Abstract
Stevia rebaudiana Bertoni is a commercially important zero calorie natural-sweetener herb which produce sweet compounds known as steviol glycosides. Rising demands of steviol glycosides by food and beverage industries has led to an increase in its cultivation in various countries. Unfortunately, stevia cultivation faces 2–25% yield penalty due to weeds which further adds to its cultivation cost. To resolve this major challenge, Agrobacterium-mediated genetic transformation of in vitro derived stevia-nodal explants using herbicide resistance gene (bar) has been optimized, for the production of stable transgenic stevia plants. Several parameters including explant type, pre-incubation duration, acetosyringone (As) concentration, Agrobacterium cell density, Agro-inoculation duration, co-cultivation duration, selection regime and plant growth regulators (PGRs) combination and concentration, have been successfully optimized. Among the two types of explants used, nodal explants showed a higher regeneration response of 82.85%, with an average of 25 shoots/explant. The best PGRs combination and concentration for shoot-induction, shoot-elongation and root-induction was found to be 6-benzyladenine (1.0 mg l−1) + naphthalene acetic acid (0.5 mg l−1), gibberellic acid (1.0 mg l−1), and half-strength MS medium, respectively. The two-step selection (phosphinothricin) regime resulted in an average transformation efficiency of 40.48% with nodal explants. Molecular characterization of putative transformants through PCR, RT-PCR, qRT-PCR and Southern-blot hybridization confirmed the presence, stability, expression as well as copy number of bar gene respectively. Compared to the non-transgenic plants, the T0 transgenic plants successfully tolerated 8 mg l−1 glufosinate ammonium sprays. Thus, the optimized protocol can be useful for the introduction of other genes (inter-kingdom transfer) into stevia genome.
Similar content being viewed by others
Introduction
Stevia rebaudiana Bertoni (family: Asteraceae) commonly known as ‘honey leaf’, ‘candy leaf’, or ‘sweet herb’ is a perennial shrub of South America.There are 154 species of genus Stevia, and among them S. rebaudiana is the only species which synthesize steviol glycosides like stevioside, dulcoside A and rebaudioside (rebaudioside A, B, C, D and E) in its leaves. The stevioside and rebaudiosides are 300–400 folds sweeter as compared to cane sugar1,2. Although a zero-calorie sweetener, stevia also regulates blood sugar level, body-weight, blood-pressure and prevents tooth-decay3. It also possesses anti-oxidant, anti-hyperglycemic, anti-hyperlipidemic, anti-diabetic4, anti-microbial, anti-inflammatory, anti-proliferative, anti-cancer and anti-diarrheal properties5 etc. According to World Health Organization the daily acceptable dose of steviol glycoside is 0–2 mg kg−1of body weight6. The increase in the acceptability of this natural sweetener and incidences of diabetes and obesity has led to a large-scale commercial production of stevia crop in developing and developed countries7.
The conventional strategies used for stevia cultivation are not reliable because of poor viability of seeds and germination percentage8. Hence, to overcome such limitations, in vitro propagation is the only remedy that can facilitate large-scale production of genetically identical stevia plants. Moreover, its cultivation faces several abiotic and biotic constraints. Among the biotic ones, the most important is that stevia is a poor competitor of weeds. The cropping season of stevia is from March to September, and it has to compete with weeds during July to September (rainy season). Several weeds belonging to the plant family Asteraceae, Poaceae, Fabaceae, Solanaceae, Malvaceae, Convolvulaceae, Caryophyllaceae, Brassicaceae, Rubiaceae, Primulaceae, and Plantaginaceae cause reduction in branching and the yield of stevia9,10,11. Therefore, implementation of a robust weed control strategy is crucial for sustainable stevia cultivation.
At present, herbicide applications and mechanical methods are being used abundantly to control excessive weed growth in fields of commercial crops. However, there are smaller number of registered herbicides reported for stevia cultivation which often leads farmers to go for mechanical, hand weeding or certain other chemical practices for weed control. But, chemical methods are more promising and cost-effective as compared to other weed control methods such as mulching (including organic or synthetic mulching), hand weeding and cover crops12. Moreover, non-selective herbicides such as paraquat, glufosinate (Basta R) and glyphosate (Roundup R) are toxic for both weeds and crop plants and cause crop injury during field application. Chemical methods include applications of different herbicides also have detrimental effects for crop plant, human being and environment. According to International Agency for Research on Cancer (IARC), two commercial herbicides, glyphosate and 2,4-D are potent human carcinogens13,14. Therefore, introduction of foreign gene (for herbicide resistance) into crop plant is highly desirable in order to control weed infestation. The bar gene encodes for phosphinothricin acetyltransferase which provide resistance against broad spectrum herbicide phosphinothricin or glufosinate ammonium. Glufosinate is marketed under various brand names (Buster R, Liberty R, Basta R and Finale R) and has some advantages over other herbicides such as, low toxicity and short half-life (low persistence)15,16. In this study, bar gene has been deployed to develop herbicide tolerant stevia plants.
Several studies have been conducted on in vitro regeneration response of stevia17,18 using various explants including nodal sections19, leaf20, inter-nodal segment21, axillary buds22, shoot-tip explants suspension cultures, hairy root and anthers22,23. However, there are very few reports on optimization of Agrobacterium-mediated transformation of stevia.
The major factors which affect the Agrobacterium-mediated transformation regime are age and nature of explants, concentrations or combinations of PGRs and physiochemical and temporal conditions of Agro-inoculation and co-cultivation24. The objectives of our study were (1) optimization of in vitro regeneration of stevia, (2) optimization of Agrobacterium-mediated genetic transformation of stevia with herbicide tolerant bar gene, (3) molecular characterization and (4) herbicide tolerance assay of the T0 transgenic plants.
