Abstract
NONO-TFE3 RCC is a subtype of Xp11.2 translocation renal cell carcinoma (RCC). So far, only a small amount of NONO-TFE3 RCC have been reported owing to lack of effective diagnosis methods. Utilizing the novel dual-fusion fluorescence in situ hybridization (FISH) probe reported here, 5 cases of NONO-TFE3 RCC were identified and were ultimately confirmed by RT-PCR. Histopathology, all 5 cases were consisted by sheets of epithelial cells and papillary architecture. The cytoplasm was abundantly clear, and nucleoli was not prominent. Besides, the nuclear palisading, subnuclear vacuoles and psammoma bodies were identified. The most distinctive features were strong positive TFE3 staining but equivocal split signals of the TFE3 probe, which might lead to the misdiagnosis of Xp11.2 translocation RCC. The median age and median tumor size of the five patients were 41.2 years and 3.6 cm, respectively. A median following follow-up of 27 months showed moderate disease progression and prognosis in NONO-TFE3 RCC patients. In conclusion, the present study demonstrates the effectiveness and reliability of the NONO-TFE3 dual-fusion FISH probe for diagnosing NONO-TFE3 RCC. Suspected cases of Xp11.2 translocation RCC showing biphasic pattern, strong positive TFE3 staining, and equivocal split signals in the TFE3 FISH assay indicated a possibility of NONO-TFE3 RCC.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) associated with Xp11.2 translocation/TFE3 gene fusion (Xp11.2 translocation RCC) is a rare but aggressive type of renal tumor. In 20161, Xp11.2 translocation RCC was identified as translocation RCC of the microphthalmia (MiTF) family of transcription factors. According to the fusion pattern, Xp11.2 translocation RCC can be divided into at least 10 subtypes, and tumors with different specific gene fusions are associated with distinctive biological processes2,3,4. For instance, ASPSCR1-TFE3 RCC, one of the most common subtypes of Xp11.2 translocation RCC, was reported to be more malignant than other subtypes5,6. NONO-TFE3 RCC, which involved in the inv(X) (p11.2; q12) chromosomal variation, accounts for 12.5% of the Xp11.2 translocation RCC patient population7,8. However, less than 20 cases of NONO-TFE3 RCC have been reported since the first cases in 19979,10. This is because most cases of NONO-TFE3 RCC were remain undiagnosed or incorrectly diagnosed.
In general, TFE3 immunohistochemical (IHC) and TFE3 break-apart fluorescence in situ hybridization (FISH) assays are the most popular methods for diagnosing Xp11.2 translocation RCC5,11. Xp11.2 translocation RCC is typically characterized by strong nuclear immunoreactivity with TFE3 antibody and a larger split signal in the TFE3 break-apart FISH assay, respectively12. However, TFE3 immunoreactivity only reflects the expression levels of TFE3 protein, and the FISH split signal merely indicates rearrangements in TFE3. Identification of fusion patterns still relies on fusion FISH assays5,13, reverse transcription-polymerase chain reactions (RT-PCR), cytogenetic karyotypic analysis10, or sequencing14. However, the fusion FISH assay represents the most inexpensive and accessible method, due to economic difficulties and complicated sample requirements of the other methods.
NONO gene, a member of the Drosophila behavior/human splicing (DBHS) protein family, is located adjacent to TFE3. However, the adjacent location of NONO and TFE3 genes result in an equivocal split signal distance of TFE3, which leads to the misdiagnosis of NONO-TFE3 RCC as non-Xp11.2 translocation RCC9,15. Previously, Argani and Xia et al.8,9 introduced their single-fusion NONO-TFE3 FISH assays. In the present study, we designed a novel NONO-TFE3 dual-fusion FISH assay and identified five cases of NONO-TFE3 RCC based on our FISH probe. Therefore, the present study was designed to determine the effectiveness of the novel assay and highlight the diagnostic potential of dual-fusion and single-fusion FISH assays.
