Abstract
Sex determination in tephritid fruit flies involves a signaling cascade of alternatively spliced genes. The Transformer (TRA) and Transformer-2 (TRA-2) complex establishes an autoregulatory loop switching sex-specific splicing of tra pre-mRNA in females. The TRA/TRA-2 complex also regulates the sex-specific splicing of downstream effector genes, doublesex (dsx) and fruitless (fru). In Ceratitis capitata, a Maleness-on the-Y (MoY) gene modulates sex-specifically spliced Cctra pre-mRNA and results in the breakdown of the Cctra autoregulatory loop in males. In this study, the tra-2 and fru genes were characterised in two key pests, Bactrocera dorsalis and B. correcta. The tra-2 genes showed high degrees of conservation among tephritids. The complex gene organisation for each of Bdfru and Bcfru were identified. There are sex-specific and non sex-specific transcripts generated by alternative promoters as found in Drosophila melanogaster and other insects. RNAi knockdown of Bdtra transcripts showed that BdTRA controls the sex-specific splicing of Bddsx and Bdfru pre-mRNAs. Developmental expression analysis shows that multiple splice variants of Bdtra and Bctra RNAs are present before and during cellular blastoderm formation and that the mature sex-specific variants become fixed later in embryogenesis. Furthermore, the BddsxM splice variants are found in early embryos at the beginning of gastulation, but BdfruM does not appear until the larval stage. We proposed that the zygotic tra loop is initiated in both female and male embryos before becoming automatised or abolished by MoY, respectively.
Similar content being viewed by others
Introduction
Sexual reproduction is a key point for the continuation of multicellular eukaryotes. Males and females evolve into sexual dimorphism through the sexual selection processes1. Sex-determination mechanisms are highly diverse, and they usually occur during early development2. The sex determination of insects is one of the most diverse and well-characterised systems3,4. Its evolutionarily divergent model constitutes three levels of sex-determining regulators4. The primary signals are variable among insect species. Most of their signaling mechanisms are still unknown, but the signals are relayed via alternative splicing of a conserved binary switch gene. This switch gene, encoded for a splicing regulator, relays similar alternative splicing messages to the highly conserved downstream effectors.
Tephritid fruit flies are in a very diverse dipteran family comprising at least 5,000 species5. Several of them are high profile pests, including Ceratitis capitata (Mediterranean fruit fly), Bactrocera dorsalis (oriental fruit fly), B. oleae (olive fruit fly), B. tryoni (Queensland fruit fly), Anastrapha ludens (Mexican fruit fly), A. obliqua (West Indian fruit fly), and A. suspensa (Caribbean fruit fly). Many sex-determining orthologues have been characterised to understand the developmental cascades which are also beneficial for determining new pest control strategies6,7,8. For instance, genetic sexing strains have been developed by employing modern genetic approaches for pest control programs using the sterile insect technique (SIT)6,9,10,11,12,13.
Recently, the primary signal Maleness-on the-Y (MoY) has been discovered in C. capitata14. The MoY orthologues were also characterised in many Bactrocera spp. such as B. dorsalis, B. oleae, and B. tryoni. Disruption or overexpression of CcMoY can feminise XY embryos or masculinise XX embryos, respectively. CcMoY modulates sex-specific splicing of transformer (tra) pre-mRNA14. In tephritid female flies, the tra and transformer-2 (tra-2) orthologues encode the TRA and TRA-2 proteins that later form a TRA/TRA-2 complex to maintain an autoregulatory loop of the female-specifically spliced tra transcript (traF)15,16,17,18,19,20,21,22,23,24,25. The Bdtra22,23,24 and Bdtra-222,23 were previously characterised in B. dorsalis. The RNAi-mediated knock-down of any one of these genes generated only males because the autoregulatory loop was disrupted. Likewise, the autoregulatory loop is disrupted by the presence of MoY orthologues in males14. The TRA/TRA-2 complex also switches on the DoublesexF protein (DSXF) by regulating female-specific splicing of dsx pre-mRNA. The switching-off mode results in DSXM because the absence of the functional TRA/TRA-2 complex allows male-specific splicing of dsx pre-mRNA15,16,17,18,19,22,23,24. DSXF and DSXM are highly conserved functional effectors in insects26,27. They are transcriptional factors with a masculinised or feminised function for downstream genes, controlling somatic sexual dimorphism26,28. The dsx orthologues studied in several tephritid fruit flies (i.e., C. capitata29, B. tryoni30, B. oleae31, B. dorsalis32,33, B. correcta33, and A. obliqua34) are conserved, suggesting similar somatic sexual differentiation among them.
In a tephritid and remotely related insects, the TRA/TRA-2 complex switches were also found to control the sex-specific splicing of the other downstream effector gene, fruitless (fru), that confers the sexual behaviors of males18,35,36,37,38. However, the fru gene has not been characterised in detail among tephritid species. The molecular mechanisms of fru in sex determination were proposed based on non-tephritid models35,36,37,38,39,40,41. The fru genes are very complex loci encoding sex-specific and non sex-specific transcriptional factor isoforms42,43. FRU proteins generally contain a dimerisation-interface domain called the BTB (Broad-complex, Tramtrack and Bric-a-brac) domain and one alternative C2H2 zinc finger domain35,36,44,45. Each of the fru orthologues has many putative transcripts derived from any one of the alternative promoter-derived 5′ sex- and non sex-specific exons, which connect to four to five common exons and then connect to any one of the 3′ alternative zinc-finger encoding exons35,36,38,39,40,41. Notably, the sex-specifically spliced transcripts are only processed from the most distal promoter (P1) derived pre-mRNA in many non-tephritid species35,36,38,39,41. In Drosophila melanogaster, the non sex-specific FRU isoforms have developmental functions. They are translated from P2 to P4 transcripts46,47,48.
In this study, we report the gene structure and expression of tra-2 and fru orthologues in B. dorsalis and B. correcta. The RNAi knockdown experiments showed that BdTRA controls the sex-specific splicing of BdtraM, BddsxM, and BdfruM transcripts in pseudomales. In addition, a comprehensive sex-determination cascade was investigated from the developmental expression profiles of MoY, tra, tra-2, dsx, and fru in single embryos, larvae, pupae, and adults. These results likely ascertained the time frames of critical sex-determining events in these Bactrocera fruit flies.
Results
B. dorsalis and B. correcta transformer-2 (tra-2) orthologues
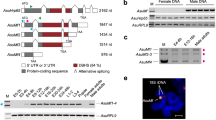
Full-length transcripts of B. dorsalis tra-2 (Bdtra-2) and B. correcta tra-2 (Bctra-2) are approximately 1.2 kilobases (kb) and 1.4 kb, respectively, in both sexes. The genomic DNA and cDNA sequences of Bdtra-2 (GenBank Acc. No. MT900512, MT900514) and Bctra-2 (GenBank Acc. No. MT900513, MT900515) were acquired from adult males and females. Both genes consist of eight exons (Fig. 1) and are structurally comparable to B. oleae (GenBank Acc. No. AJ547623), B. tryoni21, B. jarvisi21, Z. cucurbiate25, C. capitata18, Anastrepha spp.19,20, and D. melanogaster49,50. The open reading frames (ORF) of Bdtra-2 and Bctra-2 are 753 bp, encoding for a predicted polypeptide of 251 amino acids with 98% identity (Supplementary Table S1). The RNA recognition motif (RRM) region of BdTRA-2 and BcTRA-2 were identical within Bactrocera species and highly conserved across the tephritidae family (Supplementary Fig. S1). The nucleotide sequences of the Bdtra-2 are almost identical to the previously characterised Bdtra-222; however, 29 bp-longer and 88 bp-shorter sequences were found at the 5′- and 3′-UTRs, respectively.
