth when combined with other proteasome mutations. This was surprising as this tag has been used by a number of labs, including our own, and strains expressing β5-GFP show normal incorporation of this subunit into proteasomes, and exhibit normal behavior in several microscopic and biochemical assays19,20,21. We observed the relocalization of β5-GFP tagged proteasomes to cytosolic granules under a number of conditions. Some of these were consistent with proteasome storage granules (PSGs) that form under conditions of carbon starvation20,22,23. However, other properties distinguish these granules from PSGs. In all, our data suggest that cells can recognize β5-GFP proteasomes as abnormal and sequester them away from the nucleus in stressful conditions.
Results
GFP tagging of β5 results in altered proteasome localization
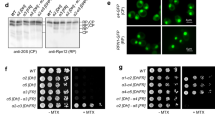
We have previously reported that autophagy of the CP and the RP occur through different pathways, a model that has been supported by subsequent data19,24,25. Our initial model was, in part, based on subcomplex localization in autophagy mutants; RP remained largely nuclear in strains deleted for ATG7 or ATG17, while CP appeared in cytosolic punctate structures19. These phenotypes were apparent when the β5 subunit of the proteasome CP was tagged with GFP19 (Fig. 1a, top panel). Unexpectedly, we observed little to no granules in atg7Δ strains where the CP subunit α1 or α6 was fused with GFP (Fig. 1a). To test if the phenotype was associated with tagging of β subunits or active site subunits specifically, we next tagged the other active site subunits, β1 and β2, as well as the non-catalytic subunit β4. The β1 and β2 active site subunits showed no granule formation in atg7Δ strains starved for nitrogen (supplementary Fig. S1), indicating this phenomenon was not specifically associated with active subunits. We did, however, observe granule formation with GFP-tagged β4, (supplementary Fig. S1). Considering β4 is next to β5 in the half CP26, this might indicate that this area of CP is particularly sensitive to tags, potentially due to conformational changes associated with efficient catalysis (see below and supplementary Fig. S2). α1-,α6- and β5-GFP tagged strains all showed normal autophagy upon nitrogen starvation in a wildtype background as observed by free GFP formation on immunoblots (Fig. 1c). Furthermore, microscopic analyses showed vacuolar localized fluorescence indicative of autophagic degradation (Fig. 1d). Thus, the difference in localization we observed in the atg7Δ background likely resulted from a tag-induced artifact under this specific condition for the GFP-tag on the β5 subunit. To further investigate this, we generated strains where we combined the different GFP tagged CP subunits in the atg7Δ background with the mCherry-tagged regulatory particle subunit Rpn1. The Rpn1-mCherry signal remains nuclear in atg7Δ cells that are starved for nitrogen and the presence of α1-GFP or α6-GFP did not change this localization (Fig. 1b). However, the presence of β5-GFP induced a relocalization of Rpn1-mCherry. As opposed to nuclear localization, Rpn1-mCherry co-localized with β5-GFP positive cytosolic puncta upon nitrogen starvation of these atg7Δ cells (Fig. 1b). This was not unique for Rpn1-mCherry and was the direct result of tagging β5 as tagging this subunit changed the localization of α1, Rpn5 and Rpn2 under this condition as well (supplementary Fig. S1). Our data show that all of these tagged subunits efficiently incorporate into active proteasomes with no obvious assembly defects (supplementary Fig. S2). In all this shows that α1-GFP, α6-GFP, β1-GFP, and β2-GFP, represent physiological CP localization upon nitrogen starvation when autophagy is disrupted. The C-terminal tagging of β5 not only changed the localization of CP, but also RP (as confirmed with base and lid tags), suggesting the tag caused a relocalization of proteasome holo-enzymes under our assay conditions.
GFP tagging of β5 results in altered proteasome localization. (a) atg7Δ strains expressing β5-GFP, α1-GFP or α6-GFP from the endogenous locus were starved for nitrogen for 24 h. before images were collected. (b) atg7Δ strains expressing Rpn1-mCherry in combination with either β5-GFP, α1-GFP or α6-GFP, were starved for nitrogen for 24 h. and imaged as in (a). (c) WT or ubp3Δ yeast expressing β5-GFP, α1-GFP or α6-GFP were starved for nitrogen for 24 h. Protein lysates were obtained using the alkaline lysis method, separated on SDS-PAGE, and immuno-blotted for GFP, and PGK1 (loading control). (d) WT or ubp3Δ yeast with β5-GFP, α1-GFP or α6-GFP tags were starved as in (a) and microscopy was performed. (e) β5-GFP atg7Δ yeast were starved for nitrogen or carbon for 24 h. Microscopy shows the β5-GFP signal largely in cytosolic granules. For recovery, cells from 10 mL of culture were harvested and transferred to 10 mL of rich media (YPD). Microscopy was performed at indicated times.