Results
FISH analysis
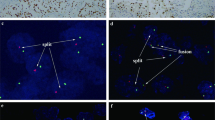
As displayed in Fig. 1a, NONO gene was labeled with red fluorescence and TFE3 gene was green fluorescence by bacterial artificial chromosomes (BACs). In general, signals were considered to be fused when yellow and abutting (equal to or less than the one signals’ diameters) green–red signals were observed. Given the small gap between gene specific probe (GSP) TFE3 and GSP NONO (nearly 21 Mb), controls might also show adjacent or partially overlapping signals. Therefore, patient gender and signal sizes needed to be taken into consideration for the evaluation of FISH results (Fig. 1b). For males, when the 2 fusion signals (2F) appeared simultaneously, the reciprocal translocation between the NONO and TFE3 genes could be confirmed (Fig. 2). For female cases, a pair of split signals (1G1R) were also required. In contrast, the presence of one fusion signal in males, and two fusion signals unaccompanied with two split signals in females, both represented adjacent TFE3 and NONO signals. By this method, the observed fusion and split signals could be accurately identified as NONO-TFE3 fusion genes. In addition, signal size was another factor used to identify the signal patterns. Fusion signals were consisted of two split signals, with sizes markedly smaller than integrated green or red signals. Therefore, there were two pairs of small split signals and one pair of large unsplit signal in females with the NONO-TFE3 gene fusion.
A schematic representation of the NONO-TFE3 dual-fusion probe. (a) Bacterial artificial chromosomes (BACs) labeled with red fluorescence covered the almost entire NONO gene, and BACs labeled with green fluorescence covered the entire TFE3 gene; (b) the principle of the NONO-TFE3 dual-fusion FISH assay.
In total, there were 5 cases were showed NONO gene and TFE3 gene fusion. All five cases showed equivocal split signals with distances of nearly one signal diameters, which might be analyzed as a false-negative diagnosis by an unexperienced pathologist. For males with NONO-TFE3 translocation (Fig. 3a,b), two fusion signals (2F) were observed in 14.1–35% of the tumor cells, and a pair of split signals as well as a fusion signal (1G1R) were observed in 13–25% cells. For NONO-TFE3 dual-fusion probe positive females (Fig. 3c,d), two fusion signals in addition to a pair of split signals (1G1R2F) were observed in 8–21% cells, and two pairs of split signals (2G2R) were observed in 19–27% cells. For negative cases (Fig. 4), such as ccRCC and pRCC, 2F and 1G1R signal patterns were observed in 1% and 22–88.35% of male tumor cells, respectively. Moreover, 1G1R2F and 2G2R signal patterns were observed in 3% and 4.71–67.3% of female tumor cells. In total, the rate of false positive detection of this fusion FISH probe was 1.8%, and the threshold value was set to 2%.
Adequate RNA was extracted from FFPE tissues from all 5 patients and subjected to RT-PCR analysis. ~ 200 bp bands extracted from two cases and a ~ 300 bp bands were reverse transcribed by the primer pairs NONO exon 9 (F) and TFE3 exon 7 (R). On the other hand, a ~ 300 bp bands and a ~ 500bps band from the remaining patient were reverse transcribed by the primer pairs NONO exon 7 (F) and TFE3 exon 8 (R). All PCR products were successfully extracted and further analyzed (Fig. 5). Finally, three fusion patterns were identified, including 3 cases (cases 1, 3 and 4) of a fusion point between exon 9 of NONO and exon 6 of TFE3, one case (cases 2) of a fusion point between exon 7 of NONO and exon 6 of TFE3, and one case (case 5) of a fusion point between exon 9 of NONO and exon 5 of TFE3 (Fig. 6).
(a)The results of reverse transcriptase–PCR showed a 500 bp targeted NONO-TFE3 transcript by the primer pairs NONO exon 7 (F) and TFE3 exon 8 (R); (b) Case 2 showed a 300 bp targeted NONO-TFE3 transcript by the primer pairs NONO exon 7 (F) and TFE3 exon 8 (R), while 3, 4 and 5 showed respectively 200 bp and 500 bp targeted NONO-TFE3 transcript by the primer pairs NONO exon 9 (F) and TFE3 exon 7 (R).
Patients
Including 8 cases of ASPSCR1-TFE3 RCC, 8 cases of PRCC-TFE3 RCC and the 13 cases of fusion partner undefined Xp11.2 translocation RCC, NONO-TFE3 RCC in our study accounted for 17.2% of the total population of Xp11.2 translocation RCC patients. The clinicopathological features of the 5 NONO-TFE3 RCC patients are listed in Table 1. Of the 5 cases of NONO-TFE3 RCC, 4 cases were initially diagnosed as Xp11.2 translocation RCC and the other one was negative to the TFE3 break-apart probe. The mean age of these patients was 41.2 years and the mean tumor size was 3.6 cm. Four patients were treated with nephron-sparing surgery (NSS), with radical nephrectomy (RN) conducted on the remaining patient diagnosed with a tumor near the pelvis. According to the 2010 American Joint Committee on Cancer TNM staging system, 4 of these patients were classified as stage I and one as stage III. Follow-up studies were conducted on all five patients from 5 to 51 months (mean, 28.2 months; median, 27 months). Four patients were found to be alive with no evidence of disease, but a 42-year-old man died 28 months after the surgery.
