Abstract
McrA is a key transcription factor that functions as a global repressor of fungal secondary metabolism in Aspergillus species. Here, we report that mcrA is one of the VosA-VelB target genes and McrA governs the cellular and metabolic development in Aspergillus nidulans. The deletion of mcrA resulted in a reduced number of conidia and decreased mRNA levels of brlA, the key asexual developmental activator. In addition, the absence of mcrA led to a loss of long-term viability of asexual spores (conidia), which is likely associated with the lack of conidial trehalose and increased β-(1,3)-glucan levels in conidia. In supporting its repressive role, the mcrA deletion mutant conidia contain more amounts of sterigmatocystin and an unknown metabolite than the wild type conidia. While overexpression of mcrA caused the fluffy-autolytic phenotype coupled with accelerated cell death, deletion of mcrA did not fully suppress the developmental defects caused by the lack of the regulator of G-protein signaling protein FlbA. On the contrary to the cellular development, sterigmatocystin production was restored in the ΔflbA ΔmcrA double mutant, and overexpression of mcrA completely blocked the production of sterigmatocystin. Overall, McrA plays a multiple role in governing growth, development, spore viability, and secondary metabolism in A. nidulans.
Similar content being viewed by others
Introduction
Most filamentous fungi produce a high number of asexual spores to survive and propagate in the environment1. Fungal spores are rapidly distributed in the air affecting humankind in a variety of ways. Spores can act as the major infectious agent in both animals and humans, and the inhaled fungal spores can cause invasive infectious diseases in immunocompromised individuals2. Fungal spores are also easily widespread in the air and germinate in the crops causing economic losses3. In addition, spore formation in some filamentous fungi is closely linked to the biosynthesis of secondary metabolites, such as mycotoxins4,5.
The genus Aspergillus is one of the most important fungal genera as some species can cause diseases in humans (A. fumigatus), produce the most potent carcinogen in nature aflatoxins (A. flavus), and are used for the food and pharmaceutical industries (A. oryzae and A. niger)3,6. Aspergillus species use asexual sporulation (called conidiation) as a main reproductive mode7. Aspergillus species produce a specialized asexual reproductive structure called conidiophore, which bears numerous chains of asexual spores called conidia8,9. A conidiophore is composed of varying cell types and the process of its production is precisely regulated by multiple positive and negative regulators10. Some of these developmental regulators are thought to be conserved in Aspergillus species, and they have been extensively studied in the model fungus A. nidulans11,12.
The asexual life cycle of A. nidulans can be divided in two stages; vegetative growth and conidiation8. During vegetative growth, fungal spores germinate, leading to the formation of the undifferentiated hyphae. In the early growth phase, the FadA- and GanB-mediated heterotrimeric G protein signaling pathways activate spore germination and vegetative growth, but repress conidiation and sterigmatocystin (ST) production7. During the early vegetative growth phase, key negative regulators such as NsdD, VosA, and SfgA cooperatively repress expression of brlA, the essential activator for the initiation of conidiation, until the acquisition of the developmental competence13,14,15,16. Under the appropriate conditions, it is speculated that these negative regulators are displaced from the brlA promoter, and upstream positive regulators induce brlA mRNA expression, thereby the fungus begins conidiation and forms conidiophores10.
BrlA is a C2H2 transcription factor (TF) that activates abaA, which in turn activates wetA17,18,19,20,21. These genes consist of the conserved central cascade of conidiation in Aspergillus species and they control expression of thousands of asexual developmental genes8,22,23,24,25. The process of the spore formation and maturation is governed by WetA and the VosA-VelB complex15,24,25,26,27,28. These regulators cooperatively control expression of spore-specific genes for conidia formation and integrity, and confer feed-back negative regulation of brlA24,25,28,29.
VosA and VelB are the velvet regulators governing multiple processes including conidiation, spore viability, secondary metabolism, and conidial trehalose biogenesis in A. nidulans26,28,29,30. Genome-wide expression and protein-DNA analyses demonstrated that VosA and VelB directly bind to the promoter regions of many genes such as fksA, tpsA, and brlA, and control their expression29,30. Further analyses have led us to define additional VosA-VelB target genes such as vadA (VosA/VelB-Activated Developmental gene)31, mtfA (Master TF A)32, sclB (Sclerotia-like B)33, and mcrA (MultiCluster Regulator A; AN8694)34.