Materials and methods
Plant material and culture conditions
Three months old stevia plants (variety GVS-16) were obtained from Green Valley Stevia industry, Pojewal, Punjab, India. Leaves, nodal segments and shoot tips from mother plants were taken as explant for in vitro callus regeneration. Explants were surface sterilized with 5% (v/v) Tween 20 for 20 min, washed with tap water, and further treated with 0.1% (w/v) bavistin for 15 min to remove fungal sporesand rinsed 3–4 times with autoclaved Milli-Q water (Merck Millipore, USA). Thereafter, the explants were treated with 0.1% (w/v) HgCl2 for 5 min and rinsed 3–4 times with autoclaved Milli-Q water, to remove any traces of HgCl2. The explants were then treated with ethanol (70%) for 60 s, followed by several rinses with autoclaved Milli-Q water. The sterilized explants were cultured on MS media25 containing 0.3% Phytagel (Sigma, USA), and different combinations and concentrations of PGRs (2,4-D, BAP, KIN, and NAA). Prior to autoclaving (at 121 °C for 15 min), the pH of MS medium was adjusted to 5.7. After autoclaving, the sterile medium was dispensed into sterile jam bottles. All the cultures were incubated at 22 ± 2 °C under white light (PFD: ~ 52 μmol m−2 s−1) for a photo-period of 16/8 h light/darkness. All experiments were repeated at least three times. Callus formation, shoot regeneration, shoot number and root number, shootlength and root length, were recorded at regular intervals and the cultures were maintained under aseptic conditions.
Rooted plantlets were washed with autoclaved Milli-Q water to remove phytagel media from roots. The plantlets were planted in small plastic pots containing sterile soilrite (soil-conditioning mixture) and regularly irrigated with Hoagland solution. The pots were shielded with transparent perforated polythene sheets to maintain moisture (80–90%) and incubated in a plant growth chamber (Conviron, USA) set at 22 ± 2 °C and 16 light/8 h dark photo-period with PFD of 80 µmol m–2 s–1. After 22–25 days of acclimatization, polythene bags were removed from the plants.
Agrobacterium strains and vector construct
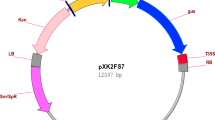
The plant expression vector pPZP200 35sde-bar-loxP harbouring herbicide tolerant bar gene driven by DECaMv35s promoter and loxP terminator, was used in the study (Fig. 5A). The enzyme phosphinothricin acetyl transferase (PAT) encoded by the bar gene inactivates phosphinothricin. The aadA2 gene encodes an antibiotic (streptomycin-spectinomycin) resistant protein. The construct was used to transform Agrobacterium tumefaciens strain GV1301. The active ingredient of the herbicide bialaphos (glufosinate ammonium) was used as selection agent.
Agrobacterium-mediated transformation and explant regeneration
Nodal sections and leaf originated callus from in vitro regenerated plants (Fig. 2A) were dissected and incubated for 2 days on pre-incubation media containing MS salts + BAP (2 mg l−1). These acclimatized explants were used for Agrobacterium-mediated transformation with pPZP200-bar-loxP constructs. Explant wounding was done with sterile needle to enhance the transformation efficiency. Different parameters such as type of explants, pre-incubation duration, acetosyringone (As) concentration, Agrobacterium cell density, Agro-inoculation duration and co-cultivation duration were optimized meticulously (Fig. 1A–F). After agroinoculation, the explants were incubated on co-cultivation medium (Fig. 2B) containing MS salts + BAP (2 mg l−1) + acetosyringone (As), at 22 °C, in dark for 3 days. Thereafter, the explants were washed twotimes with liquid MS media fortified with cefotaxime (250 mg l−1), followed by incubation on MS media containing 250 mg l−1 cefotaxime. After 8–10 days, explants were subjected to first selection media (SIM-1) having MS salts + BAP (1 mg l−1) + NAA (0.5 mg l−1), cefotaxime (250 mg l−1) + glufosinate ammonium (2 mg l−1) for 22–30 days. The regenerated shoots with a pair of vegetative leaves were identified, excised into segments and were placed on the second selection medium (SIM–2) (Fig. 2C) having the same constituents as SIM–1, except 4 mg l−1 of glufosinate ammonium, and incubated for 22–30 days. The independent shoots that regenerated on SIM-2 were transferred to culture-tubes containing elongation medium (SEM) having MS salts + GA3 (1 mg l−1), and incubated for 15–20 days (Fig. 2D). The elongated shoots were incubated on rooting medium (RIM) containing ½ strength MS medium, for 15–20 days (Fig. 2E). The rooted plantlets were transferred to plastic containers filled with sterile soilrite (soil conditioning mixture) and irrigated with Hoagland solution. The containers were bagged with perforated polythene bags and incubated in a plant growth chamber (Conviron, USA) set at 80% relative humidity, for 15 days of acclimatization (hardening) in soilrite (Fig. 2F). The acclimatized plantlets were planted in plastic pots (12 in.) filled with soil:sand:farmyard manure (3:1:1) and transferred to glasshouse maintained at 24 ± 1 °C under natural light, for normal vegetative and reproductive growth phases (Fig. 2G). The Fig. 4 represents an outline of the optimized protocol for stevia transformation. The percentage transformation efficiency using nodal segments and calluses was determined by the formula as shown below
Optimization of Agrobacterium-mediated nuclear transformation of stevia. (A) Effect of explant type, (B) Concentration of acetosyringone (µm), (C) O.D600 of Agrobacterium co-cultivation medium, (D) Pre-incubation duration (days), (E) Agro-inoculation duration (min) and (F) Co-cultivation duration (days).