Morphology and IHC
All 5 cases displayed a distinctive biphasic pattern in papillary architecture accompanied with sheets of epithelial cells (Fig. 7). The predominantly papillary architecture was lined by columnar cells with clear cytoplasm (Fig. 7b). The nuclear palisading (nuclear palisading means a special pathological phenomenon that the nuclei are arranged as palisade) and subnuclear vacuoles were identified, and nucleoli was not prominent (WHO/ISUP grade 1 or 2; Fig. 7e). Psammoma bodies or calcification were observed in 4 cases (Fig. 7c). Three cases were associated with a predominantly papillary architecture, and the other two showed equal prevalence of papillary architecture and sheets of epithelial cells. Immunohistochemical assays showed that all 5 cases were immunoreactive for TFE3 (Fig. 7f), RCC, CD10, P504S, and Ki-67; but negative for Vim, CA-IX (7 g), and CK7 (Fig. 7h). One out of the five cases had CD117-positive cells, with the other cases being completely negative (Supplementary Table 1).
The typical morphologically and IHC feature of NONO-TFE3 RCC. (a–c) In cases 1, tumor have predominantly papillary architecture, which features epithelioid cells with clear to finely granular eosinophilic cytoplasm. The neoplastic abundant clear cytoplasm, and nuclei were oriented toward the luminal surface and were round and uniform in shape, resulting in the appearance of secretory endometrioid subnuclear vacuolization. Focal psammoma bodies were also observed (arrows). (d, e) In cases 4, tumor have predominantly nested to papillary architecture, The neoplastic cells lined with high columnar cells with distinct cell borders, a flocculent eosinophilic cytoplasm, and WHO/ISUP grade 2 nuclei; (f) the neoplastic cells demonstrate moderate nuclear labeling for TFE3; (g) the neoplastic cells demonstrate negative labeling for CA-IX; (h) the neoplastic cells demonstrate negative labeling for CK-7.
Discussion
Xp11.2 translocation RCC is a rare tumor which mainly occurs in children and young adults. Xp11.2 translocation RCC has been reported to have an incidence of 30, 15, and 1.5% in pediatrics, young adults (younger than 45 years), and adults, respectively16. The first cases of Xp11.2 translocation RCC involved in t(X; 1) (p11; q21) were reported by de Jong et al. in 198717. However, this rare but aggressive renal tumor did not attract much attention until 2004, when it was classified as a distinctive type of RCC by WHO18. In the following decades, distinctive features of Xp11.2 translocation RCC have been increasingly characterized. Nowadays, ASPSCR1, PRCC, and SFPQ are recognized as relatively common gene fusion partners associated with Xp11.2 translocation renal cell carcinoma8,19. However, the involvement of NONO, CLTC, LUC7L3, and RBM10 have only been discussed in case reports8,20,21,22.
FISH is currently the most convenient and effective method for identifying gene alteration by using custom BAC probes on FFPE tissue sections. Utilizing ASPSCR1-TFE3 and PRCC-TFE3 dual-fusion FISH assays previously designed by us5,13, 8 cases each of ASPSCR1-TFE3 RCC and PRCC-TFE3 RCC were identified (Table 1). In the present study, 5 cases of Xp11.2 translocation RCC were found to be positive to the NONO-TFE3 dual-fusion FISH assay and the diagnosis was eventually confirmed by PCR. Gene sequencing of the PCR products revealed three NONO-TFE3 fusion points. These are exons 1–7 of NONO and exons 6–10 of TFE3, exons 1–9 of NONO and exons 5–10 of TFE3, and exons 1–9 of NONO and exons 6–10 of TFE3. The former two fusion points have been reported earlier9. However, the present study is the first report of the last fusion point. Of the 5 NONO-TFE3 RCC cases, one (20%) was previously identified as ccRCC owing to equivocal split signals of the TFE3 gene. A possible remediation is that the possibility of Xp11.2 translocation RCC should not be eliminated if a case is moderate or strong positive to TFE3 stanning but negative to TFE3 break-apart assay, and other complementary approaches should be adopted to further clarify and confirm the diagnosis.