Molecular structure of tra-2 genes from (a) B. dorsalis and (b) B. correcta. Both genes contain eight exons. Boxes and lines represent exons and introns, respectively. The white and yellow parts in the exons are untranslated and translated regions, respectively. The potential start codons (ATG) are indicated. Small black squares mark the potential stop codons.
B. dorsalis and B. correcta fruitless (fru) orthologues
A classical PCR-based approach was combined with available bioinformatics tools and genomic databases to predict the putative exon–intron organisation of the fru gene. The 5′-, 3′-RACEs and RT-PCRs were performed to assemble long transcripts of B. dorsalis fru (Bdfru) (GenBank Acc. No. MT900516–MT900519, MT900524–MT900528) and B. correcta fru (Bcfru) (GenBank Acc. No. MT900520–MT900523, MT900529–MT900533) genes. Both orthologues consist of three components (i.e., alternative P1 to P4 derived exons, the middle common exons, and four alternative 3′ ending exons (Fig. 2a and Supplementary Fig. S2). For B. dorsalis, the genomic sequence is available in the database; cDNA sequences of Bdfru transcripts were aligned against the genomic scaffold (GenBank Acc. No. NW_011876307) to analyse the gene organisation. The results suggested that the Bdfru gene structure is very large in size, spanning approximately 330 kb of genomic sequence (Fig. 2b). The genome sequence of B. correcta is not available. The putative Bcfru structure was thus predicted based on cDNA sequence and conceptual amino acid sequence alignments of Bcfru and Bdfru transcripts, putative donor–acceptor exon sequences, and conceptual amino acid position. The results showed the identical pattern of putative donor–acceptor nucleotide sequences on exons and conceptual amino acid sequences. This suggested that the predicted exon structure of Bcfru gene is similar to that of Bdfru gene (Fig. 2c and Supplementary Fig. S2). However, the availability of B. correcta genomic sequences would bring certainty to the proposed Bcfru gene organisation.
Gene organisation of B. dorsalis fru (Bdfru) and B. correcta fru (Bcfru) genes. The schematic drawings show (a) all possibilities for alternative splicing of fru transcripts and gene organisation of (b) Bdfru and (c) Bcfru (not to scale). The fru transcripts contain one of the four alternative 5′ exon blocks (i.e., sex-specific, U1, U2–U4, and U3 blocks), the common exons (i.e., C1 to C5 exons), and four alternative 3′ ends (i.e., ZnF A to ZnF D exons). P1 signifies the most distal promoter; its transcripts begin with a male-specific exon (M) and potential female-specific exon (F) as shown in dark gray and light gray boxes, respectively. P2 to P4 express non sex-specific transcripts which start with untranslated exons (U1–U4) as shown in the white boxes. The five common exons (yellow boxes) are the BTB domain coding exons (C1–C2) and the connecting exons (C3–C5). The zinc-finger encoding exons (ZnF A, ZnF B, ZnF C, and ZnF D) are in the green boxes. Boxes and lines represent exons and introns, respectively. Two putative translational start codons (ATG) are presented in the M and C1 exons. The potential translational in-frame stop codons are indicated by small black squares. The white and coloured parts in the exons represent untranslated and translated regions, respectively. The lengths of exons and putative introns are shown in black and gray numbers, respectively. The connection between F and common exons (dashed line) is still hypothetical because the RT-PCR experimental results were not positive.
At the 5′ portions of the Bdfru and Bcfru genes, five putative exons (male-specific exon (M) and untranslated exons (U1, U2, U3, and U4)) were found and spliced to the middle common exons in four different patterns (Fig. 2a and Supplementary Fig. S2). The genomic location of the four alternative 5′ exons are located many kb apart. This suggested that there are at least four putative promoters. The most distal promoter of Bdfru transcripts begin with an M exon, which is located 292 kb upstream of the C1 exon and appears to be a male-specifically spliced transcript (P1 transcripts) (Fig. 2b). The other three patterns are non sex-specific transcripts as P2 transcripts begin with the U1 exon located 249 kb upstream of the C1 exon, P3 transcripts begin with the U2 exon located 57 kb upstream of the C1 exon, and P4 transcripts begin with the U3 exon located 49 kb upstream of the C1 exon. The common C1–C2 and C3–C5 exons encode for the BTB domain and its connecting polypeptides, respectively (Fig. 2a). Additional complexity arises from the presence of four alternative 3′ end exons, which are zinc-finger encoding exons (ZnF A, ZnF B, ZnF C, and ZnF D) (Fig. 2a). The 3′ RACE and RT-PCR approaches were used to analyse the connection between each of the P1 to P4 derived exons and the alternative zinc-finger exons of Bdfru and Bcfru (Supplementary Table S2). The alternative ZnF A, ZnF B, and ZnF C exons were part of the P1 transcripts whereas the P2 and P3 transcripts consisted of all possible alternative zinc-finger exons. The P4 transcripts included the alternative ZnF A, ZnF C, and ZnF D exons. In addition, the M exon provides an extra start codon in the ORF resulting in an additional 64 amino acids at the amino terminal end of male-specifically spliced P1 transcripts (Supplementary Fig. S3). The other four exons (U1 to U4) are untranslated regions; therefore, ORFs of the P2, P3, and P4 transcripts may be translated with the putative start codon located in the BTB coding exon (C1).
In silico analysis of the female-specific exon and its cis-regulatory elements in Bdfru and Bcfru transcripts
To identify the sex-specific transcripts that were potentially derived from the P1 promoter, the forward primer located on the M exon and the reverse primer located on the common BTB exon C2 were used. A longer amplified product was expected to be detected in female flies due to the presence of a female-specific exon as reported in the other dipteran insects35,36,37,38,39,40,41. By contrast, in B. dorsalis and B. correcta, the amplified RT-PCR product was only observed in males. The bioinformatics approach was then used to identify the putative female-specific (F) exon and its cis-regulatory elements from the genomic scaffold. The 1.9 kb region downstream of the M exon was available in the B. dorsalis genomic scaffold (GenBank Acc. No. NW_011876307.1). The putative TRA/TRA-2 binding sites and multiple in-frame stop codons were in silico identified in this region (Supplementary Fig. S4). Both components were found to be clustered in an approximately 400 bp stretch adjacent to the M exon. PCR was performed using genomic DNA templates to verify this 400-bp region in B. dorsalis and B. correcta. The putative TRA/TRA-2 binding sites and multiple in-frame stop codons were found in B. dorsalis as expected and also in B. correcta (Supplementary Fig. S4a and S4b). The putative TRA/TRA-2 binding sites of Bdfru and Bcfru genes were comparable to the consensus sequence of TRA/TRA-2 binding sites located in the female-specific exon of the fru genes from the other insect species (Supplementary Fig. S4c). The in silico analysis suggested that the 400-bp region has the structural features of the putative F exon of Bdfru and Bcfru transcripts.