We previously reported that nitrogen starvation-induced autophagy of the CP, as monitored with a β5-GFP tag, was reduced upon deletion of UBP319. Surprisingly, we did not observe this to the same extent when using α1-GFP or α6-GFP to monitor CP as evident from the higher levels of free GFP detected on immunoblots for the latter two (Fig. 1c, compare lane 4, 8, and 12). Consistently, we observed substantial vacuolar fluorescence after nitrogen starvation for α1-GFP ubp3Δ and α6-GFP ubp3Δ strains upon microscopic analyses, while the vacuolar fluorescence for nitrogen starved β5-GFP ubp3Δ was strongly reduced (Fig. 1d). In all, our data indicate that Ubp3 is not as important for normal CP autophagy as we initially concluded. Nevertheless, we did observe some reduction in free GFP derived from α1-GFP or α6-GFP upon UBP3 deletion (Fig. 1c).This suggests that Ubp3 does play a modest role in CP autophagy, although we cannot exclude a minor defect in lysosomal targeting resulting from tag-induced effects. Consistent with this, Ubp3 appears to be important for proteasome autophagy under specific conditions including carbon starvation of blm10Δ cells25. In our hands, the GFP tag on β5 appears to increase proteasome dependence on Ubp3 for autophagy.
Proteasomes relocalize from cell nuclei to cytosolic puncta termed proteasome storage granules (PSGs) under conditions of carbon limitation22. To determine if the β5-GFP puncta had properties similar to proteasome storage granules (PSGs), we monitored their dynamics following a switch from minimal media lacking carbon or nitrogen to rich media (yeast extract, peptone, dextrose (YPD)). Upon reintroduction of carbon, PSGs dissipate within minutes and proteasomes re-localize to the nucleus quickly14,20,21,27. Under carbon starvation, our β5-GFP strain showed similar PSG dynamics, indicating this tag does not induce different behaviors for proteasomes in our strain background or with our assay conditions. Thus, β5-GFP granules that form under carbon starvation behave as normal PSGs and likely represent physiological granules (Fig. 1e). Unlike PSGs, granules induced upon nitrogen starvation in the β5-GFP tagged atg7Δ strain persisted up to 3 h following transfer to rich media (Fig. 1e). Thus, the granules that form in the atg7Δ strain under nitrogen starvation are distinct from PSGs and are putatively regulated by different factors.
GFP tagging of β5 results in less CP activity than untagged proteasomes
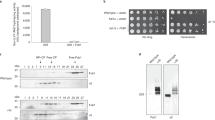
Although both CP and RP are present in PSGs, these subcomplexes are not proposed to be associated as they can be targeted to these structures independently21,22,25. Our data suggest that the whole 26S proteasome is present in the β5-GFP induced granules, as the β5-GFP tag caused RP to colocalize in those granules (Fig. 1b). Therefore, we reasoned that there were likely structural differences present in the 26S complex when β5 is tagged. To test this, we generated yeast expressing Rpn1-GFP or β5-GFP with a protein-A tag on Rpn11 for affinity purifications. When we purified these proteasomes from rich media, we did not detect any alterations in proteasome composition, indicating that all proteasome subunits were incorporated regardless of the subunit tagged (Fig. 2a). This is consistent with our previous analyses showing that these tagged subunits are efficiently incorporated into 26S proteasomes19. However, one possible explanation for translocation of 26S complexes in doubly tagged strains involves artificial 26S stabilization through the β5-GFP tag. Therefore, to test for differences in stability between 26S complexes, we removed nucleotide from the proteasome purifications using the enzyme apyrase in the presence or absence of the proteasome inhibitor MG132 (Fig. 2b). 26S proteasomes are unstable in the absence of ATP or ADP and proteasome inhibitors can stabilize 26S complexes in the absence of these nucleotides28. However, both β5-GFP and Rpn1-GFP tagged proteasome displayed RP-CP dissociation upon ATP depletion as reflected by the increased amount of free CP (Fig. 2b lanes 2 and 5). Thus, it does not appear that the β5-GFP proteasomes are more stable than Rpn1-GFP proteasomes. However, the data did seem to indicate a difference in proteasome activity. To pursue this further we analyzed whole cell lysate where β5-GFP tagged CP appeared to be less active that untagged CP (supplementary Fig. S2). One caveat here is that Rpn4 dependent upregulation of proteasomes might complicate the analysis, particularly because we observed a synergistic effect of combining β5-GFP and a deletion of RPN4 (Fig. 3a,b). While purifications have the disadvantage of introducing an additional tag, upon purification, this is removed by Tev protease cleavage and we can ensure equal proteasome amounts. . We tested the relative activity of these purified proteasomes by comparing the activity present in β5-GFP proteasomes to a dilution series of purified Rpn1-GFP proteasomes (Fig. 2c). Here, we observed that, indeed, β5-GFP proteasomes are less active than Rpn1-GFP proteasomes; 5 μg of purified Rpn1-GFP material appeared to contain an amount of 26S proteasomes equal to the β5-GFP containing prep based on the Coomassie stained gel; however, the activity of the β5-GFP proteasomes was reduced about twofold as it resembled the activity of 2.5 μg of the Rpn1-GFP proteasome preparation. Thus, the β5-GFP tag likely interferes with the catalytic activity of this subunit. In sum, it appears that β5-GFP tagged proteasomes are less active than proteasomes with an untagged β5 .