Stevia transformation and in vitroregeneration of nodal-explants, (A) explant (nodal segments) preparation for agro-inoculation (bar = 1.0 cm), (B) explants incubated on co-cultivation media (bar = 1.0 cm), (C) regenerated explants subjected to antibiotic selection (bar = 1.8 cm), (D) shoot elongation in SEM (bar = 1.8 cm), (E) rooting in RIM (bar = 1.0 cm), (F) hardening of in vitro raised plantlets (bar = 2.5 cm) and (G) green-house grown acclimatized transgenic stevia plants (bar = 4.5 cm).
Molecular characterization of transgenic plants
Genomic DNA isolation from T0 transgenic stevia plants was performed using DNeasy Plant Maxi kit (Cat # 68163, Qiagen, Germany). PCR analysis was done to check the transgene integration and Southern-blot hybridization was performed to check the copy number of transgene26. PCR analysis of 100 ng genomic DNA was done by using gene-specific set of internal primers, forward-5ʹCGCCGATGGTTTCTACAAAGA3ʹ and reverse-5ʹTCAATGACCGCTGTTATGCG3ʹ, for the amplification of 146 bp region of bar gene with GeneAmp PCR system 9700 (PE Biosystems, USA). PCR amplification cycle used was: initial denaturation: 95 °C (5 min), denaturation: 95 °C (10 s), annealing: 50 °C (30 s), and extension: 72 °C (1 min). For Southern blotting, ~ 10 μg genomic DNA of T0 transgenic stevia leaves was digested overnight (12–14 h) with EcoRI (Cat #R0101L, New England Biolabs, USA) run on 0.7% gel and transferred on BioBond-plus nylon membrane (Sigma, USA). After pretreatment, the Southern-blot was hybridized for 24 h at 58 °C with α[32P] dCTP labeled 552 bp bar gene-probe (BRIT, Mumbai, India), followed by three stringent washings. The blots were exposed to Fuji screen for forty eight hours and then analyzed on a phosphoimager (Typhoon Trio+, Sweden).
Reverse transcriptase PCR (RT-PCR) analysis
RNA from leaves of transgenic (T0) plants was extracted with TRI reagent (Cat #T9424; Sigma, USA) according to manufacturer’s instructions. For RNA quantification, ND-1000 spectrophotometer (NanoDrop Technologies Inc., USA) was used. DNase (Cat #AMPD1; Sigma, USA) treatment was given to each RNA sample to remove any DNA contamination. cDNA was synthesized from RNA using Power ScriptRT (Cat #RR037B; Takara, Japan) using 5 µg of plant total RNA. Reverse transcription reaction consisted of pre-treatment of 25 μl reaction mixture at 50 °C (10 min), initial denaturationat 95 °C (5 min), denaturation at 95 °C (10 s), followed by and annealing and extension at 60 °C (30 s). Amplification of cDNA fragments was done using specific primers that amplifies 200 bp region of bar gene. These amplified gene fragments were then electrophoresed on agarose gel (1%) and analyzed using a gel-documentation system (Bio-Rad, USA).
Quantitative real-time PCR (qRT-PCR)
SYBR green premix was used to perform qRT-PCR on synthesized cDNA. β-actin gene of Stevia was used as a control (endogenous) for qRT-PCR analysis. Specific primers for β-actin gene were, forward-5ʹTCTTGATCTTGCTGGTCGTG3ʹ, and reverse-5ʹGCGGTTTCAAGTTCTTGCTC3ʹ and bar gene specific primers were forward-5ʹGTTTCACCACGTCATCAACG3ʹ and reverse-5ʹTGCCAATTTCCATGTTTGAA3ʹ giving an amplicon size of 150 and 200 bp, respectively. Quantitative PCR was used to analyze the relative quantity of bar gene transcript in putative transgenic plants with Quantifast SYBR green PCR kit (Cat. #204054; Qiagen, Germany) using StepOne real-time PCR machine (Applied Biosystems, USA). The reaction mixture (10 μl) comprised of primers (5 pmol), cDNA (1/10-fold diluted), SYBR green (5 μl of 1×) and MQ water (2.5 μl). The experiment was performed using three technical as well as biological replicates.
Comparison of morphological charactersand chlorophyll content
After shifting the transgenic plants to green-house (Fig. 2G), morphological characters (height of plant, no. of leaves and branches) and chlorophyll content of transgenic and control plants were recorded. For chlorophyll estimation fresh leaves (100 mg) were crushed in acetone (80%), centrifuged at 10,000 rpm for 10 min, at 4 °C. Absorbance of the supernatant was recorded spectrophotometrically (UV-2700 UV–Vis Spectrophotometer, Shimadzu, Japan) at 663 and 645 nm for chlorophyll a and b, respectively. The experiments were performed three times and in triplicates.
Herbicide tolerance assay
Herbicide tolerance (glufosinate) assay was performed to check the efficacy of transgenic stevia plants for herbicide tolerance as compared to non-transgenic plants. To perform this assay, the wild type (non-transgenic control) stevia plants were divided into five groups, each having five plants (4 test + 1 control). Each group was sprayed (using hand sprayer) with different concentrations of glufosinate (50 ml of 2, 4, 6, and 8 mg l−1) under green-house conditions to find the minimum lethal dose for stevia plants. It was observed that 8 mg l−1 of glufosinate was the minimum lethal dose for stevia plants. Thereafter, five healthy T0 transgenic and non-transgenic control (wild type) stevia plants each were sprayed separately with 8 mg l−1 glufosinate in green-house. The experiment was repeated three times and the observations were recorded after 12 days of spray.