Our previous study presented that the expression of mcrA in conidia requires both VosA and VelB30. The mcrA gene encodes a putative TF with a Zn(II)2Cys6 domain, which acts as a multicluster negative regulator of fungal secondary metabolism34. Overexpression of mcrA caused repression of secondary metabolite production, whereas deletion of mcrA induced the production of novel secondary metabolites in A. nidulans34. Independent to this study, we have been characterizing the mcrA gene as a direct target of the VosA-VelB heterodimer in conidia and present new findings in the present report. Briefly, the deletion of mcrA resulted in the decreased production of conidia, a rapid loss of conidial viability, and a reduced amount of conidial trehalose, but increased the levels of the conidial β-(1,3)-glucan and secondary metabolites such as ST. Conversely, overexpression of mcrA led to a near complete shut-down of secondary metabolites and the fluffy-autolytic phenotypes. Further genetic studies led us to conclude that McrA represses secondary metabolism downstream of FadA-mediated signaling pathway. We present a genetic model depicting the roles of McrA coordinating cellular and metabolic development in A. nidulans.
Results
Expression and the role of McrA in asexual development of A. nidulans
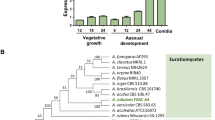
Previous study described that mRNA levels of mcrA in conidia were drastically low by the lack of VelB or VosA, and a promoter region of mcrA contains a putative VosA response element (VRE), suggesting that McrA is a potential target of the VosA-VelB complex in A. nidulans30. The mcrA ORF composed of 1,453 bp with four exons predicted to encode a 399 aa-length protein that contains a GAL4-like Zn(II)2Cys6 domain at the C-terminus. To begin to investigate its function, the levels of mcrA mRNA in the life cycle were investigated. As shown in Figs. 1A and S1A, mcrA mRNA was detectable throughout the life cycle and was high at 48 h after asexual developmental induction.
Expression and role of mcrA in asexual development. (A) Levels of mcrA mRNA during A. nidulans life cycle. C = conidia. The time (hr) of incubation in liquid submerged culture (Vege) and post asexual (Asexual) or sexual (Sexual) induction. Equal loading of total RNA was validated using ethidium bromide staining of rRNA. (B) Phenotypic analyses of WT (FGSC4), ΔmcrA (TMK19), and C’mcrA (TMK20) strains. All strains were point inoculated onto solid MMG and incubated at 37 °C for 3 d, the bottom panels show close-up views of the middle of the plates (bar = 100 μm). (C) Quantitative analysis of conidia formation of the strains shown in (B). The numbers of conidia per plate were counted in triplicates (***p < 0.001). (D) Levels of brlA mRNA during the life cycle of WT and ΔmcrA strains. C = conidia. The time (hr) of incubation in liquid submerged culture (Vege) and post asexual developmental induction (Asexual) is shown. Equal loading of total RNA was confirmed using ethidium bromide staining of rRNA.
To investigate the roles of mcrA, we generated the mcrA deletion (ΔmcrA) mutant and the ΔmcrA complemented (C’mcrA) strains, which reintroduced a wild-type copy of the mcrA gene back into the other locus. Wild type (WT), ΔmcrA, and complemented strains were point-inoculated onto a minimal medium (MM) with 1% glucose (MMG) agar and their phenotype was checked. In comparison to WT and complemented strains, the ΔmcrA mutant strain produces abnormal conidiophores and brown colony (Fig. 1B). The deletion of mcrA resulted in a reduced number of conidia and reduced levels of brlA compared to WT and complemented strains (Figs. 1C,D and S1B). These results suggest that McrA plays a key role in asexual development in A. nidulans.
McrA is required for conidial integrity
As the expression of mcrA was activated by VosA-VelB, which controls the conidial viability and integrity, we hypothesized that McrA may play a role in spore survival. To test this hypothesis, WT, ΔmcrA, and C’ strain conidia were collected from 2, 5, 8, 10 and 20 days grown colonies and checked for the viability. As shown in Fig. 2A, the ΔmcrA mutant conidia rapidly lost viability starting from day 5. As trehalose is a key component conferring the long-term spore viability, conidial trehalose amount was tested in the ΔmcrA mutant conidia. The amount of trehalose in ΔmcrA conidia was about twofold less than that of WT or C’ strains (Fig. 2B). Since the absence of vosA increased the levels of β-(1,3)-glucan in conidia, we investigated the β-(1,3)-glucan levels in the ΔmcrA mutant conidia, and found that β-(1,3)-glucan levels in the ΔmcrA conidia were about twofold higher than those of WT and C’ conidia (Fig. 2C). This was corroborated by the finding that ΔmcrA mutant conidia exhibited elevated mRNA levels of fksA, a gene encoding a β-1,3-glucan synthase (Fig. 2D). These results indicate that VosA-VelB-activated McrA is necessary for the governing the integrity of conidia.