β5-GFP tagged proteasomes have reduced proteolytic activity. (a) Yeast strains with indicated tags were grown in YPD overnight. Cells were lysed and the protein A affinity purification tag on Rpn11 (Rpn11-ProA) enabled proteasome purification with IgG affinity resin. After elution, samples were separated by SDS-PAGE and gel stained using Coomassie brilliant blue (CBB). (b) Proteasomes purified in (a) were treated with apyrase (to deplete ATP and ADP) in the presence or absence of the proteasome inhibitor MG132. DMSO was used as a vehicle control (−). Samples were separated by native gels and peptidase activity was visualized using LLVY-AMC in gel activity assay in the presence (top) or absence of 0.02% SDS (to visualize free CP) and stained with CBB (right). (c) A sample of 10 μg β5-GFP purified proteasomes was compared to a dilution series of Rpn1-GFP purified proteasomes using native gel analysis and visualized using LLVY-AMC activity in the presence of 002% SDS (left) and CBB (right).
GFP tagging of β5 results in growth phenotypes in yeast. (a) Colonies derived from a tetrad dissection where one spore contained both the β5-GFP tag together with a deletion of RPN4 (indicated with square). Images from plate 2 days (left) and 4 days (right) post dissection. The dramatically slower growth was only observed for β5-GFP rpn4Δ (n = 4). Note changed colony shape for 2nd to 4th colonies was caused by re-streaking. (b) Indicated strains were grown to log phase in rich media. The equivalent of 1 mL of cells at OD600 1 was harvested, washed and resuspended in sterile water. Fourfold serial dilutions were transferred onto YPD plates, plates lacking arginine, or lacking arginine and containing indicated concentration of canavanine using a pin array. Plates were grown at 30 °C or the indicated temperature. (c) as (b) for indicated strains and conditions. (d) β4-protein A tagged cells expressing α1-CFP, α2-CFP or β2-YFP were grown to log phase in YPD media, then switched to carbon starvation media. Microscopy was performed at indicated times. (e) WT and NAS2 HSM3 double deletion strains expressing Rpn1-GFP or β5-GFP were grown at 37 °C in YPD for indicated time and analyzed by microscopy. Quantification of the percentage of cells with granules and percent unbudded were performed manually using FIJI.
GFP tagging of β5 results in growth phenotypes in yeast
Phenotypically, reduced proteolytic activity commonly presents as sensitivity to high temperature or the arginine analog canavanine, as these conditions induce proteotoxic stress. However, these phenotypes are not always apparent due to upregulation of proteasomes by the transcription factor Rpn4. Rpn4 levels increase under conditions of reduced proteasome activity, because Rpn4 is also a proteasome substrate. As a result, any compromised proteasome function resulting from mutated subunits is often masked by Rpn4-dependent transcriptional upregulation of proteasome genes8,10,11. Our efforts to generate a β5-GFP tagged strain, also harboring a deletion of RPN4, using standard yeast transformation methods were not successful. On the other hand, a deletion of RPN4 was readily obtained in the Rpn1-GFP background. To test if this indicated a synthetic lethality between β5-GFP and rpn4Δ, we generated a diploid strain harboring β5-GFP and rpn4Δ single mutations. While tetrad dissections of this diploid showed that cells with both β5-GFP and a deletion of RPN4 were viable, we did observe very slow growth, exclusively for those cells (Fig. 3a, colony on the left). Next, we tested the growth of the single and double mutants under conditions of proteolytic stress. Strains deleted for RPN4 are known to show slow growth and some sensitivity to proteolytic stress12,29, which was exacerbated by the β5-GFP tag (Fig. 3b). Furthermore, β5-GFP alone caused growth defects in the presence of high levels of canavanine. These phenotypes were specific to the β5-GFP tagged strains, as they were not observed with the Rpn1-GFP tag. This was consistent with our observations of reduced proteasome activity for β5-GFP tagged proteasomes (Fig. 2c).