Residual phytotoxic effect
Residual phytotoxic effect of glufosinate on soil was studied by spraying the potted soil with three different concentration of glufosinate i.e. 0.25, 0.50, and 1.0% (v/v) under greenhouse condition. After five days of spray, ten seeds each of indicator plants i.e. corn and cucumber were sown into these pots. The experiment was performed in triplicates and the soil of control plant was sprayed with water as an experimental control. Seed germination percentage was recorded after 10 days of sowing. The plantlet height was recorded with the help of a measuring scale after 20 days of sowing.
Statistical analysis
Different parameters (percentage of callus formation, number of shoots and roots, were examined using three replicates for each treatment. The values given in tables were expressed as mean ± SD of three replicates i.e. n = 3. Mean and standard deviation were calculated using SPSS software, version 14.0 (SPSS Inc. USA).
Results and discussion
Callus induction
In this study, we used different concentrations of 2,4-D (1–3 mg l−1), KIN (1–2 mg l−1) and BAP (1–3 mg l−1) to obtain callus from different explants viz. leaf, nodes and shoot tips of in vitro raised Stevia plants. Callus was initiated from leaf-discs after 4–5 weeks on culture media while, the other explants responded after 6–7 weeks. Hence, leaf discs were most efficient in callus formation and maximum callus induction was achieved on MS2 medium [2,4-D (2 mg l−1) and KIN (1 mg l−1)] (Supplementary Table 1). Significantly higher callus induction was reported with leaf explants cultured on MS1, MS2 and MS3 media as compared to nodes and shoot tips. The trend observed for callus induction in different media was MS2 > MS3 > MS1 > MS6 > MS5 > MS4 > MS9 > MS7 > MS8.
Leaf discs were found most efficient for callus formation while shoot tips were found least effective. Various researches have been carried out to find the callusing potential of different explants of stevia including leaves27, anthers, cell suspension, flower28, nodes and roots27. Our results are in consonance with that of Janarthanam et al., who found that leaf (juvenile) explants respond better with respect to callus formation (MS media fortified with 2.22 µM BAP and11.31 µM 2, 4‐D) as compared to nodal explants29. 80% of callus formation was achieved with leaf segments as compared to 60% with nodal segments. Shooting was achieved from these calluseson MS media supplemented with 1.34 µM NAA and 4.44 µM BAP after 28 days of incubation. In a report by Sairkar et al., leaf discs were found highly efficient for callus formation as compared to nodal segments when incubated on MS media fortified with 1 mg l−1 KIN and 2 mg l−1 2,4-D. Subculturing of callus was done after interval of 25–30 days30.
Shoot regeneration
In this study it has been found that less number of shoots was produced from callus in comparison to direct shoot regeneration from explants (Supplementary Table 2) (Fig. 3). Although, leaf explants derived-callus cultured on MS3 media [BAP (1 mg l−1) and NAA (0.5 mg l−1)] showed maximum shoot regeneration (4 ± 1.00), the nodal sections cultured on MS6 media [BAP (1 mg l−1) and NAA (0.5 mg l−1)] exhibited maximum direct shoot regeneration (25 ± 3.2). Direct regeneration of shoots, without any callusing stage is more effective as compared to indirect regeneration. Shoots regenerated through callus are generally asynchronous while directly regenerated shoots are homogeneous, diploid and true-to-type. Significantly higher numbers of shoots were obtained directly from nodal sections cultured on MS4, MS5 and MS6 media as compared to rest of the explants. In a study, a combination of 1.5 mg l−1 BAP with 0.5 mg l−1 KIN was reported to efficiently induce multiple shoot regeneration from nodal explants28. In a study by Debnath, maximum shoot formation was observed with nodal sections and shoot tips cultured on MS media fortified with IAA (1.13 mg l−1) and BAP (2.0 mg l−1)31. In a report by Singh and Dwivedi, they mentioned that the nodal explant showed maximum shoot regeneration (98%) response as compared to shoot tips (55%) and inter-nodal segments (15%). Bud regeneration was reported earlier (in 5.50 days) using nodal section than other explants32. Undoubtedly, nodal sections have been the explants of choice for direct shoot regeneration using MS medium fortified with different concentrations of PGRs viz. 1.0 mg l−1 BAP and 0.25 mg l−1 IAA33, 0.5 mg l−1 BAP and 2.0 mg l−1 KIN34. Our report is in agreement with above reports that nodal sections are the choice of explant for efficient and early shoot regeneration.
Shoot elongation and rooting
The regenerated shoots were cut and further sub-cultured on SEM containing MS media supplemented with various concentrations of GA3 (0.5–3.0 mg l−1). It has been observed that 1.0 mg l−1 of GA3 exhibited maximum significant shoot elongation as compared to other concentrations of GA3 within 15 days of incubation (Supplementary Fig. 1). According to Sreedhar et al., MS medium fortified with IBA 4.92 µM and 30 g sucrose was most efficient for shoot elongation35. GA3 was used for shoot elongation in stevia regeneration by Giridhar et al.36. It has been reported in their study that 0.05 µM of GA3 was most efficient for maximum shoot elongation36. Sivaram and Mukundan reported that rooting medium (MS media fortified with 4.90 µM IBA) also acted as shoot elongation medium37.
The elongated shoots (~ 2 cm) were transferred to root-induction medium (RIM). In our study, maximum number of roots (9 ± 2.0) was reported from shoots (5–7 cm) regenerated from nodal section, cultured on half-strength MS media devoid of PGR (Supplementary Table 3). No. of roots and root length was found significantly higher in directly regenerated shoots from nodal sections cultured on MS5 and ½ MS media. A comparison between number of shoots and roots originated from directly regenerated shoots and callus regenerated shoots is presented in Supplementary table 2 and 3, respectively. Various research groups have reported root-induction in in vitro regenerated shoots, using different combinations of growth hormones. Singh and Dwivedi reported the maximum rooting response with ¼ MS media augmented with 1.0 mg l−1 of IBA + 50 mg l−1 of activated charcoal. This media combination produced an average 11 number of roots per shoot32. Sreedhar et al. reported best rooting response in shoots incubated on ½ MS fortified with 4.92 μM IBA and 15 g dm−3 sucrose35.