RT-PCR experiments were subsequently performed to prove the existence of the F exon (Supplementary Fig. S5). The cDNA templates were prepared from male and female adult heads of both species. Three forward primers (located on the M exon, putative junction of the M and F exons, and the putative F exon) were individually paired with a reverse primer (located on the putative F exon) (Supplementary Fig. S5a). The RT-PCR experiment showed a female-specific band when these primer combinations had been used (Supplementary Fig. S5b). This result suggested the existence of an F exon which is adjacent to 3′ end of the M exon. The connection between the F exon, and the common exon C2 could not be validated by the use of a C2-specific reverse primer in RT-PCR experiments (Supplementary Fig. S5d). Taken together, the in silico analysis and RT-PCR experiments suggested that the female-specific Bdfru and Bcfru transcripts contain potential in-frame stop codons that result in prematurely truncated female-specific ORF.
Bdtra gene regulated sex-specifically spliced Bddsx and Bdfru transcripts
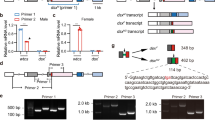
The endogenous Bdtra transcripts were knocked down in preblastoderm embryos by the microinjection of Bdtra dsRNA. The brown- and white-pupae genetic sexing strain (Salaya1) of B. dorsalis was used to identify adult pseudomales (males emerging from white pupae) (Fig. 3). Male-specifically spliced Bdtra, Bddsx, and Bdfru transcripts were detected in the same adult pseudomales although the female-specific isoforms of the Bdtra and Bddsx were still observed in contrast to the sex-specific patterns observed in the male- and female-wild-types as well as the Bdtra dsRNA treated brown-pupae males (Fig. 4). Bdtra-2 transcripts were detected in all cases. All of the adult pseudomales had male genitalia and mostly abnormal testes (Fig. 3). The results suggested that Bdtra RNAi can change the expression of Bddsx and Bdfru and generate masculinisation of the female embryos.
Expression analysis of Bdtra dsRNA treated and wild-type B. dorsalis. RT-PCRs of Bdtra, Bdtra-2, Bddsx, and Bdfru were performed to analyse the switch of splicing patterns from female to male resulting from the masculinisation by Bdtra dsRNA injection. The expression of Bdgapdh is used as a positive control. The expression patterns of the wild-type brown-pupae (BP) males and white-pupae (WP) females were compared with the Bdtra dsRNA treated brown-pupae males and white-pupae pseudomales. The primers used in this experiment are shown in Supplementary Tables S5 and S6. The full-length gels are presented in Supplementary Fig. S9.
The expression profiles of sex-determining genes during early embryonic, larval, pupal, and adult stages in the males and females of B. dorsalis and B. correcta
The early development stages of B. dorsalis and B. correcta embryos were very similar when observed from 0 to 9 h after egg laying (h AEL) (Supplementary Fig. S6). The pole cells and syncytial blastoderm appeared at 3 and 4 h AEL, respectively. Subsequently, the cellular blastoderm seemed to be completed by 7 to 8 h AEL.
The expression profiles of MoY, tra, tra-2, dsx, fru, and slow as molasses (slam) were studied in unfertilised eggs and during early embryonic development (1 to 12 h AEL) as well as in larval, pupal, and adult stages (Fig. 5). Gene- and/or sex-specific transcript primers were used for RT-PCR from single embryos, larvae, pupae, and adults. The embryos, larvae, and pupae were sexed by PCR using genomic DNA templates with Y-specific primers (Supplementary Table S6).
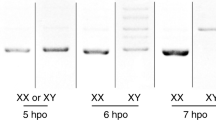
Expression analysis of sex-determining genes during the early stages of embryogenesis and other developmental stages in (a) B. dorsalis and (b) B. correcta. The cDNA samples were prepared from unfertilised eggs, embryos (1 to 12 h AEL), larvae, pupae, and adults. The presence of the slam gene indicates the zygotic transcription period in embryogenesis. Five sex-determining genes, MoY, tra, tra-2, dsx, and fru, were studied by RT-PCR analysis. In B. dorsalis (a), the male- and the female-specifically spliced transcripts (traM and traF) are 980 bp and 626 bp, respectively. In B. correcta (b), the male- and the female-specifically spliced transcripts (traM and traF) are 951 bp and 599 bp, respectively. For the dsx gene, the female-specifically spliced dsx transcript (dsxF) is 677 bp whereas the male-specific dsx transcript (dsxM) is 484 bp in both species. The male-specifically expressed fru transcript (fruM) appears in male samples at a length of 506 bp. Unexpected bands are the result of non-specific binding of primers. The gapdh gene was used as a positive control. The negative control was setup in the absence of RTase. The primers used in this experiment are shown in Supplementary Table S5 and S6. The full-length gels are presented in Supplementary Fig. S10.
In B. dorsalis, the expression of Bdslam began in 2 h AEL zygotes and continued throughout the adult stage (Fig. 5a). The slam seemed to be the first zygotically expressed gene. The presence of female-specifically spliced Bdtra (BdtraF) and Bddsx (BddsxF), and Bdtra-2 transcripts in unfertilised eggs shows that these transcripts are contributed maternally. The continued presence of the transcripts in developing embryos indicates zygotic activation of transcription of these genes. The expression of the MoY transcript was only observed in male samples beginning from 3 h AEL embryos, barely visible in larvae, and disappeared in pupae and adults. The combination of BdtraF, zygotic male-specifically spliced Bdtra transcripts (BdtraM), and their intermediate transcripts were firstly observed in both male and female samples at 5 h AEL. The intermediate transcripts consisted of three possible partial sequences of male-specific exon(s) (Supplementary Fig. S7). This observation became more obvious at 6 to 7 h AEL. This suggested that the activation of the zygotic tra loop had occurred in both sexes. In 8 h AEL embryos, the same combinations of the BdtraF and M and intermediate transcripts was observed in males. Conversely, the BdtraF transcripts were only a major product in female embryos, suggesting higher efficacy of the zygotic tra loop. Sex-specific splicing patterns of the Bdtra gene arose in 9 and 10 h AEL embryos. The female-specifically spliced pattern continued to at least 12 h AEL and in larvae, pupae, and adults. The male-specifically spliced counterpart also followed this trend, except that the mixture of BdtraF and M and intermediate transcripts reappeared once in the 11 h AEL male zygotes. The expression of the zygotic male-specifically spliced Bddsx transcript (BddsxM) appeared at the beginning of gastrulation (8 to 10 h AEL) and was detected at all later stages in males. The sex-specific splicing pattern of the Bddsx gene was established in the larvae, pupae, and adults. The male-specifically spliced Bdfru transcript (BdfruM) derived from the P1 promoter was observed in male samples from larval, pupal, and adult stages, but not in the embryonic stage. Notably, a non-specific RT-PCR amplicon was observed and sequenced. It was found to be against predicted B. dorsalis protein peanut and E3 ubiquitin-protein ligase genes in embryos when BLAST was used. The expression profiles of the other Bdfru transcripts derived from the P2, P3, and P4 promoters were also absent in the unfertilised eggs and embryos when tested in 4, 8, and 12 h AEL embryos (Supplementary Fig. S8a). Unlike the male-specifically spliced P1 transcripts, the other Bdfru transcripts were bisexually expressed in larvae, pupae, and adults. Nonetheless, all Bdfru transcript isoforms appeared during development later than the other sex-determining genes. All sex-determining orthologous genes from B. correcta showed similar expression profiles to those of the B. dorsalis species (Fig. 5b, Supplementary Fig. S7, and S8a). However, the appearance of Bcslam and BcdsxM occurred an hour after those of the B. dorsalis.