To test if these properties were associated with any other tagged CP subunits or specifically associated with β5, we compared growth under these conditions for GFP tagged β5, α1, α4, or α6. We did not observe any compromised growth when strains contained C-terminal GFP tags on subunits α1, α4, or α6. This shows that the tagging the α1, α4, or α6 CP subunits did not lead to any obvious defects in proteasome function. Indeed, even in a rpn4Δ background, α1-GFP showed no increased sensitivity (Fig. 3c). However, when we performed tetrad dissections to combine these different fluorescently tagged CP subunits with a C-terminal truncation of Rpn11 which has similarities to the Rpn11 mutation found in mpr1-1 or rpn11-m1 strains30,31, we observed synthetic lethality for the Rpn11 truncation with GFP-tagged α1, α2, and β5 (not shown). Only the combination of Rpn11 truncation with α4-GFP was well tolerated by cells. With respect to localization, we have observed altered behavior when either α1 or α2 were tagged with CFP in combination with a protein A tag on β4. As mentioned previously, proteasomes exit the nucleus and localize to PSGs in the cytoplasm during carbon starvation; however, strains with an α1- or α2-CFP tag, in combination with a β4 protein A tag, failed to form granules following 24 h of starvation (Fig. 3d). Like β5-GFP, β4-GFP caused mislocalization of proteasomes in an ATG7 deleted background following nitrogen starvation. This combined with the observation that β4-proA affects granule formation under carbon starvation supports the idea of this area of the CP being particulary sensitive to tagging. Interestingly, proteasomes with a β4 protein A tag in combination with β2-YFP formed granules similarly to what has previously been described, suggesting β4 protein A alone may not be responsible for failure in granule formation upon carbon starvation but more of cumulative effect of where the proteasome is tagged. In all, it appears that certain combinations of tagged or mutant proteasome subunits can affect both cell viability and proteasome localization following certain stress conditions. More specifically, the commonly used GFP-tag on the active site subunit β5 in yeast results in phenotypes typically observed in strains with compromised proteasome function.
Besides these various mutation combinations and disruption of RPN4, proteasome chaperone mutations are also known to affect cell viability under conditions of protein stress4,8,9. To determine if a GFP tag on β5 or Rpn1 would exacerbate these phenotypes, we generated strains harboring a deletion of the regulatory particle chaperones NAS2 and HSM3 in a background expressing Rpn1-GFP or β5-GFP. Under logarithmic growth at 37 °C, the majority of GFP fluorescence in the WT Rpn1-GFP and β5-GFP strains was nuclear (Fig. 3e). Upon deletion of NAS2 and HSM3, the fluorescent signal in cells appears more diffuse and cytoplasmic for both Rpn1-GFP and β5-GFP, perhaps suggesting that interfering with proteasome assembly reduces the efficiency of proteasome nuclear import. Indeed, it has been shown that excess Rpt subunits were sequestered into cytosolic puncta when proteasome assembly was compromised9. The cytosolic enrichment of GFP in our NAS2 and HSM3 deletion mutants might result from Rpn1 and β5 being incorporated into proteasomes at reduced rates. When these strains were grown for 24 h at 37 °C, to induce proteolytic stress, there was no apparent growth defect resulting from the introduction of the Rpn1-GFP tag. The nas2Δ hsm3Δ cells with β5-GFP, on the other hand, appeared to be dying (see below). Microscopic analyses showed that for the Rpn1-GFP tagged strains, localization remained largely the same at both 4 h and 24 h of growth at 37° C. For the strain lacking NAS2 and HSM3, both timepoints showed less nuclear and more cytoplasmic fluorescence. In contrast, 24 h of growth at 37° C showed significant differences for the β5-GFP fluorescent signal. In the wildtype background, β5-GFP relocated from the nucleus to cytosolic puncta. Here, 38% of cells with β5-GFP tagged formed granules compared to 6% for Rpn1-GFP tagged cells. These granules appear similar to those observed under nitrogen starvation in a β5-GFP atg7Δ strain. In the β5-GFP tagged hsm3Δ nas2Δ strain, the fluorescence signal was close to background levels, and interestingly, the majority of cells failed to complete budding (97% were unbudded compared to only 10% in Rpn1-GFP tagged hsm3Δ nas2Δ). Many cells also appeared to be dead or dying. In summary, mutations that compromise proteasome levels and cell viability under stress are exacerbated when combined with a β5-GFP tag.
Properties of β5-GFP granule formation
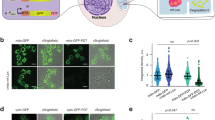
As the β5-GFP granules formed upon nitrogen starvation in strains that were defective in autophagy (by a deletion of ATG7, ATG17, or UBP3), we reasoned these granules might represent stalled autophagic cargo. To analyze this, we first monitored how quickly β5-GFP granules formed upon nitrogen starvation in β5-GFP atg7Δ cells. Granule formation was relatively slow as we did not detect granules at 4 h (Fig. 4a). At 8 h we detected granules, but they were not as abundant or intense as compared to 24 h post starvation initiation. Autophagosomes, on the other hand, form within minutes following starvation and autophagic bodies can be detected as early as 2 h inside vacuoles32,33. Furthermore, Atg8, a protein found in autophagosomes, did not accumulate in the granules we observed (Fig. 4b). The latter was consistent with the published literature as Atg7 is required in this process34,35. Thus, β5-GFP specific granules that formed after nitrogen starvation in strains defective for autophagy are likely not failed cargo since autophagosomes cannot form in this condition. Instead, the β5-GFP granules could be similar to granules that form upon proteasome inhibition and encompass aggregated proteasomes36. As this pathway depends on Hsp42, we deleted HSP42 in the β5-GFP atg7Δ background and monitored granule formation (Fig. 4c). Since we observed no difference in the appearance or amount of β5-GFP granules that formed following nitrogen starvation, the β5-GFP granules that formed following nitrogen starvation do not represent an intermediate in the Hsp42-dependent pathway of inactive proteasome autophagy.