Optimization of Agrobacterium-mediated transformation
To find the effect of explants type on genetic transformation of stevia, two types of explants (nodal sections and callus) were subjected to Agro-inoculation (O.D600 = 0.6). Young nodal sections (0.5 cm) exhibited a high regeneration response of 69.92%, in comparison to a low response of 31.43% with callus explants (Fig. 1A). Different concentrations (50, 100, 200 and 300 µM) of acetosyringone (As) were used to find their effect on transformation efficiency. Supplementation of 100 µm acetosyringone (As) gradually increased the percentage of responding explants to 72.5% while, a lower or a higher concentration (than 100 µM) resulted in reduction of percentage regeneration response (Fig. 1C). Different cell densities (0.2, 0.4, 0.6 and 0.8 O.D600 of Agrobacterium culture) were used to evaluate their effect on transformation efficiency Maximum regeneration response observed at O.D600 was 0.6, while at higher O.D600, Agrobacterium contamination was observed on explants (Fig. 1D). Maximum regeneration response (62%) was achieved with a pre-incubation duration of 2 days (Fig. 1B), Agro-inoculation duration of 20 min (55% regeneration response) (Fig. 1E) and co-cultivation duration of 2 days (55% regeneration response) (Fig. 1F). Incubation with Agrobacterium enhances the transformation process due to active cell division and formation of vir-inducing compounds which enhance the binding of Agrobacterium cells on the surface of newly synthesized cell wall38.
The optimizations for Agrobacterium-mediated transformation and regeneration of stevia are (1) preferred explant type: nodal sections; (2) acetosyringone (As) concentration: 100 μm; preincubation duration: 2 days; (3) Agrobacterium cell density (OD600): 0.5–0.6; (4) agro-inoculation duration: 20 min; and (5) co-cultivation duration: 2 days; (6) shoot-induction medium (SIM): [MS Basal + BAP (1.0 mg l−1) + NAA (0.5 mg l−1)]; (7) shoot elongation medium (SEM): [MS Basal + GA3 (1.0 mg l−1)]; and (8) root-induction medium (RIM): half-strength MS Basal. After co-cultivation, the explants were cultured on two step selection regime (MS media containing 2 and 4 mg l−1 of glufosinate ammonium). A high transformation efficiency of 40.48 ± 0.72% was achieved with nodal sections (Table 1) as compared to 27.94 ± 5.75% with the callus explants (Table 2). The parameters (for shoot regeneration, elongation and rooting) that were optimized for in vitro regeneration of stevia were in consonance with the regeneration after transformation. Figure 4 represent the outline of optimized protocol for stevia transformation using nodal/callus explants.
Molecular characterization of putative transformants
Genomic DNA and total RNA from randomly selected nine putative transformants (TR1-TR9) were used for bar gene integration and expression analyses by PCR, RT-PCR, Southern-blot hybridization and qRT-PCR. PCR result of the nine putative transformants showed amplification of anticipated 146 bp region of bar gene which was similar to positive control (plasmid DNA developed with gene-specific primers) (Fig. 5A,B). However, no such amplification was observed with untransformed control plantlets. RT-PCR analysis of the nine promising transformants also revealed amplification of the expected fragment of 200 and 150 bp, which verify the presence of bar and actin gene transcript in the transgenic plants (Fig. 5C,D). However, the band intensities of the c-DNA amplification product differed in each plant. The TR1 exhibited highest band-intensity while the TR9 exhibited the lowest. The nine T0 transformants were also subjected to qRT-PCR analysis. The bar gene expression levels of the nine T0 transgenic plants was in consonance with the respective band intensities obtained during RT-PCR analysis. TR9 having the lowest expression level, was taken as reference-control for qRT-PCR result analysis. The fold change in expression of bar gene was calculated in terms of 2−ΔΔCT method and plotted on graph39. Figure 5E shows ~ 47-fold increase in bar gene expression in TR1 transgenic stevia plant as compared to the control (TR9).
Molecular characterization of T0 transgenic stevia plants. (A) T-DNA region of pPZP200 vector harbouring bar gene driven by DECaMv35s promoter, (B) PCR amplification of 146 bp of bar gene, (C) RT-PCR analysis of nine randomly selected transformants showing an amplicon size of 200 bp (bar gene), (D) RT-PCR analysis of nine randomly selected transformants showing an amplicon size of 150 bp (actin gene), (E) relative fold change in expression of bar gene in T0 transgenic plants with respect to TR9 (low expressing transgenic plant taken as reference). C control/wild type, TR T0 transgenic plants, M 100 bp ladder and (F) southern hybridization analysis of six T0 transgenic plants probed with 552 bp bar gene. + C: 552 bp bar gene (positive control); − C: wild type (negative control).
Southern hybridization analysis of six highly expressing T0 transgenic events revealed the transgene copy number. Genomic DNA from non-transgenic and T0 transgenic plants (TR1, TR2, TR3, TR4, TR6 and TR7) was digested with EcoRI and subsequently hybridized with 552 bp of bar-gene-probe. The hybridization pattern of six T0 transgenic plants revealed single and double copy integration that ranged in sizes from 3.5 to 20.5 kb, but the non-transgenic plant (control) did not show hybridization with the gene probe (Fig. 5F).