Discussion
The molecular organisations of Bdtra-2 and Bctra-2 are almost identical to both the previously characterised Bdtra-222,23 and orthologues from other tephritid fruit flies18,19,20,25, including Bactrocera spp.21. TRA-2 is a constitutive component required for TRA/TRA-2 complex for turning on the sex-determining-autoregulatory loop that generates female-specifically spliced tra transcripts in tephritids18,19,20,22. The TRA-2 proteins have two conserved RS-rich regions and an RRM domain which were proposed to be under the purifying selection process19,25 (Supplementary Table S1 and Fig. S1). This suggested the roles of TRA-2 in spliceosome formation49,51,52.
This work is the first study of fru genome organisation with various splicing patterns and expression profiles during development in tephritid fruit flies. The Bdfru and Bcfru orthologues are also one of the longest and most complex genes. The Bdfru gene spans approximately 330 kb. There are probably many transcripts with conserved splicing patterns, as in many insect species: D. melanogaster35,36, Anopheles gambiae39, Aedes aegypti41, Musca domestica38, and Nasonia vitripennis40. Alternative promoters can generate various 5′ exons that connect to the continuous array of four or five common exons before splicing with one of the different 3′ zinc-finger encoding exons.
The overall deduced amino acid sequences of the BdFRU and BcFRU are generally conserved, as in D. melanogaster. The BTB domain is the most conserved characteristic of all FRU proteins53,54. It is encoded near the beginning of the C1 exon. This BTB domain is an evolutionarily conserved protein–protein interaction domain found throughout the evolutionary lineages from Drosophila to mammal44. Many alternative zinc-finger domains for different FRU proteins are less conserved. On the other hand, a high degree of polymorphism was found in amino acid sequences encoded by the male-specific exon. This male-specific sequence is still highly conserved within the family (i.e., tephritidae). This suggests the strong adaptive divergence of sexual behaviors and may confer male-specific transcriptional regulatory roles of FRUM in different insect taxa.
The P1 derived primary transcripts of Bdfru and Bcfru are sex-specifically spliced at the 5′ exons as in the case of D. melanogaster35,36, An. gambiae39, Ae. Aegypti41, M. domestica38, and N. vitripennis40. The male- and female-specific exons are adjacent. TRA/TRA-2 binding sites located in the female-specific exon suggested an alternative splicing mechanism which selected the 5′ splice site located at the end of the female-specific exon. The binding of TRA/TRA-2 complex hinders the formation of the spliceosome complex at the male-specific splice site in D. melanogaster37. The female-specific FRU protein was truncated because of its in-frame stop codons in the female-specific exon36,37,38,39,40,41,45,46. Transcripts connecting the female-specific exon and the common C1 exon could not be confirmed by RT-PCR experiments because the female-specifically spliced transcript may be expressed at a low level. It was found that the expression level of the fruF transcripts were lower than the fruM transcripts in D. melanogaster46 and Ae. Aegypti41.
We propose that Bdfru and Bddsx are regulated by Bdtra in the sex-determination pathway as previously proposed in the Medfly15,18. This is based on the presence of TRA/TRA-2 binding sites in their female-specific exons22,23,24,32,33 (in this study) and the sex-determining gene splicing analysis in the pseudomales made from Bdtra RNAi. The same regulatory roles of Bctra to the Bcfru and Bcdsx genes can be implicated based on the conserved sequences and gene structures24,33 (Fig. 2) and developmental expression profiles (Fig. 5).
The expression profiles of sex-determining genes in B. dorsalis and B. correcta begin with maternal traF and tra-2 transcripts. The maternal-to-zygotic transition of these genes may have occurred at 2–3 h AEL as indicated by the expression of the zygotic cellularisation slam gene. In B. jarvisi and C. capitata, the expression of the orthologs of the slam gene were detected in embryos as early as 4 h AEL21,55. The Y-linked BdMoY and BcMoY genes also started to express around the 3 h AEL in male embryos. In C. capitata, the CcMoY transcription was also detected between 2–3 h AEL14. All of these MoY orthologues continued to express in males during the embryonic stages, less so in the larvae, and disappeared in the pupae and adults14 (in this study).
The expressions of zygotic Bdtra and Bctra genes were firstly indicated by the appearance of the BdtraM and BctraM transcripts at 5 h AEL for both male and female embryos. However, these transcripts were found in heterogeneity, i.e., all together with BdtraF and BctraF and their intermediate transcripts. The heterogeneous pattern of RNA splicing was equally extended from 5 to 8 h AEL in both sexes. This finding is consistent with the expression pattern of Cctra during the same developmental period55. It is hypothesised that the activation of the zygotic tra loop occurred in all embryos during this period. For the XX embryos, the accumulation of TRA/TRA-2 complex level above a certain threshold would generate a tra-autoregulatory loop. TRA and TRA-2 are serine/arginine (SR) type splicing regulators which operate under threshold dependency56. The presence of BdtraF and BctraF and the absence of BdtraM, BctraM, and their intermediate transcripts suggested that the level of TRA/TRA-2 complex was high enough to complete the automatisation of the tra loop from the 9 h AEL developmental point onward in females. In the case of the XY embryos, the non-autonomous zygotic tra loop was later disrupted by the suppression of the MoY gene. The appearance of mostly BdtraM and BctraM transcripts suggested the breakdown of the tra loop in males beginning at 9 h AEL. This finding is supported by a new discovery of an autosomal derived microRNA, miRNA-1-3p, that is required for male determination of B. dorsalis57. The miRNA-1-3p is involved in the suppression of Bdtra transcripts by targeting 3′ UTR. The relative expression of this miRNA-1-3p is also significantly increased and male-biased during the same critical period of the Bdtra loop breakdown process, i.e., from 6 to 9 h post oviposition at 28 °C. The microRNA is an example of intermediate male determiners. Further bioinformatics and molecular studies that shed light on how MoY relays the instruction to disrupt the tra loop will be focused on the period of 5 to 9 h AEL in B. dorsalis and B. correcta embryos.