Properties of β5-GFP granule formation. (a) atg7Δ strains expressing β5-GFP were starved for nitrogen as in Fig. 1a and analyzed microscopically at indicated times. (b) atg7Δ yeast expressing β5-mCherry and GFP-Atg8 were starved for nitrogen and analyzed by microscopy at the indicated times. (c) WT, atg7Δ, hsp42Δ and atg7Δhsp42Δ yeast expressing β5-GFP were starved for nitrogen as above or grown in rich media at 37 °C for 24 h. Quantification of the percentage of cells with granules was performed manually using FIJI. (d) β5-GFP expressing yeast with atg7Δ or atg7Δsnx4Δ were starved for nitrogen for 24 h. Quantification of number of cells with nuclear GFP was carried out using FIJI.
Next, we examined the role of Snx4/Atg24, a sorting nexin that plays a role in proteasome autophagy upon nitrogen starvation24. Interestingly, α4-GFP atg8Δ cells used by Nemec et al. formed cytoplasmic punctate structures similar to our observations with a β5-GFP atg7Δ strain. These granular structures required the presence of Snx4, which led to the model that Snx4 triggered proteasome cytosolic agglomeration prior to autophagic engulfment. When we compared nitrogen starved β5-GFP atg7Δ with β5-GFP atg7Δ snx4Δ cells, we noticed that deletion of SNX4 largely prevents granular structures and the fluorescent signal remained predominantly nuclear (Fig. 4d). Considering the role of Snx4 in proteasome autophagy, these β5-GFP granules are possibly stalled autophagy cargo. These cargo granules must be a step prior to autophagosome formation because they can be observed in ATG7 and ATG17 deleted strains, which do not form autophagosomes. While these granules are not associated with autophagosomes, they perhaps are a prerequisite for proteasome CP autophagy. Another possibility is that these proteasomes are recognized as faulty, and Snx4 facilitates their cytosolic retention in granules. This seems more likely since other proteasome GFP reporters remained nuclear when autophagy was blocked. That said, Snx4 is also required for autophagy of proteasomes when the tag does not induce granular localization upon autophagy impairment. Here, Rpn2 and Rpn5 were not reported to be in cytosolic granules prior to autophagy, but required Snx4 for autophagic degradation24. In all, it appears that β5-GFP proteasomes are regulated by different machinery in yeast than proteasomes with other subunits tagged due to a synthetic effect of the tag under these specific conditions.
Discussion
Efforts by several labs in recent years have revealed a surprising complexity in the localization of proteasomes under different stress conditions in a number of organisms9,19,20,24,25,37,38. Monitoring the dynamic nature of proteasome assembly, proteasome autophagy, and PSG formation has relied on fluorescent tags fused to particular subunits. However, due to its complex structure, numerous conformations, and the many regulators that bind the complex under different conditions1, particular care should be taken in deciding which proteasome subunits should be tagged and where. For instance, the C-terminal regions of the six base Rpt subunits should never be tagged as these C-termini are involved in crucial interactions with CP4,39,40,41. We have found that tagging the β5 subunits of the proteasome core particle, which has been used in several studies and often appears fully functional compared with untagged β5, altered 26S proteasome localization under specific conditions of cellular stress. We report that not only does this tag cause mis-localization of 26S proteasomes, but that proteasomes tagged at this subunit have reduced proteolytic activity compared to proteasomes that are tagged on the RP subunit Rpn1.