Comparison of morphological characters and chlorophyll content
The transgenic plants did not exhibit any significant difference with the control in terms of morphological characters and chlorophyll content (Supplementary Table 4). Average plant height observed in wild type was 73 cm while it was 71 cm in transgenic plants. The leaf count in wild type and transgenic plants was 215 and 211, respectively. Moreover, average chlorophyll content was 7.85 mg g−1 and 7.32 mg g−1 in wild type and transgenic plants, respectively.
Herbicide tolerance assay
Herbicide tolerance assay was performed by spraying the wild type stevia (non-transformed) and T0 transgenic plants with 8 mg l−1 glufosinate ammonium, in green-house. The wild type stevia plants showed symptoms of chlorosis (Fig. 6A), phytotoxicity and defoliation after 4th day of herbicide spray and even death after 12th day. On the other hand the transgenic plants did not show such symptoms and remained healthy (Fig. 6B–E).
Herbicide tolerance assay of transgenic stevia plants. (A) control (wild type) plants sprayed with glufosinate ammonium (8 mg l−1), (B) T0 transgenic plants sprayed with glufosinate ammonium (8 mg l−1). Leaf morphology after glufosinate spray (C) control leaf after four days of spray, (D) control leaf after twelve days of spray and (E) transgenic leaf after twelve days of spray. Bar = 5 mm.
Various reports are available regarding the application of glufosinate on crop plants and weed species. Manickavasagam et al. performed herbicide resistance trials on transgenic (herbicide resistant) sugarcane cultivars Co92061 and Co67140. In this study in vitro regenerated non-transformed sugarcane plants were sprayed with different concentrations (0.5, 2.5 and 5.0 g l−1) of glufosinate ammonium to find the lethal dose of herbicide. Glufosinate @ 2.5 g l−1 with an average dose of 6.25 mg plant−1 was observed as lethal dose. This lethal dosage was then sprayed on transgenic plants under greenhouse conditions. Observations were recorded after 30 days to select the transgenic plants. Herbicide resistant sweet potato (Ipomoea batatas L.) cultivar “Yulmi” was sprayed and painted with 0.5% glufosinate herbicide (@ 900 mg l−1) under greenhouse condition to estimate their efficacy for herbicide resistance. It was found that control plants showed extensive leaf necrosis while transgenic plants remained green without any symptom of leaf necrosis41. In a study by Abdeen and Miki, it has been reported that glufosinate spray on Arabidopsis plants led to inhibition of photosynthesis and ultimately plant death, after 6–48 h of spray. While, the transgenic Arabidopsis harboring bar gene survived under the experimental conditions42. Two Chinese rice cultivars (HD297T-31, HD297T-523) were also transformed with bar gene through Agrobacterium mediated transformation, for making them resistant to glufosinate herbicide. Transgenic HD297T-31 exhibited almost 100% resistantce to glufosinate while, HD297T-523 showed moderate resistance43. In our study, glufosinate adversely affected the wild type (non-transgenic plants), while the transgenic shoots survived on 4 mg l−1 glufosinate concentration. Herbicide tolerance assay with T1 transgenic jute (Corchorus sp.) plants was carried out to analyze their herbicide resistance potential. It was found that control plants died after 12 h of glufosinate spray (0.25%) while transgenic plants successfully recovered from herbicide stress after 7th day of spray44. Our results of herbicide tolerance assay are in consonance with these reports.
Residual phytotoxic effect
No harmful phyto-toxic effect of glufosinate was found on seed germination of both the indicator plants. The difference between parameters (seed germination and seedling length) of control and treated pots was found non-significant. The corn seed germination percentage of 92.78% was observed in water treated pots (control) and 91–92% in glufosinate treated pots. Similarly, with cucumber seed germination percentage of 93.28% was observed in water treated pots and 90–91% in glufosinate treated pots. The corn seedling length of 33.43 cm was recorded in water treated pots and 32–33 cm in glufosinate treated pots. The cucumber seedling length of 7.34 cm was recorded in water treated pots and 6–7 cm in glufosinate treated pots. Moreover, no phytotoxic effect was observed on seedlings of both the indicator plants (Supplementary Table 5).
Conclusions
Glufosinate (Basta) and glyphosate (Roundup) are two most commonly used broad-spectrum herbicides for weed control in agricultural fields. However, extensive use of a single herbicide induces the weeds to develop resistance against them. At present, nearly 38 weed species has developed resistance to glyphosate, which has been distributed to 37 countries17. These resistant weeds are a major challenge to efficient weed control strategies. However, these weeds can be controlled by the strategic application of glufosinate45.
In addition to herbicide tolerance, bar gene has also been used as a selective marker in many plant genetic transformation experiments46,47. In a recent report, cotyledon explants from seven cultivars of soybean were used to introduce bar gene as a selective marker. With a transformation efficiency of 14.71%, transgenic soybean expressing bar gene was successfully developed48. In our study, much higher transformation efficiency (40.48 ± 0.72) was achieved with nodal sections of stevia plants. This suggests the feasibility of using them for high-frequency Agrobacterium-mediated transformation with novel genes of diverse origin.To the best of our knowledge, it is the first report on Agrobacterium-mediated nuclear transformation of stevia for herbicide resistance trait. This robust transformation protocol can be useful in stevia crop-improvement programs.