In 2017, Xia et al.9 introduced a single-fusion FISH assay for the identification of NONO-TFE3 translocation. Given the adjacent location of NONO and TFE3 genes, only two BAC clones, RP11-934E1 (labeled with 5-fluorescein-dUTP and located centromeric to the NONO gene locus) and RP11-416B14 (labeled with 5-ROX-dUTP and located telomeric to the TFE3 gene locus), were used in their study. Fusion of TFE3 and NONO genes led to overlap of the separated signals. The experimental design of the single-fusion assay ensures rapid analysis of signal patterns (fusion or split). However, other issues could not be suitably clarified by this assay. First, signals consisting of single BAC clones were weak and less obvious, which made data acquisition difficult and increased the possibility of a false-negative diagnosis. Second, it was hard to distinguish target signals from noises, especially for samples with dirty backgrounds, which also increased the difficulty in diagnosis. Third, although single-fusion events represent gene translocation, these may not always balance translocation, which was an important character of Xp11.2 translocation RCC. For example, the fusion of the ASPSCR1 and TFE3 genes could occur not only in ASPSCR1-TFE3 RCC, but also in alveolar soft part sarcoma (ASPS)23,24, a stromal tumor involving an unbalanced der (17) t(X; 17) (p11; q25) translocation. As a control, both TFE3 and NONO gene assays in our study consisted of more than 5 BAC clones, which improved signals due to increased signal to noise ratio (SNR). On the other hand, we modified the inter-spot spacing standard as one signal diameter, which was effective in distinguishing fusion signals from split signals. In addition, the dual-fusion assay includes both NONO-TFE3 fusion and TFE3-NONO fusion, and therefore, represents an intuitive and direct response to genetic changes in NONO-TFE3 RCC. Easy and accurate diagnosis by the dual-fusion assay required information on patient gender, the number of split and fusion signals, and signal size.
By utilizing the NONO-TFE3 dual-fusion FISH probe, 5 cases of NONO-TFE3 RCC were diagnosed, accounting for 17.2% of the total Xp11.2 translocation RCC population. In summary, the most distinctive morphological features of the 5 NONO-TFE3 RCC cases were sheets of epithelial cells and papillary architecture, which were consisted with previous report8,9. The cytoplasm was abundantly clear (or flocculent in eosinophilic cells); and the nuclei were rounded and uniform in shape and were oriented toward the luminal surface. Immunohistochemical assays on NONO-TFE3 RCC showed a strong immunoreactivity (3+++) to TFE3 antibody. According to Green et al.6,25, biphasic morphological pattern and strong TFE3 immunoreactivity were important features of NONO-TFE3 RCC. They proposed that NONO-TFE3 RCC should be taken into consideration for patients with biphasic morphological pattern and strong TFE3 immunoreactivity, combined with equivocal split signals. In our study, all of these features were verified in all 5 cases of NONO-TFE3 RCC. Taken together, IHC and break-apart FISH probe could serve as precursor methods, and suspected cases could be further evaluated by the NONO-TFE3 dual-fusion FISH for final confirmation.
In general, Xp11.2 translocation RCC tends to occur in patients aged between 20 to 50 years, and Xp11.2 translocation RCC is believed to be 2 to 3 times more prevalent in females compared to males2,8,9,19. However, our data and previous reports seem to support male predominance in NONO-TFE3 RCC. Compared with ASPSCR1-TFE3 RCC and PRCC-TFE3 RCC (Supplementary Table 2), patients of NONO-TFE3 RCC were younger and had smaller tumor sizes. In terms of prognosis, NONO-TFE3 RCC were reported as one of the most moderate subtypes among all Xp11.2 translocation RCC patients2,9. Even our data showed that NONO-TFE3 RCC patients had a longer survival than ASPSCR1-TFE3 RCC and PRCC-TFE3 RCC in a short time follow-up study. However, there was a case at the pT1aM0N0 stage, where the patient was diagnosed with peritoneal metastasis at 10 months and dead at 28 months postoperation. The other 4 patients were alive with no evidence of disease progression after their initial resection with a median follow-up of 20.5 months. Given the low number of cases, it’s hard to believe that any of these data are statistically significant. The physiological characteristics of NONO-TFE3 RCC need further investigation with a larger population and long-term follow-up.