Considering that the β5 subunit contains the active site residues responsible for a major part of the proteolytic activity of the CP42, the reduced activity might not be completely surprising. To become proteolytically active, β5 needs to undergo autocatalytic pro-peptide processing. If this is compromised it could result in less chymotrypsin-like activity and an accumulation of a higher molecular weight β5-GFP on immunoblots of total lysate or purified proteasomes due to the presence of propeptide. While we observed a higher molecular weight species on western blots when blotting nitrogen starved cells for GFP (supplementary Fig. S4, black arrow), we did not detect β5-propeptide associated with these bands when using a specific antibody. We however cannot exclude the possibility that the pro-peptide form is below our detection limit under these assay conditions. Another possibility that may explain the observed reduction in activity is that tagging β5 with GFP results in less efficient CP assembly. While this could explain the growth phenotypes we observed, it would not explain the reduced activity for assembled purified proteasomes. The β5 active site is located on the inner surface of the barrel shaped CP, while the C-terminus, where GFP is fused, is outfacing. Based on the general thought that CP does not undergo dramatic conformational changes during the hydrolytic process, GFP tagging might not be expected to compromise function as long as it does not interfere with the ability of CP to interact with other complexes. As the latter does not appear to be the case, at least for the ability to bind RP, the tagging of β5 might impact subtle conformational changes that do occur in CP. For example, the engagement of the active site with an inhibitor is known to cause allosteric changes at the CP alpha ring where RP binds28,43. If the additional mass at the C-terminus of β5 (or β4) changes such dynamics, or changes relative abundance of the different forms of the 26S proteasome that have been observed44, it might change the fate of 26S complexes, uniquely in this background, under particular conditions. Such a subtle change might explain why β5 acts similar to other tagged proteasomes in one condition and is uniquely recognized in others. The regulation and localization of β5-tagged proteasomes described here may expose a pathway that recognizes aberrant or inactive proteasomes under particular conditions.
In conclusion, our data show that tags that appear innocuous based on several analyses can still interfere with particular functions. We think this is particularly the case for hubs of protein–protein interactions where different factors might associate with a complex under different conditions. Furthermore, it is important to realize that complexes with critical function in the cell are likely to have compensatory mechanisms in place that might be activated in tagged strains where detrimental effects of a tag may be masked. Thus, for important multi-subunit complexes, the use of a number of tags on different subunits might be critical. For the proteasome specifically, we did not observe tag induced phenotypes for the various assays we conducted with α1, α4,, α6, β1, or β2-GFP tagged CP as reporters. However, depending on the research question being addressed, tags that function well in one condition might not be ideal in another, in which case it is important to use multiple tags to show the same effects.
Materials and methods
Yeast strains
All strains used in this study are reported in supplementary table S1. Our background strains are DF5 derived haploids SUB61(Matα) or Sub62(MatA); (lys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1)45. Standard PCR based procedures (primers and plasmids presented in supplementary table S2) were used to delete specific genes from the genome or introduce sequence at the endogenous locus that resulted in the expression of C-terminal fusions of GFP, mCherry, or CFP46,47,48. Yeast dissections were performed using standard practices and desired strains were confirmed by drug selection as well as PCR based methods. GFP-atg8 expressing strains were generated by transformation with integrating BS-Ura3-GFP-Atg8, a gift from Zhiping Xie (Addgene plasmid #69194)49.
Yeast growth conditions
Overnight cultures of yeast were diluted to an OD600 of 0.5 and grown in yeast peptone dextrose (YPD) medium to an OD600 1.5 (approximately 4 h). To induce starvation, cultures growing logarithmically were centrifuged, washed with the respective starvation medium, re-inoculated at an OD600 of 1.5, and incubated at 30 °C with constant shaking. 37 °C cultures were grown in YPD medium. For proteasome purifications, 1.5 L of YPD were used to grow strains with a protein A tag on Rpn11. Cultures were harvested by centrifugation, washed with sterile water and frozen at − 80 °C until proteasome purifications were performed. Yeast phenotypes were assayed by growing cultures to log phase and harvesting 1 OD600. Pellets were washed and resuspended in sterile water. This suspension was serially diluted (fourfold), stamped to assay plates, and incubated at 30 °C unless otherwise indicated.
Protein lysates and electrophoresis
For western blotting, 2 OD600 of cells were collected at indicated times following starvation and stored at − 80 °C. The alkali lysis method was used as previously described, and 10 μL of lysate was loaded on SDS-PAGE50. Following electrophoreses, samples were transferred to PVDF membranes and immuno-blotted using antibodies against GFP (1:500; Roche, #11814460001), Pgk1 (1:10,000; Invitrogen, #459250) and β5-propeptide (1:2000, a generous gift from Dr. John Hanna at Harvard Medical School) followed by incubation with horseradish-peroxidase conjugated :secondary antibodies. Horseradish-peroxidase activity was visualized using the Immobilon Forte Western HRP substrate (Millipore) and images were acquired sing the G-box imaging system (Syngene) with GeneSnap or GeneSys software (version 1.7.2., https://www.syngene.com/). Data shown are representative of consistently observed trends from at least three independent biological replicates.