The Bddsx and Bcdsx genes are maternally transcribed. The BddsxF knockdown experiment resulted in an interruption of yolk protein (yp) expression in adults and led to a significant reduction of ovary size, number of oocytes, and malformation of reproductive organs32. The presence of BddsxF and BcdsxF transcripts were observed in both sex embryos up to at least 12 h AEL. DSXF may be used in the expression of genes related to the nourishment of embryos such as the yp gene58. A similar finding showed that the BjdsxF transcript was also expressed in both sex embryos at sometime between 1 to 9 h AEL21. In the medfly, the CcdsxF transcription was maternally derived and also detected at 4 h AEL in all embryos. However, the CcdsxF disappeared from 5 to 9 h AEL and reappeared as a pattern of sex-specifically spliced CcdsxM and CcdsxF transcripts in male and female embryos, respectively, at 10 h AEL55. This indicated that the zygotic expression of the Ccdsx gene had begun around this time. The first zygotic expression of Bddsx and Bcdsx genes were also in evidence at 8 and 9 h AEL, respectively, when the BddsxM and BcdsxM transcripts were detected. In B. jarvisi, a high number of dsxM transcripts was detected after 6 h AEL21.
If the presence of BddsxF and BcdsxF transcripts were zygotically expressed in male embryos during the period of 9 to 12 h AEL, it would result from the binding between TRA/TRA-2 complex proteins and the dsx pre-mRNA. However, there is evidence that the availability of functional TRA/TRA-2 complex protein is not enough to generate BdtraF and BctraF transcripts from their pre-mRNAs for the same duration. This suggests that TRA/TRA-2 binding complexity at the respective pre-mRNAs of the tra and dsx orthologues are different. Ruiz et al.17 proposed, due to the putative cis-regulatory elements on sex-specific exons of tra and dsx pre-mRNAs, that TRA plays a dual role in the sex-determination pathway of tephritids. The putative TRA/TRA-2 complex binding sites on the female-specific exon of dsx pre-mRNA suggested the requirement of simpler components for the formation of an enhancing complex. In contrast, additional TRA-2 and SR protein(s) may be required for binding at the putative silencing elements on the male-specific exon(s) of tra pre-mRNA17.
The first appearance of BddsxM and BcdsxM transcripts in cellular blastoderm embryos (from 7 to 9 h AEL, supplementary Fig. S6) suggested the presence of both effector proteins, DSXM and DSXF. This indicated that sex-specific transcription of DSX-regulated genes could begin in the embryo. However, the sex-specifically spliced patterns were distinctively observed in larval, pupal, and adult stages where sexual dimorphism are well developed and require regulation of later sexual differentiation genes.
The fru gene is the other downstream effector required for later sexual differentiation. The developmental expression of the P1 derived Bdfru and Bcfru transcripts were detected later in the larval, pupal, and adult stages similar to that of D. melanogaster36 and Ae. aegypti41. In D. melanogaster males, default splicing of P1 transcripts are expressed in a small fraction of neurons36,46,59,60,61. This results in functional FRUM proteins playing transcriptional factor roles which determine male courtship and orientation59,60,62. This suggests that the FRUM effector influences male development of neurons and sexual behaviors after the embryonic stage. The P2 to P4 derived Bdfru and Bcfru transcripts were also detected in a similar developmental stages to the P1 transcript. They may have a non sex-specific developmental function as found in D. melanogaster36,46,47,48. Further functional studies of the fru genes could be complex because there are a lot of fru transcript variants. However, the function of the fruM transcripts would be significantly related to the sex-determination pathways, sexual development of the insect brain, and sexual behavior.
The molecular study of Bdtra-2 and Bdfru genes have made it possible to investigate every player in the general sex-determination cascades of tephritid fruit flies4: BdMoY14, Bdtra22,23,24, Bdtra-222,23 (also in this work), Bddsx32,33 and Bdfru in B. dorsalis. The developmental expression profile analyses can outline the following critical time points. The presence of maternal Bdtra transcripts could provide BdTRA proteins to initiate the autoregulatory loop. The Bdtra-2 transcript is continuously expressed throughout development as found in a previous study22. The Y-linked male-determiner, BdMoY, starts expressing instructions from 3 h AEL until the larval stage. The zygotic tra loop activation becomes apparant in both sex embryos at 5 h AEL. The first male-specific splicing pattern of Bdtra suggests that the BdMOY protein has completed the process of Bdtra loop disruption at 9 h AEL. In addition, the Bdtra autoregulatory loop is fully functional due to the disappearance of BdtraM and intermediate transcripts from 8 to 9 h AEL in female embryos. When the binary Bdtra is switched OFF, it transduces the first signal to the BddsxM effector at 8 h AEL although the appearance of the BddsxF transcript is present in male embryos until at least 12 h AEL. The expression of the BdFRUM effector is not apparent in embryos but only in later development. The comparative studies of all sex-determining orthologues from B. correcta imply similar sex-determination schemes24,33 (in this study). Detailed study of sex-determination cascades can be focused on more specific times in embryonic and sexual development of tephritid fruit flies. Fundamental mechanisms (e.g., how MoY breaks down the tra loop; how the autoregulatory loop of zygotic tra is established; how tra switches ON and OFF sex-specific splicing of dsx and fru pre-mRNAs; and how fru + neurons program male sexual behaviors) can be explained with better resolution. Novel intermediates and accessory proteins required for sex-determination can be screened and validated using bioinformatics and genome editing approaches. Comprehensive understanding of sex-determination may lead to new genetic tools and the improvement of insect pest control strategies in the high profile key pests, B. dorsalis and B. correcta63.
Methods
Fruit fly strains
The laboratory strains, B. dorsalis (Phayathai1 strain) and B. correcta (Phayathai2 strain), were used for the isolation of fru and tra-2 genes and the expression analyses of sex-determining genes in different developmental stages. The B. dorsalis Salaya1 genetic sexing strain was used for the RNAi experiment of Bdtra. This strain has sexual phenotypes of brown/white pupae, with males and females emerging from brown and white pupae, respectively64. The fruit flies were reared at 25 °C, with 13 L: 11 D cycles, and around 70% relative humidity (RH) at Regional R&D Training Center for Insect Biotechnology, Mahidol University.
Egg and embryo collection
Two different fruit fly cages were set up for harvesting unfertilised eggs and embryos from sexually mature (14-day old) virgin and mated females, respectively. The induction of egg oviposition was done by putting a pretested oviposition chamber containing guava juice into the fruit fly cages for one hour. The experimental batches of eggs and embryos were harvested from each new oviposition chamber after being left in the cage for 10 min. All samples were incubated at 25 °C and 70% RH. The first batch of eggs was incubated for 48 h to validate virginity and fertilisation of the females from embryo hatching. A series of egg batches were subsequently and independently collected. Each of these egg batches was incubated according to only one specific time point (i.e., representing individuals taken every hour from 1 to 12 h AEL).
Embryo preparation for RNA isolation
Collected embryos were dechorionated in 3% sodium hypochlorite solution and washed in DEPC-treated water. Single embryos were individually transferred to a microcentrifuge tube (1.5 ml) before immediate grinding in 100 μl TRIzol reagent (Invitrogen, USA). The homogenates were kept at − 80 °C until the RNA isolation step.