In summary, we reported a newly designed NONO-TFE3 dual-fusion FISH assay and confirmed the accuracy of this novel probe for detecting NONO-TFE3 RCC. Five cases of NONO-TFE3 RCC were identified utilizing this probe and their clinicopathological characteristics were explored. The equivocal split signals make the diagnosis of NONO-TFE3 RCC difficult. However, this feature could be regarded as a diagnostic marker to provide additional insights to pathologists. In a short-term study, NONO-TFE3 RCC showed a moderate prognosis compared to that in ASPSCR1-TFE3 RCC and PRCC-TFE3 RCC. These findings enhance our understanding of NONO-TFE3 RCC.
Materials and methods
Case selection
The current study was conducted in accordance with the ethical principles of the Helsinki Declaration II. Written informed consent was obtained from both adult patients and legal guardians of pediatric patients before the clinical investigations were performed. This study was approved by the Institutional Review Board of the Nanjing Drum Tower Hospital. 13 confirmed and 5 suspected cases of Xp11.2 translocation RCC, reported from January 2007 to October 2018, were included in this study. In addition, 15 randomly chosen cases each of clear cell renal cell carcinoma (ccRCC) and papillary renal cell carcinoma (pRCC) and UOK 109 cells10 (generously gifted by Dr. W. Marston Linehan, National Cancer Institute, Bethesda, MD) were used as controls. All 13 cases of unclassified Xp11.2 translocation RCC were identified by the TFE3 break-apart FISH probe, but were excluded from ASPSCR1-TFE3 RCC and PRCC-TFE3 RCC, by previously reported dual-fusion FISH assays reported by us5,13. The 5 suspected cases showed moderate (2+) to strong (3+) immunoblotting response to the TFE3 antibody but equivocal split signals to the TFE3 probe. All the hematoxylin and eosin (HE) stained tissue sections and IHC sections were reviewed by two experienced pathologists (XHP and SFC). In each case, blocks containing the largest proportion of tumor tissue were selected for subsequent FISH assays. Clinicopathologic features of patients including epidemiological data, clinical manifestations, operation strategies, pathological findings, and prognostic information were recorded and evaluated.
Reverse transcription–PCR
Total RNA was extracted with the E.Z.N.A. FFPE RNA Kit (Omega Bio-Tek, Guangzhou, China) and complementary cDNA strands were generated using the HiScript Reverse Transcriptase (Vazyme, Nanjing, China). Two primer pairs were designed to detect the fusion transcripts. The PCR primers used were as follows: NONO exon 9 (F), ACCTGCCACTATGATGCC; TFE3 exon 7 (R), TTCTTCTGCCGTTCCTTC; NONO exon 7 (F), CGGCAGCAAGAAGAAATG; and TFE3 exon 8 (R), GGTCACTGGACTTAGGGATG. PCR amplification was performed in a volume of 20 μl under the following conditions: 95 °C for 10 min; followed by 35 cycles of 95 °C for 45 s, 60 °C for 45 s, and 72 °C for 60 s; followed by a final 10-min extension at 72 °C.
PCR products were separated in 1.2% agarose gels and extracted using the TIANgel Midi Purification Kit (Tiangen Biotech, Beijing, China), and subsequent sequence analyses were performed on a BigDye Terminator and ABI Basecaller (Applied Biosystems, Grand Island, NY, USA). The resulting PCR product sequences were compared by using the BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Dual-fusion dual-color probe design and development
Dual-fusion dual-color probe design
Dual-fusion dual-color probe were designed as previously described5,13. Suitable BACs for the dual-fusion dual-color probe were selected from https://genome.ucsc.edu/. CTD-3090M11, RP11-485H23, RP11-624G23, CTD-2350P17, and RP11-402P6, labeled with tetramethyl rhodamine-5-dUTP (showing red fluorescence), were located on the long arm of the X chromosome and covered the NONO gene. CTD-2311N12, RP11-416B14, CTD-2522M13, CTD-2516D6, CTD-2312C1, CTD-2248C21, and RP11-959H17, labeled with fluorescein-12-dUTP (showing green fluorescence), were located on the short arm of the X chromosome and covered the entire TFE3 gene (Fig. 1a).