For native gel analyses, proteasomes were affinity purified using IgG resin as previously described18,51. Equal protein was loaded on gel and separated by electrophoresis (90 V, 2.5–3 h, 4 °C). The gels were then incubated with suc-LLVY-AMC (100 μg/mL) and imaged using a Syngene G-Box (UV excitation, 440 nm band-pass emission filter) to visualize peptidase activity of proteasome complexes. Samples for SDS-PAGE were prepared by mixing equal protein with 1/5 volume of 6X SDS sample buffer (10% SDS, 40% glycerol, 60 mM DTT, 345 mM Tris–HCl [ph. 6.8] 0.005% bromophenol blue). Apyrase treatment of these purified proteasomes was performed first by diluting samples in proteasome lysis buffer (50 mM Tris (pH 8.0), 5 mM MgCl2, 1 mM ATP, 1 mM EDTA), then adding 30 milliunits/μL apyrase and incubating for 45 min at 30 °C. Proteasome inhibitor (10 μM MG132) or DMSO as a control were added at the same time as apyrase. Samples were then run on native gels and visualized as described above. To monitor native proteasome composition and activity from whole cell lysate, the cryogrinding method was used as previously describe51. Briefly, 50 OD600 of yeast cultures were collected, washed, and frozen dropwise in liquid nitrogen. These pellets were ground using mortar and pestle in the presence of liquid nitrogen then resuspended in lysis buffer (50 mM Tris (pH 7.5), 5 mM MgCl2, 1 mM ATP, 1 mM EDTA). Equal volumes of lysate were loaded on native gels. Following electrophoreses, native gels were imaged for GFP (excitation with blue LED and 525 nm long pass emission filter) and mCherry (with green LED and 605 nm long pass emission filter) using G:Box mini from Syngene. The gels were then incubated with suc-LLVY-AMC (100 μg/mL) and imaged on the G:Box mini to visualize peptidase activity of proteasome complexes (UV excitation, 440 nm long pass emission filter).
Fluorescence microscopy
All microscopy was done with live yeast cells where proteasome subunits were fluorescently tagged (Rpn1-GFP, Rpn5-GFP, Rpn2-mCherry, α1-GFP, α6-GFP, β1-GFP, β2-GFP, β4-GFP, β5-GFP, or β5-mCherry) at their endogenous locus with expression driven by the endogenous promotor. After indicated treatments, approximately 2 ODs of cells were pelleted, washed with PBS, then resuspend in 30 μL of PBS, and 3 μL mounted on 1% soft agar slides as described by Muller52 (https://www.youtube.com/watch?v=ZrZVbFg9NE8). All imaging by fluorescence microscopy was done within 10 min following the wash step to avoid changes in localization due to prolonged incubation on slides. Images were acquired at room temperature using a Nikon Eclipse TE2000-S microscope at 600 × magnification with a Plan Apo 60 ×/1.40 objective equipped with a Retiga R3 camera (Q-Imaging). Images were collected using Metamorph software (Molecular Devices) and analyzed using FIJI.
Abbreviations
- CP:
-
Core particle
- RP:
-
Regulatory particle
References
Bard, J. A. M. et al. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018).
Finley, D., Chen, X. & Walters, K. J. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem. Sci. 41, 77–93 (2016).
Tomko, R. J. & Hochstrasser, M. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 82, 415–445 (2013).
Lee, S.Y.-C., De la Mota-Peynado, A. & Roelofs, J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J. Biol. Chem. 286, 36641–36651 (2011).
De La Mota-Peynado, A. et al. The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J. Biol. Chem. 288, 29467–29481 (2013).
Gomes, A. V. Genetics of proteasome diseases. Scientifica (Cairo). 2013, 637629 (2013).
Schmidt, M. & Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 (2014).
Roelofs, J. et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 (2009).
Nahar, A., Fu, X., Polovin, G., Orth, J. D. & Park, S. Two alternative mechanisms regulate the onset of chaperone-mediated assembly of the proteasomal ATPases. J. Biol. Chem. 294, 6562–6577 (2019).
Dohmen, R. J., Willers, I. & Marques, A. J. Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim. Biophys. Acta Mol. Cell Res. 1773, 1599–1604 (2007).
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U. S. A. 98, 3056–3061 (2001).
Wang, X., Xu, H., Ju, D. & Xie, Y. Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics 180, 1945–1953 (2008).
Enenkel, C., Lehmann, A. & Kloetzel, P.-M. GFP-labelling of 26S proteasomes in living yeast: insight into proteasomal functions at the nuclear envelope/rough ER. Mol. Biol. Rep. 26, 131–135 (1999).
Laporte, D., Salin, B., Daignan-Fornier, B. & Sagot, I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181, 737–745 (2008).
Enenkel, C. Using native gel electrophoresis and phosphofluoroimaging to analyze GFP-tagged proteasomes. Methods Mol. Biol. 832, 339–348 (2012).
Wilkinson, C. R. M. et al. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 17, 6465–6476 (1998).
Kuo, C.-L., Collins, G. A. & Goldberg, A. L. Methods to rapidly prepare Mammalian 26S proteasomes for biochemical analysis. Methods Mol. Biol. 1844, 277–288 (2018).