The role of mcrA in conidia. (A) Spore viability of WT (FGSC4), ΔmcrA (TMK19), and C’mcrA (TMK20) strains grown at 37 °C for 2, 5, 8, 10 and 20 days. (B) The amount of trehalose (μg) per 107 spores from the 2 day-old colonies of WT, ΔmcrA and C’mcrA strains. (measured in triplicates, ***p < 0.001) (C) The amount of β-glucan (pg) per 104 spores in 2-day-old conidia of WT, ΔmcrA and C’mcrA strains (measured in triplicates, ***p < 0.001). (D) Levels of fksA mRNA in the WT, ΔmcrA and C’mcrA conidia. (measured in triplicates, ***p < 0.001).
The absence of mcrA leads to elevated secondary metabolism
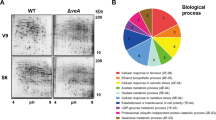
Previously, Oakley and colleagues have shown that the McrA is a negative regulator of the production of secondary metabolites in A. nidulans34. We confirmed that the absence of mcrA alters the patterns of secondary metabolites and increases ST production in stationary cultures (Fig. 3A,B). We then tested the secondary metabolite patterns in conidia using the high-performance liquid chromatography (HPLC) and found that the ΔmcrA conidia showed about fourfold enhanced production of ST and an unknown metabolite compared to those of WT and complemented strains (Fig. 3C–E). We then tested mRNA levels of aflR encoding an essential Zn(II)2Cys6 TF for the activation of the ST gene cluster and found that the ΔmcrA conidia exhibited higher levels of aflR mRNA than those of WT and C’ conidia (Fig. 3F). Overall, these results imply that McrA is a key negative regulator of secondary metabolite production in both hyphae and conidia.
The effects of ΔmcrA on production of secondary metabolites. (A) Thin-layer chromatography (TLC) of secondary metabolites from WT, ΔmcrA (TMK19), and C’mcrA (TMK20) strains under dark for 3 days stationary culture with mycelia. (B) Amount of ST in conidia of WT, ΔmcrA (TMK19), and C’mcrA (TMK20) strains by HPLC analysis (measured in triplicates, ***p < 0.001). (C) Secondary metabolites in asexual spores by HPLC analysis. Asexual spores (2Χ108) of WT, ΔmcrA (TMK19), and C’mcrA (TMK20) strains were collected and extracted with chloroform, processed, and subjected to HPLC analysis. The blue arrow designates ST. The green arrow designates unknown metabolites (measured in triplicates). (D) Amount of ST in conidia of WT, ΔmcrA (TMK19), and C’mcrA (TMK20) conidia (measured in duplicate, ***p < 0.001). (E) Amount of unknown metabolite in conidia of WT, ΔmcrA (TMK19), and C’mcrA (TMK20) strains (measured in duplicate, *p < 0.05). (F) Levels of aflR mRNA in the WT, ΔmcrA, and C’mcrA conidia.
Pleiotropic effects caused by overexpression of McrA
To further test the potential role of mcrA in conidiation and secondary metabolism, mcrA overexpression (OE) strains by expressing an ectopic copy of mcrA under the alcA promoter (alcA(p)::mcrA) were generated and checked for their phenotypes. First, we checked ST accumulation under non-inducing (MMG) and inducing (MM with 100 mM threonine, MMT) conditions and found that there were no differences between WT and OEmcrA for ST production in MMG. However, when cultured in MMT, OEmcrA caused the total blockage of ST and other metabolites’ production (Fig. 4A). The previous study described that OEmcrA leads to a reduction of fungal growth that might be related to a reduction of secondary metabolite production34. We then cultured WT and OEmcrA strains onto solid MMG and solid MMT and found that OEmcrA led to the fluffy-autolytic phenotype with about tenfold reduction in conidia formation, whereas growth and development of WT and OEmcrA strains were similar in MMG (Fig. 4B,C). We then examined whether OEmcrA-caused fluffy-autolytic phenotypes were coupled with accelerated cell death using the alamarBlue reduction assay and found that, OEmcrA led to dramatically reduced cell viability starting at day 3 compared to WT (Fig. 4D). Taken together, these results imply that McrA plays an important role not only in the production of ST and other secondary metabolites, but also in fungal growth, conidiation, autolysis, and cell death.