FISH experimental procedure
FISH were performed as previously described9,11. 3 mm thick sections from formalin-fixed, paraffin-embedded (FFPE) tumor tissue blocks were first heated overnight in an incubator maintained at 65 °C. Then, sections were deparaffinized by treatment with xylene for 30 min, followed by washing with absolute ethanol for 10 min. Sections were rehydrated in 100, 90, and 70% ethanol and double distilled water, in turn, for 3 min. This was followed by boiling these sections in sterilized purified water for 20 min and subsequent air drying. Sections were then digested with 10 μl pepsin (4 mg/mL, 0.02 M HCl; Sigma-Aldrich, Beijing, China) at 37.5 °C for 5 to 10 min and washed with 2X saline–sodium citrate (SSC) for 5 min. Subsequently, these sections were dehydrated in 70, 90, and 100% ethanol, in turn, for 3 min and air dried. Hybridization was performed with the newly designed FISH probe, denatured at 85 °C for 5 min, and incubated overnight (10–18 h) at 37 °C under lucifugal conditions. The sections were then immersed in 2X SSC for 10 min; and in 0.1% NP-40 and 2X SSC for 5 min at 37 °C. Then, the sections were immersed in 70% ethanol for 3 min and air dried. The nuclei were counterstained with 10 μl of 4′,6-diamidino-2-phenylindole (DAPI). After hybridization, all slides were stored in the dark.
FISH evaluation
Evaluation of FISH results was conducted with an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan) fitted with 3 filters (DAPI/FITC/Texas Red) and the FISH analysis software (Imstar, Paris, France). The FISH assay is effective only when clear FISH signals are observed in > 100 nonoverlapping nuclei.
References
Moch, H., Cubilla, A. L., Humphrey, P. A., Reuter, V. E. & Ulbright, T. M. The 2016 WHO classification of tumours of the urinary system and male genital organs-part a: renal, penile, and testicular tumours. Eur. Urol. 70, 93–105. https://doi.org/10.1016/j.eururo.2016.02.029 (2016).
Wang, X. T., Xia, Q. Y., Zhou, X. J. & Rao, Q. Xp11 Translocation renal cell carcinoma and the mesenchymal counterparts: an evolving concept with novel insights on clinicopathologic features, prognosis, treatment, and classification. Crit. Rev. Oncog. 22, 481–497. https://doi.org/10.1615/CritRevOncog.2017020558 (2017).
Marchionni, L. et al. MicroRNA expression profiling of Xp11 renal cell carcinoma. Hum. Pathol. 67, 18–29. https://doi.org/10.1016/j.humpath.2017.03.011 (2017).
Pivovarcikova, K. et al. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am. J. Surg. Pathol. 41, 138–140. https://doi.org/10.1097/Pas.0000000000000730 (2017).
Chen, X. et al. Newly designed break-apart and ASPL-TFE3 dual-fusion FISH assay are useful in diagnosing Xp11.2 translocation renal cell carcinoma and ASPL-TFE3 renal cell carcinoma: a STARD-compliant article. Medicine 94, e873. https://doi.org/10.1097/MD.0000000000000873 (2015).
Ellis, C. L. et al. Clinical heterogeneity of Xp11 translocation renal cell carcinoma: impact of fusion subtype, age, and stage. Mod. Pathol. 27, 875–886. https://doi.org/10.1038/modpathol.2013.208 (2014).
Kauffman, E. C. et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat. Rev. Urol. 11, 465–475. https://doi.org/10.1038/nrurol.2014.162 (2014).
Argani, P. et al. TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am. J. Surg. Pathol. 40, 723–737. https://doi.org/10.1097/PAS.0000000000000631 (2016).
Xia, Q. Y. et al. Xp112 translocation renal cell carcinoma with NONO-TFE3 gene fusion: morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod. Pathol. 30, 416–426. https://doi.org/10.1038/modpathol.2016.204 (2017).
Clark, J. et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 15, 2233–2239. https://doi.org/10.1038/sj.onc.1201394 (1997).
Qu, Y. et al. Diagnosis of adults Xp11.2 translocation renal cell carcinoma by immunohistochemistry and FISH assays: clinicopathological data from ethnic Chinese population. Sci. Rep. 6, 21677. https://doi.org/10.1038/srep21677 (2016).
Argani, P. et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am. J. Surg. Pathol. 31, 1149–1160. https://doi.org/10.1097/PAS.0b013e318031ffff (2007).