RNA and DNA isolation from adult samples
Genomic DNA was individually extracted according to Baruffi et al.65. Total RNA was isolated from 2-day old flies using TRIzol reagent according to the manufacturer′s instructions.
RNA and DNA isolation from single embryos, larvae and pupae
Parallel RNA and genomic DNA extractions were carried out when using single embryos, larvae and pupae. Total RNA was isolated from each sample using TRIzol reagent as per the instructions from the manufacturer. During the RNA isolation, genomic DNA was extracted from the interphase and the organic phase of the TRIzol/chloroform mix21. The RNA precipitation required an additional 10 µg of RNase-free glycogen (Thermo Scientific, USA) for single embryos. The resuspended RNA solution was evaluated using NanoDrop spectrophotometer (Thermo Scientific, USA).
Isolation of tra-2 and fru orthologues
For B. dorsalis and B. correcta tra-2 genes, the primers were designed from the conserved CDS of Bactrocera spp. and C. capitata tra-2 genes (GenBank Acc. No. are presented in Supplementary Table S3) to achieve the full transcript of Bdtra-2 and Bctra-2 genes. One microgram of total RNA from adult males and females was converted to the first-strand cDNA using SMARTer RACE 5′/3′ Kit (Takara, USA) according to the protocol of the manufacturer. One-tenth of 5′- and 3′-RACE-Ready cDNA samples were subsequently used as templates for RT-PCR and 5′ and 3′ RACEs with gene-specific primers. Semi-nested RT-PCRs were performed to derive a more specific amplification according to SMARTer RACE 5′/3′ Kit. The negative controls were also performed by excluding reverse transcriptase. The tra-2 gene structures were assembled based on PCR experiments using three pairs of PCR primers that were derived from Bdtra-2 and Bctra-2 full-transcripts and genomic DNA templates. The long Bdtra-2 and Bctra-2 cDNA and genomic DNA sequences were deposited to GenBank (Supplementary Table S3). The exon/intron junctions were deduced from these sequence alignments. A list of PCR primers in the tra-2 studies can be found in Supplementary Table S4.
Total RNA from freshly emerged adult heads of both sexes was used in the study of Bdfru and Bcfru transcripts. Several gene-specific primer pairs were used to amplify Bdfru and Bcfru cDNA based on conserved sequences that encoded the BTB domain and four zinc-finger domains of the fru orthologues. These primers were designed from predicted fru transcripts in the transcriptome of many tephritid species and M. domestica fru transcripts (GenBank Acc. Nos. are presented in Supplementary Table S3). Bdfru and Bcfru common (C1 to C4) exon-encoded sequences were identified from the gene-specific RT-PCR using primers derived from the BTB coding sequence and its 3′ adjacent (Supplementary Fig. S2b and S2d). Four alternative 3′ exon-encoded cDNA sequences were amplified from the zinc-finger domain (ZnF A, ZnF B, ZnF C, and ZnF D) derived primers (Supplementary Fig. S2b and S2d). Four alternative 5′ exon sequences were isolated from 5′ RACE experiments using reverse primers derived from the homologous sequence of the C3 exon and the male-specific exon of Mdfru and Ccfru, respectively (Supplementary Fig. S2a and S2c). Similarly, the four alternative 3′ exon sequences were amplified from the 3′ RACE experiments using forward primers derived from the homologous sequence of the C4 exon of Mdfru (Supplementary Fig. S2a and S2c). The 5′- and 3′-RACE methods were done as previously described. The male-specific exon was identified by RT-PCR using a forward primer derived from the male-specific Ccfru sequence and gene-specific reverse primer derived from a homologous sequence of the C2 exon of Mdfru. The putative female-specific exon was amplified using specific primers derived from the available 1.9 kb genomic sequence adjacently downstream to the male-specific exon in the B. dorsalis genomic scaffold (GenBank Acc. No. NW_011876307). Intron/exon junctions of all isolated Bdfru transcripts were analysed by blasting them to the available B. dorsalis genomic scaffold (GenBank Acc. No. NW_011876307). The cDNA sequences of long Bdfru and Bcfru transcripts were deposited to GenBank (Supplementary Table S3). All primers used in the fru gene studies are described in Supplementary Table S4.
RT-PCR analyses of sex-determining genes during early embryonic development and later stages
Sexual identification of single embryo, larval, and pupal samples were molecularly identified by genomic PCR using Y-specific primers which were designed based on sequences such as the MoY orthologue14. A glyceraldehyde-3-phosphate dehydrogenase (gapdh) gene-specific PCR was used to control false negative samples. Up to 300 ng of total RNA from single embryos, larvae, pupae, and adults were reverse transcribed into the first strand cDNA using ImProm-II Reverse Transcriptase (Promega, USA), following the manufacture′s instructions. One-tenth of the initial reverse transcription reaction was used as a template for RT-PCR using gene-specific primers. For each sample, the negative control of RT-PCR was performed by excluding the reverse transcriptase. The gapdh positive control system was also carried out in the RT-PCR. Slam, a zygotically expressed cellularisation gene, was used as an early developmental marker21,55. Therefore, the 2 to 3 h AEL embryos or older without detectable slam were discarded. Relative locations of gene-specific primers to each sex-determining gene were schematically plotted in Supplementary Table S5. These primer sequences are listed in Supplementary Table S6. All RT-PCR experiments gave the same electrophoretic banding patterns when they were repeated using cDNA templates of different individuals.
Bdtra RNAi experiment
Approximately 1.1 kb dsRNA of the Bdtra gene was synthesised by in vitro transcription with T7 RNA polymerase using the MEGAscript Kit (Ambion, USA). Embryos were prepared from the B. dorsalis Salaya1 genetic sexing strain, whose sexual phenotypes can be separated by pupal colour dimorphism64. One μg/μl of Bdtra dsRNA was microinjected at the posterior end of precellular blastoderm embryos. The RNAi-treated individuals were reared until adult emergence. Wild-type male and female adults emerged from brown and white pupae, respectively. The white-pupae males were therefore categorised as pseudomales24. The genitalia and testes of the wild-type males and pseudomales were compared under a microscope. The sex-specific splicing pattern of Bdtra, Bdtra-2, Bddsx, and Bdfru were carried out using the previously described RT-PCR conditions in two pseudomales (randomly selected). The sex-specific splicing patterns of these genes were compared to the wild-type males and females as well as the RNAi-treated wild-type males.
Molecular cloning and DNA sequencing of PCR amplification products
The specific amplicon was purified with the GF-1 AmbiClean Kit (Vivantis, Malaysia). The products were subsequently cloned into the pGEM-T Easy vector (Promega, USA) according to the manufacturer′s instructions. The recombinant plasmids were transformed into E. coli (DH5α) competent cells and screened for positive colonies. Plasmids were extracted using the GF-1 Plasmid DNA Extraction Kit (Vivantis, Malaysia). All sequencing reactions were performed on both strands using the sequencing service from Macrogen Inc., Seoul, Korea.
DNA sequence analysis
Query DNA sequences were searched for homology using blast against the NCBI database for gene identification. Genomic DNA and cDNA sequences were aligned using the ClustalX2 program66 and Unipro UGENE Version 1.14.067. For sequence analysis of Bdfru, the genomic scaffold (GenBank Acc. No. NW_011876307) was used to predict the gene structure. GenBank accession numbers of the references and the newly isolated sequences are in Supplementary Table S3.