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- aadA2 gene:
-
Aminoglycoside nucleotidyltransferase gene
- As:
-
Acetosyringone
- BAP:
-
6-Benzylaminopurine
- Bar:
-
Bialaphos
- CaMv35sDE :
-
CaMv35s Promoter with duplicated enhancer
- GA3:
-
Gibberellic acid
- IAA:
-
Indole-3-acetic acid
- IBA:
-
Indole-3-butyric acid
- KIN:
-
Kinetin
- MS:
-
Murashige and Skoog
- NAA:
-
α-Napthalene acetic acid
- Nos:
-
Nopaline synthase
- PAT:
-
Phosphinothricin acetyl transferase
- PCR:
-
Polymerase chain reaction
- PGR:
-
Plant growth regulator
- PFD:
-
Photon flux density
- qRT-PCR:
-
Quantitative real-time PCR
- RIM:
-
Root induction medium
- RT-PCR:
-
Reverse transcriptase PCR
- SEM:
-
Shoot elongation medium
- SIM:
-
Shoot induction medium
References
Chatsudthipong, V. & Muanprasat, C. Stevioside and related compounds: Therapeutic benefits beyond sweetness. Pharma. Ther. 121(1), 41–54 (2009).
Yadav, A. K., Singh, S., Dhyani, D. & Ahuja, P. S. A review on the improvement of stevia [Stevia rebaudiana (Bertoni)]. Can. J. Plant Sci. 91(1), 1–27 (2011).
Suresh, V., Fetricia, J. P., Saranya, V., Sarithra, S. & Tamilselvan, K. Uses of stevia (Stevia rebaudiana). J. Med. Plant. 6(2), 247–248 (2018).
Ahmad, U. & Ahmad, R. S. Anti diabetic property of aqueous extract of Stevia rebaudiana Bertoni leaves in Streptozotocin-induced diabetes in albino rats. BMC Comp. Alter. Med. 18(1), 179 (2018).
Alavala, S. et al. Stevioside, a diterpenoid glycoside, shows anti-inflammatory property against Dextran Sulphate Sodium-induced ulcerative colitis in mice. Eur. J. Pharmacol. 15(855), 192–201 (2019).
Benford, D. J., DiNovi, M. & Schlatter, J. Safety evaluation of certain food additives: Steviol glycosides. WHO Food Additives Series World Health Org. Joint FAO/WHO Expert Comm. Food Additives (JECFA) 54, 140 (2006).
Das, K., Dang, R., Khanam, S. & Rajasekharan, P. E. In vitro methods for production of steviosides from stevia. Ind. J. Nat. Prod. 2, 14–15 (2005).
Jagatheeswari, D. & Ranganathan, P. Studies on Micropropagation of Stevia rebaudiana Bert. Int. J. Pharm. Biol. Archit. 3, 315–320 (2012).
Taak et al. Comparative assessment of mulching and herbicide treatments for weed management in Stevia rebaudiana (Bertoni) cultivation. S. Afr. J. Bot. 000(2020), 1–9 (2020).
Harrington, K. C., Southward, R. C., Kitchen, K. L. & He, X. Z. Investigation of herbicides tolerated by Stevia rebaudiana crops. Crop Hort. Sci. 39, 21–33 (2011).
Kumar, R. et al. Effect of plant spacing and organic mulch on growth, yield and quality of natural sweetener plant Stevia and soil fertility in western Himalayas. Int. J. Plant Prod. 8(3), 311–334 (2014).
Ghorai, A. K. Integrated weed management in jute (Corchorusolitorius). Ind. J. Agric. 53, 149–151 (2008).
EFSA (European Food Safety Authority). Conclusion on the peer review of the pesticide risk assessment of the active substance glyphosate. EFSA J. 13(11), 4302 (2015).
Bai, S. H. & Ogbourne, S. M. Glyphosate: Environmental contamination, toxicity and potential risks to human health via food contamination. Environ. Sci. Pollut. Res. 23, 18988–19001 (2016).
Duke, S. O. Taking stock of herbicide-resistant crops ten years after introduction. Pest Manag. Sci. 61, 211–218 (2005).
Heap, I. & Duke, S. O. Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 74, 1040–1049 (2018).
Pande, S. S. & Gupta, P. Plant tissue culture of Stevia rebaudiana (Bertoni): A review. J. Pharmaco. Phytother. 5(1), 26–33 (2013).
Gantait, S., Arpita, D. & Nirmal, M. Stevia: A comprehensive review on ethnopharmacological properties and in vitro regeneration. Sug. Tech. 17(2), 95–106 (2014).
Singh, P., Dwivedi, P. & Atri, N. In vitro shoot regeneration of Stevia rebaudiana through callus and nodal segments. Int. J. Agric. Environ. Biotechnol. 5(2), 101–108 (2012).
Ali, A., Gull, I., Naz, S. & Afghan, S. Biochemical investigation during different stages of in vitro propagation of Stevia rebaudiana. Pak. J. Bot. 42(4), 2827–2837 (2010).
Thiyagarajan, M. & Venkatachalam, P. Large scale in vitro propagation of Stevia rebaudiana Bert. for commercial application: Pharmaceutically important and antidiabetic medicinal herb. Ind. Crops Prod. 37(1), 111–117 (2012).
Das, A., Gantait, S. & Mandal, N. Micropropagation of an elite medicinal plant: Stevia rebaudiana Bert. Int. J. Agric. Res. 6, 40–48 (2011).
Anbazhagan, M., Kalpana, M., Rajendran, R., Natarajan, V. & Dhanavel, D. In vitro production of Stevia rebaudiana Bertoni. Emir. J. Food Agric. 22(3), 216–222 (2010).
Koul, B., Srivastava, S., Amla, D. V. & Sanyal, I. Establishment and optimization of Agrobacterium-mediated transformation and regeneration of tomato (Solanum lycopersicum L.). Int. J. Biosci. 4(10), 51–69 (2014).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum 15(3), 473–497 (1962).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, New York, 2001).
Gupta, P., Sharma, S. & Saxena, S. Micropropagation of Stevia rebaudiana (natural sweetener) using KIN for steviol glycoside production. Res. J. Biotechnol. 5, 63–67 (2010).