Leggett, D. S., Glickman, M. H. & Finley, D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. in Ubiquitin-Proteasome Protocols 057–070 (Humana Press, 2005).
Waite, K. A., De La Mota-Peynado, A., Vontz, G. & Roelofs, J. Starvation induces proteasome autophagy with different pathways for core and regulatory particle. J. Biol. Chem. 291, 3239–3253 (2016).
Gu, Z. C. et al. Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol. Biol. Cell 28, 2479–2491 (2017).
Weberruss, M. H. et al. Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 32, 2697–2707 (2013).
Enenkel, C. The paradox of proteasome granules. Curr. Genet. 64, 137–140 (2018).
van Deventer, S., Menendez-Benito, V., van Leeuwen, F. & Neefjes, J. N-terminal acetylation and replicative age affect proteasome localization and cell fitness during aging. J. Cell Sci. 128, 109–117 (2014).
Nemec, A. A., Howell, L. A., Peterson, A. K., Murray, M. A. & Tomko, R. J. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J. Biol. Chem. 292, 21466–21480 (2017).
Marshall, R. S. & Vierstra, R. D. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. Elife 7, e34532, 1–38 (2018).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 (1997).
Peters, L. Z., Karmon, O., Miodownik, S. & Ben-Aroya, S. Proteasome storage granules are transiently associated with the insoluble protein deposit in Saccharomyces cerevisiae. J. Cell Sci. 129, 1190–1197 (2016).
Kleijnen, M. F. et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14, 1180–1188 (2007).
Ju, D., Wang, L., Mao, X. & Xie, Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 321, 51–57 (2004).
Rinaldi, T., Ricci, C., Porro, D., Bolotin-Fukuhara, M. & Frontali, L. A Mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol. Biol. Cell 9, 2917–2931 (1998).
Rinaldi, T. et al. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol. Biol. Cell 19, 1022 (2008).
Baba, M., Takeshige, K., Baba, N. & Ohsumi, Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913 (1994).
Xie, Z., Nair, U. & Klionsky, D. J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 (2008).
Tanida, I. et al. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367–1379 (1999).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Marshall, R. S., McLoughlin, F. & Vierstra, R. D. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 16, 1717–1732 (2016).
Li, J., Breker, M., Graham, M., Schuldiner, M. & Hochstrasser, M. AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLOS Genet. 15, e1008387 (2019).
Cohen-Kaplan, V. et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 113, E7490–E7499 (2016).
Smith, D. M. et al. Docking of the Proteasomal ATPases’ carboxyl termini in the 20S proteasome’s α ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007).
Stadtmueller, B. M. et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J. Biol. Chem. 285, 13–17 (2010).
Park, S., Tian, G., Roelofs, J. & Finley, D. Assembly manual for the proteasome regulatory particle: the first draft. Biochem. Soc. Trans. 38, 6–13 (2010).
Jäger, S., Groll, M., Huber, R., Wolf, D. H. & Heinemeyer, W. Proteasome β-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J. Mol. Biol. 291, 997–1013 (1999).
Haselbach, D. et al. Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs. Nat. Commun. 8, 15578 (2017).
Eisele, M. R. et al. Expanded coverage of the 26S proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Rep. 24, 1301–1315.e5 (2018).
Finley, D., Özkaynak, E. & Varshavsky, A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046 (1987).
Goldstein, A. L. & McCusker, J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Hailey, D. W., Davis, T. N. & Muller, E. G. D. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 351, 34–49 (2002).
Li, D. et al. A fluorescent tool set for yeast Atg proteins. Autophagy 11, 954–960 (2015).
Kushnirov, V. V. Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 (2000).
Roelofs, J., Suppahia, A., Waite, K. A. & Park, S. Native gel approaches in studying proteasome assembly and chaperones. Methods Mol. Biol. (Clifton, N.J.) 1844, 237–260 (2018).
Sundin, B. A., Chiu, C.-H., Riffle, M., Davis, T. N. & Muller, E. G. D. Localization of proteins that are coordinately expressed with Cln2 during the cell cycle. Yeast 21, 793–800 (2004).
Acknowledgements
We thank the Roelofs lab members for their support and valuable discussions. This work was supported by grants from the National Institutes of Health (National Institute of General Medical Science: K-INBRE program P20GM103418 and R01GM118660 to J.R.).
Author information
Authors and Affiliations
Contributions
Conceptualization, K.A.W. and J.R.; Methodology, K.A.W.; Investigation, K.A.W. and A.B.; Writing—Original Draft, K.A.W. and J.R.; Writing—Review & Editing, K.A.W., A.B. and J.R.; Funding Acquisition, J.R., Supervision, J.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Waite, K.A., Burris, A. & Roelofs, J. Tagging the proteasome active site β5 causes tag specific phenotypes in yeast. Sci Rep 10, 18133 (2020). https://doi.org/10.1038/s41598-020-75126-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75126-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.