Morphological development of early embryos
The morphology of live embryos was observed during early development (0 to 9 h AEL) under differential interference contrast using an inverted confocal laser scanning microscope (Olympus FV1000). The collection of developing embryos, at each time point, was as previously described. The live embryos were gently dechorionated using 3% sodium hypochlorite solution for 5 min and then transferred in a drop of 1X phosphate buffer saline to a glass slide before observation and photography.
Ethics statement
This manuscript is a part of a research project which has been approved by Mahidol University-Institute Animal Care and Use Committee (no. MU-IACUC 2018/007). This research also complies with Biosafety Guildeline for Modern Biotechnology from Institutional Biosafety Committee under Pathogens and Animal Toxins Act, B.E. 2558 (2015); (Exempt2019-003).
Data availability
All relevant data are included in the manuscript and the Supplementary Information files.
References
Darwin, C. Principle of sexual selection. In The Descent of Man and Selection in Relation to Sex Vol. 1 (ed. Darwin, C.) 253–320 (John Murray, London, 1871).
Bachtrog, D. et al. Sex determination: Why so many ways of doing it?. PLoS Biol. 12, e1001899. https://doi.org/10.1371/journal.pbio.1001899 (2014).
Sánchez, L. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52, 837–856 (2008).
Bopp, D., Saccone, G. & Beye, M. Sex Determination in insects: Variations on a common theme. Sex. Dev. 8, 20–28 (2014).
Qin, Y., Paini, D. R., Wang, C., Fang, Y. & Li, Z. Global establishment risk of economically important fruit fly species (Tephritidae). PLoS ONE 10, e0116424–e0116424. https://doi.org/10.1371/journal.pone.0116424 (2015).
Saccone, G. et al. New sexing strains for Mediterranean fruit fly Ceratitis capitata: Transforming females into males. In Area-Wide Control of Insect Pests: From Research to Field Implementation (eds Vreysen, M. et al.) 95–102 (Springer, Dordrecht, 2007).
Koukidou, M. & Alphey, L. Practical applications of insects’ sexual development for pest control. Sex. Dev. 8, 127–136 (2014).
Raphael, K. A. et al. Australian endemic pest tephritids: Genetic, molecular and microbial tools for improved sterile insect technique. BMC Genet. 15, S9. https://doi.org/10.1186/1471-2156-15-S2-S9 (2014).
Fu, G. et al. Female-specific insect lethality engineered using alternative splicing. Nat. Biotechnol. 25, 353–357 (2007).
Dafa’alla, T., Fu, G. & Alphey, L. Use of a regulatory mechanism of sex determination in pest insect control. J. Genet. 89, 301–305 (2010).
Ant, T. et al. Control of the olive fruit fly using genetics-enhanced sterile insect technique. BMC Biol. 10, 51. https://doi.org/10.1186/1741-7007-10-51 (2012).
Ogaugwu, C. E., Schetelig, M. F. & Wimmer, E. A. Transgenic sexing system for Ceratitis capitata (Diptera: Tephritidae) based on female-specific embryonic lethality. Insect Biochem. Mol. 43, 1–8 (2013).
Leftwich, P. T. et al. Genetic elimination of field-cage populations of Mediterranean fruit flies. Proc. R. Soc. B Biol. Sci. 281, 20141372. https://doi.org/10.1098/rspb.2014.1372 (2014).
Meccariello, A. et al. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science 365, 1457. https://doi.org/10.1126/science.aax1318 (2019).
Pane, A., Salvemini, M., Bovi, P. D., Polito, C. & Saccone, G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129, 3715 (2002).
Lagos, D., Koukidou, M., Savakis, C. & Komitopoulou, K. The transformer gene in Bactrocera oleae: The genetic switch that determines its sex fate. Insect Mol. Biol. 16, 221–230 (2007).
Ruiz, M. F. et al. The gene transformer of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. PLoS ONE 2, e1239. https://doi.org/10.1371/journal.pone.0001239 (2007).
Salvemini, M. et al. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 53, 109–120 (2009).
Sarno, F. et al. The gene transformer-2 of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. BMC Evol. Biol. 10, 140. https://doi.org/10.1186/1471-2148-10-140 (2010).
Schetelig, M. F., Milano, A., Saccone, G. & Handler, A. M. Male only progeny in Anastrepha suspensa by RNAi-induced sex reversion of chromosomal females. Insect Biochem. Mol. 42, 51–57 (2012).
Morrow, J. L., Riegler, M., Frommer, M. & Shearman, D. C. A. Expression patterns of sex-determination genes in single male and female embryos of two Bactrocera fruit fly species during early development. Insect Mol. Biol. 23, 754–767 (2014).
Liu, G., Wu, Q., Li, J., Zhang, G. & Wan, F. RNA-mediated knock-down of transformer and transformer 2 to generate male-only progeny in the oriental fruit fly, Bactrocera dosalis (Hendel). PLoS ONE 10, e0128892. https://doi.org/10.1371/journal.pone.0128892 (2015).
Peng, W., Zheng, W., Handler, A. M. & Zhang, H. The role of the transformer gene in sex determination and reproduction in the tephritid fruit fly, Bactrocera dorsalis (Hendel). Genetica 143, 717–727 (2015).
Laohakieat, K., Aketarawong, N., Isasawin, S., Thitamadee, S. & Thanaphum, S. The study of the transformer gene from Bactrocera dorsalis and B. correcta with putative core promoter regions. BMC Genet. 17, 34–34. https://doi.org/10.1186/s12863-016-0342-0 (2016).
Lui, G. Q. & Wan, F. H. The gene transformer 2 of Bactrocera fruit flies and its evolvement in insects. J. Environ. Entomol. 4, 742–751 (2015).
Shukla, J. N. & Nagaraju, J. Doublesex: A conserved downstream gene controlled by diverse upstream regulators. J. Genet. 89, 341–356 (2010).
Geuverink, E. & Beukeboom, L. W. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex. Dev. 8, 38–49 (2014).
Burtis, K. C. & Baker, B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997–1010 (1989).
Saccone, G. et al. Drosophila Sex-lethal and doublesex homologous genes in Ceratitis capitata: Searching for sex-specific genes to develop a medfly transgenic sexing strain. In Proceedings of the First Research Coordination Meeting: Enhancement of the Sterile Insect Technique Through Genetic Transformation Using Nuclear Techniques, Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, 30 Sep–4 Oct (eds Franz, G. & Hendrichs, J.) 16–32 (Vienna, Austria, 1996).
Shearman, D. C. A. & Frommer, M. The Bactrocera tryoni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 7, 355–366 (1998).
Lagos, D., Ruiz, M. F., Sánchez, L. & Komitopoulou, K. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determining Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348, 111–121 (2005).
Chen, S. L., Dai, S. M., Lu, K. H. & Chang, C. Female-specific doublesex dsRNA interrupts yolk protein gene expression and reproductive ability in oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Biochem. Mol. Biol. 38, 155–165 (2008).