Xiong, L. et al. PRCC-TFE3 dual-fusion FISH assay: a new method for identifying PRCC-TFE3 renal cell carcinoma in paraffin-embedded tissue. PLoS ONE 12, e0185337. https://doi.org/10.1371/journal.pone.0185337 (2017).
Just, P. A. et al. Identification by FFPE RNA-Seq of a new recurrent inversion leading to RBM10-TFE3 fusion in renal cell carcinoma with subtle TFE3 break-apart FISH pattern. Genes Chromosomes Cancer 55, 541–548. https://doi.org/10.1002/gcc.22356 (2016).
Wang, X.-T. et al. Xp11 neoplasm with melanocytic differentiation of the prostate harbouring the novelNONO-TFE3gene fusion: report of a unique case expanding the gene fusion spectrum. Histopathology 69, 450–458. https://doi.org/10.1111/his.12949 (2016).
Cheng, X., Gan, W., Zhang, G., Li, X. & Guo, H. Clinical characteristics of XP11.2 translocation/TFE3 gene fusion renal cell carcinoma: a systematic review and meta-analysis of observational studies. BMC Urol. 16, 40. https://doi.org/10.1186/s12894-016-0154-6 (2016).
de Jong, B., Molenaar, I. M., Leeuw, J. A., Idenberg, V. J. & Oosterhuis, J. W. Cytogenetics of a renal adenocarcinoma in a 2-year-old child. Cancer Genet. Cytogenet. 21, 165–169 (1986).
Lopez-Beltran, A., Scarpelli, M., Montironi, R. & Kirkali, Z. 2004 WHO classification of the renal tumors of the adults. Eur. Urol. 49, 798–805. https://doi.org/10.1016/j.eururo.2005.11.035 (2006).
Wang, X. T. et al. RNA sequencing of Xp11 translocation-associated cancers reveals novel gene fusions and distinctive clinicopathologic correlations. Mod. Pathol. 31, 1346–1360. https://doi.org/10.1038/s41379-018-0051-5 (2018).
Argani, P. et al. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am. J. Surg. Pathol. 27, 750–761 (2003).
Argani, P., Zhang, L., Reuter, V. E., Tickoo, S. K. & Antonescu, C. R. RBM10-TFE3 renal cell carcinoma: a potential diagnostic pitfall due to cryptic intrachromosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am. J. Surg. Pathol. 41, 655–662. https://doi.org/10.1097/PAS.0000000000000835 (2017).
Malouf, G. G. et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin. Cancer Res. 20, 4129–4140. https://doi.org/10.1158/1078-0432.CCR-13-3036 (2014).
Argani, P. et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am. J. Pathol. 159, 179–192. https://doi.org/10.1016/S0002-9440(10)61684-7 (2001).
Chen, X. C. et al. The value of ASPL-TFE3 dual-color single-fusion FISH assay in diagnosing alveolar soft part sarcoma. Int. J. Clin. Exp. Pathol. 9, 10581–10586 (2016).
Green, W. M. et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am. J. Surg. Pathol. 37, 1150–1163. https://doi.org/10.1097/PAS.0b013e31828a69ae (2013).
Acknowledgements
Patient derived UOK cell line was received from the Tumor Cell Line Repository of the Urologic Oncology Branch at the National Cancer Institute (NCI). In particular, we express our sincere gratitude to Dr. W. Marston Linehan from the National Cancer Institute for providing us with the UOK 109 cell line.
Funding
This research was supported by the National Natural Science Foundation of China (81572512), Nanjing SCI-TECH Development Project (201803025), and Beijing Ronghe Medical Development Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
L.N. and G.W. designed the work and drafted the manuscript. Z.W. and P.X. reviewed hematoxylin and eosin sections to determine the presence of representative areas of the original samples. S.Q. and C.S. preformed the FISH. L.N. and P.X. designed primers and preformed the PCR. Q.F., X.L., Z.X., L.X., Z.G., and G.H. collected data from the Xp11.2 translocation RCC patients. S.Q. and Z.W. were analyzed the data and revising the manuscript. G.W. and L.D. supervised the study and helped in obtaining the funding. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, N., Guo, W., Shi, Q. et al. The suitability of NONO-TFE3 dual-fusion FISH assay as a diagnostic tool for NONO-TFE3 renal cell carcinoma. Sci Rep 10, 16361 (2020). https://doi.org/10.1038/s41598-020-73309-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-73309-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.