Ahmed, M. B. et al. An efficient method for in vitro clonal propagation of newly introduced sweetener from plant Stevia rebaudiana Bertoni in Bangladesh. Am. Eurasian J. Sci. Res. 2(2), 121–125 (2007).
Janarthanam, B., Gopalakrishnan, M., Lakshmi, S. G. & Sekar, T. Plant regeneration from leaf derived callus of Stevia rebaudiana Bertoni. Plant Tiss. Cult. Biotechnol. 19, 133–141 (2009).
Sairkar, P., Shukla, N. P. & Mehrotra, N. N. Mass production of an economically important medicinal plant Stevia rebaudiana using in vitro propagation techniques. J. Med. Plants Res. 3(4), 266–270 (2009).
Debnath, M. Clonal propagation and antimicrobial activity of an endemic medicinal plant Stevia rebaudiana. J. Med. Plant Res. 2, 45–51 (2008).
Singh, P. & Dwivedi, P. Two-stage culture procedure using thidiazuron for efficient micropropagation of Stevia rebaudiana, an anti-diabetic medicinal herb. 3 Biotech. 4(4), 431–437 (2013).
Laribi, B., Rouatbi, N., Kouki, K. & Bettaieb, T. In vitro propagation of Stevia rebaudiana (Bert.)—A non-caloric sweetener and antidiabetic medicinal plant. Int J. Med. Arom. Plants 2, 333–339 (2012).
Mehta, J. et al. Micropropagation of an anti-diabetic plant-Stevia rebaudiana Bertoni, (Natural Sweetener) in Hadoti region of south-east Rajasthan, India. J. Bio. Sci. 1, 37–42 (2012).
Sreedhar, R. V. et al. Direct organogenesis from leaf explants of Stevia rebaudiana and cultivation in bioreactor. Biol. Plant. 52(2), 355–360 (2008).
Giridhar, P., Sowmya, K. S., Ramakrishna, A. & Ravishankar, G. A. Rapid clonal propagation and stevioside profiles of Stevia rebaudiana Bertoni. Int. J. Plant Dev. Biol 4(1), 47–52 (2010).
Sivaram, L. & Mukundan, U. In vitro culture studies on Stevia rebaudiana. In Vitro Cell. Develop. Biol. Plant. 39(5), 520–523 (2003).
Sunilkumar, G., Vijayachandra, K. & Veluthambi, K. Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Sci. 141, 51–58 (1999).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR & the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Manickavasagam, M. et al. Agrobacterium-mediated genetic transformation and development of herbicide-resistant sugarcane (Saccharum species hybrids) using axillary buds. Plant Cell Rep. 23(3), 134–143 (2003).
Choi, H. J., Chandrasekhar, T., Lee, H. Y. & Kim, K. M. Production of herbicide-resistant transgenic sweet potato plants through Agrobacterium tumefaciens method. Plant Cell Tiss. Org. Cult. 91(3), 235–242 (2007).
Abdeen, A. & Miki, B. The pleiotropic effects of the bar gene and glufosinate on the Arabidopsis transcriptome. Plant Biotechnol. J. 7, 266–282 (2009).
Tian, X. et al. Transformation of upland rice with the bar gene and selection for resistance to the herbicide Basta. Euphytica 1, 151–167 (2015).
Majumder, S., Datta, K., Sarkar, C., Saha, S. C. & Datta, S. K. The development of Macrophominaphaseolina (fungus) resistant and glufosinate (herbicide) tolerant transgenic jute. Front. Plant Sci. 9, 920 (2018).
Barnes, E. R., Knezevic, S. Z., Sikkema, P. H., Lindquist, J. L. & Jhala, A. J. Control of glyphosate-resistant common ragweed (Ambrosia artemisiifolia L.) in glufosinate-resistant soybean [Glycine max (L.) Merr]. Front. Plant. Sci. 8, 1455 (2017).
Vasconcelos, M. et al. Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 164, 371–378 (2003).
Miki, B. & McHugh, S. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J. Biotechnol. 107, 193–232 (2004).
Yang, X. F., Yu, X. Q., Zhou, Z., Ma, W. J. & Tang, G. X. A high-efficiency Agrobacterium tumefaciens mediated transformation system using cotyledonary node as explants in soybean (Glycine max L.). Acta. Physiol. Plant 8(3), 60 (2016).
Acknowledgements
We are thankful to the administration of Lovely Professional University, Punjab, India and National Agri-Food Biotechnology Institute (NABI), Department of Biotechnology, Govt. of India, for the infrastructural support. We thankfully acknowledge Dr. Samir Sawant, Sr. Principal Scientist, Council of Scientific and Industrial Research-National Botanical Research Institute, Lucknow, India, for providing the modified bar gene construct.
Author information
Authors and Affiliations
Contributions
B.K. conceived the idea and P.T. performed the experiments. B.K. drafted the manuscript. S.T. co-supervised the overall experimental work and helped in data compilation. All the authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taak, P., Tiwari, S. & Koul, B. Optimization of regeneration and Agrobacterium-mediated transformation of Stevia (Stevia rebaudiana Bertoni): a commercially important natural sweetener plant. Sci Rep 10, 16224 (2020). https://doi.org/10.1038/s41598-020-72751-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72751-8
This article is cited by
-
Agrobacterium tumefaciens-Mediated Plant Transformation: A Review
Molecular Biotechnology (2023)
-
In vitro regeneration and Agrobacterium-mediated genetic transformation of Dragon’s Head plant (Lallemantia iberica)
Scientific Reports (2022)
-
Development of Herbicide-Resistant Transgenic Stevia (Stevia rebaudiana Bertoni) as an Effective Weed-Management Strategy in Stevia Cultivation
Sugar Tech (2021)
-
Biotechnological interventions of in vitro propagation and production of valuable secondary metabolites in Stevia rebaudiana
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.