Permpoon, R., Aketarawong, N. & Thanaphum, S. Isolation and characterization of Doublesex homologues in the Bactrocera species: B. dorsalis (Hendel) and B. correcta (Bezzi) and their putative promoter regulatory regions. Genetica 139, 113–127 (2011).
Ruiz, M. F. et al. The gene doublesex of the fruit fly Anastrepha obliqua (Diptera, Tephritidae). Genetics 171, 849–854 (2005).
Ito, H. et al. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. USA 93, 9687–9692 (1996).
Ryner, L. C. et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87, 1079–1089 (1996).
Heinrichs, V., Ryner, L. C. & Baker, B. S. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell Biol. 18, 450–458 (1998).
Meier, N. et al. Genetic control of courtship behavior in the housefly: Evidence for a conserved bifurcation of the sex-determining pathway. PLoS ONE 8, e62476–e62476. https://doi.org/10.1371/journal.pone.0062476 (2013).
Gailey, D. A. et al. Functional conservation of the fruitless male sex-determination gene across 250 Myr of insect evolution. Mol. Biol. Evol. 23, 633–643 (2005).
Bertossa, R. C., van de Zande, L. & Beukeboom, L. W. The Fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc finger domains. Mol. Biol. Evol. 26, 1557–1569 (2009).
Salvemini, M. et al. The orthologue of the fruit fly sex behaviour gene Fruitless in the mosquito Aedes aegypti: Evolution of genomic organisation and alternative splicing. PLoS ONE 8, e48554. https://doi.org/10.1371/journal.pone.0048554 (2013).
Sato, K. & Yamamoto, D. The mode of action of Fruitless: Is it an easy matter to switch the sex?. Genes Brain Behav. 19, e12606. https://doi.org/10.1111/gbb.12606 (2020).
Sato, K., Goto, J. & Yamamoto, D. Sex mysteries of the fly courtship master regulator fruitless. Front. Behav. Neurosci. 13, 245 (2019).
Zollman, S., Godt, D., Privé, G. G., Couderc, J. L. & Laski, F. A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 91, 10717–10721 (1994).
Usui-Aoki, K. et al. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat. Cell Biol. 2, 500–506 (2000).
Lee, G. et al. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J. Neurobiol. 43, 404–426 (2000).
Anand, A. et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics 158, 1569–1595 (2001).
Song, H. J. et al. The fruitless gene is required for the proper formation of axonal tracts in the embryonic central nervous system of Drosophila. Genetics 162, 1703–1724 (2002).
Amrein, H., Hedley, M. L. & Maniatis, T. The role of specific protein–RNA and protein–protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell 76, 735–746 (1994).
Mattox, W., Palmer, M. J. & Baker, B. S. Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 4, 789–805 (1990).
Kohtz, J. D. et al. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124 (1994).
Wu, J. Y. & Maniatis, T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 (1993).
Davis, T., Kurihara, J., Yoshino, E. & Yamamoto, D. Genomic organisation of the neural sex determination gene fruitless (fru) in the Hawaiian species Drosophila silvestris and the conservation of the fru BTB protein-protein-binding domain throughout evolution*. Hereditas 132, 67–78 (2000).
Stogios, P. J., Downs, G. S., Jauhal, J. J. S., Nandra, S. K. & Privé, G. G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82–R82. https://doi.org/10.1186/gb-2005-6-10-r82 (2005).
Gabrieli, P. et al. Sex and the single embryo: Early development in the Mediterranean fruit fly, Ceratitis capitata. BMC Dev. Biol. 10, 12–12. https://doi.org/10.1186/1471-213X-10-12 (2010).
Mayeda, A., Screaton, G. R., Chandler, S. D., Fu, X. D. & Krainer, A. R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell. Biol. 19, 1853–1863 (1999).
Peng, W. et al. miRNA-1-3p is an early embryonic male sex-determining factor in the oriental fruit fly Bactrocera dorsalis. Nat. Commun. 11, 932. https://doi.org/10.1038/s41467-020-14622-4 (2020).
Hoy, M. A. Genetic systems, genome evolution, and genetic control of embryonic development in insects. In Insect Molecular Genetics 4th edn (ed. Hoy, M. A.) 103–175 (Academic Press, Cambridge, 2019).
Manoli, D. S. et al. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436, 395–400 (2005).
Stockinger, P., Kvitsiani, D., Rotkopf, S., Tirián, L. & Dickson, B. J. Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807 (2005).
Cachero, S., Ostrovsky, A. D., Yu, J. Y., Dickson, B. J. & Jefferis, G. S. X. E. Sexual dimorphism in the fly brain. Curr. Biol. 20, 1589–1601 (2010).
Demir, E. & Dickson, B. J. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121, 785–794 (2005).
Lutrat, C. et al. Sex sorting for pest control: It’s raining men!. Trends Parasitol. 35, 649–662 (2019).
Isasawin, S., Aketarawong, N. & Thanaphum, S. Characterization and evaluation of microsatellite markers in a strain of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae), with a genetic sexing character used in sterile insect population control. Eur. J. Entomol. 109, 331–338 (2012).
Baruffi, L. et al. Polymorphism within and between populations of Ceratitis capitata: Comparison between RAPD and multilocus enzyme electrophoresis data. Heredity 74, 425–437 (1995).
Larkin, M. A. et al. Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Okonechnikov, K., Golosova, O., Fursov, M. & Ugene Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 28, 1166–1167 (2012).
Acknowledgements
This research was partially supported by research Contract No. 18795 granted by the Coordinated Research Projects from International Atomic Energy Agency (IAEA) to S.T. and N.A. All authors are participants in the a ‘Setting up Regional R&D Training Center for Insect Biotechnology’ project supported by joint research funding between the Faculty of Science and National Research University (NRU) funding of Mahidol University. K.L. was partially supported by a Ph.D. scholarship (2016–2017) from the Faculty of Science, Mahidol University. This study is part of the Ph.D. dissertation of K.L. under the supervision of S.T. at the Department of Biotechnology, Faculty of Science, Mahidol University. We gratefully appreciate Dr. Nidchaya Aketarawong for her assistant in dissection and microscopic experiments, and drafting manuscript. We gratefully acknowledge Prof. Anna R. Malacrida for providing male specific DNA sequences of B. dorsalis. The authors thank Mr. Robert Bachtell Eastland for his English editing service.
Author information
Authors and Affiliations
Contributions
K.L. and S.T. conceived and designed the research project. K.L. conducted the molecular experiments. S.I. and K.L. carried out the microinjection, microdissection, and microscopic investigation. All authors interpreted the data and discussed the results. K.L. and S.T. wrote the manuscript with intellectual inputs from all authors. All the authors have read, revised, and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Laohakieat, K., Isasawin, S. & Thanaphum, S. The transformer-2 and fruitless characterisation with developmental expression profiles of sex-determining genes in Bactrocera dorsalis and B. correcta. Sci Rep 10, 17938 (2020). https://doi.org/10.1038/s41598-020-74856-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-74856-6
This article is cited by
-
Next-generation genetic sexing strain establishment in the agricultural pest Ceratitis capitata
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.