Overexpression of mcrA causes pleiotropic effects. (A) Amount of ST production in WT (FGSC4) and OEmcrA (THS41; alcA(p)::mcrA) strains. The supernatant of each strain following 3 days of stationary culture was extracted using chloroform and subjected to TLC analysis. (B) WT (FGSC4) and OEmcrA (THS41; alcA(p)::mcrA) strains were point inoculated onto solid MMG (non-inducing) or onto solid MMT (inducing) including 0.5% YE medium. Photographs of the cultures at day 5 are also shown. The bottom panels indicate close-up views of the middle of the plates (bar = 100 μm). (C) Quantitative analysis of conidiospore formation of the strains shown in (B). The numbers of conidia per plate were counted in triplicates (**p < 0.01; ***p < 0.001). (D) Relative AB reduction rates of WT and OEmcrA (THS41) strains grown under submerged culture conditions at 37 °C. The percent of alamarBlue (AB) reduction represents the fungal cell viability. The mean values were represented by a bar graph.
Genetic epistasis between McrA and FlbA
Previous studies found that the FadA Gα-dependent signaling pathway activates vegetative growth while inhibiting development and ST biosynthesis, and that this signaling is attenuated by the regulator of G-protein signaling (RGS) protein FlbA35,36,37. Loss of flbA function leads to the fluffy-autolytic phenotype coupled with the lack of ST production, accelerated cell death and autolysis (Fig. 5A)38. As FlbA functions quite upstream of vegetative signaling, we envisioned that McrA might act downstream of FlbA-controlled pathway and exert the fluffy-autolytic phenotype and the blockage of ST production. To test this genetic epistasis, we generated the ΔflbA ΔmcrA double mutant and compared its phenotypes with those of WT and individual single mutant. As shown in Fig. 5B, the ΔflbA ΔmcrA double mutant exhibited a decrease in colony diameter which is similar to the ΔflbA mutant and exhibited partial autolysis in the edge of the colony. In addition, asexual development was not restored in the ΔflbA ΔmcrA double mutant compared to the ΔflbA or ΔmcrA single mutant (Fig. 5C). Likewise, while delayed 1 day compared to the ΔflbA mutant, the ΔflbA ΔmcrA double mutant could not regain cell viability (Fig. 5D). These results imply that mcrA is not essential but adequate to cause the fluffy-autolytic phenotype and accelerated cell death, and that McrA is likely independent to the FlbA-controlled pathway in fungal growth and autolysis. Contrarily, the ΔflbA ΔmcrA double mutant produced ST at some levels, whereas the ΔflbA mutant failed to produce any ST (Fig. 5E). These results indicate that McrA is indeed a key negative regulator of ST biosynthesis, and that McrA acts downstream of the FlbA-attenuated signaling pathway repressing ST production.
Genetic position of McrA action. (A) WT (FGSC4), OEmcrA (THS41; alcA(p)::mcrA), and ΔflbA (TMK15) strains were point inoculated onto solid MMG (non-inducing) or solid MMT (inducing) including 0.5% YE. Photographs of the cultures at day 2 and 5 are shown. (B) WT, ΔmcrA (TMK19), ΔflbA (TMK15), and ΔmcrA ΔflbA (TMK14) strains were point inoculated onto solid MMG. Photographs of the cultures at day 5 are shown. (C) Quantitative analysis of conidiospore formation of the strains shown in (B). The numbers of conidia per plate were counted in triplicates. (D) Relative AB reduction rates of WT, ΔmcrA (TMK19), ΔflbA (TMK15) and ΔmcrA ΔflbA (TMK14) strains grown under submerged culture conditions at 37 °C. The percent of alamarBlue (AB) reduction represents the fungal cell viability. The mean values were represented by a bar graph, respectively. (E) ST analysis by TLC. WT, ΔmcrA (TMK19), ΔflbA (TMK15), and ΔmcrA ΔflbA (TMK14) strains were stationary cultured in liquid MMG at 37 °C for 3 days, and extracted with chloroform and subjected to TLC. ST standard (15 μg) was loaded as a positive control. The arrow indicates ST.
Discussion
Conidial formation and maturation are regulated by three key transcription factors WetA, VosA and VelB15,25,26. These are highly conserved regulators that control the expression of spore-specific genes in Aspergillus species25,27. Genome-wide expression and protein-DNA analyses identified certain target genes for these TFs25,30. In addition, follow-up studies identified the functions of some target genes and these results provide some clues to elaborate on how these TFs can control spore formation and maturation29. For example, the VosA-VelB complex controls the expression of fksA, mtfA, sclB, and vadA, thereby fine-tuning β-glucan synthesis, secondary metabolism, oxidative response, and conidial pigmentation29,31,32,33,39,40,41. This study further expands the VosA/VelB-mediated regulatory networks involving McrA in A. nidulans. In conidia, deletion of mcrA leads to decreased conidial viability and trehalose amount but increased β-glucan and ST levels. The ΔmcrA mutant conidia exhibited an intermediate phenotype compared to the ΔvosA mutant and WT conidia. In addition, previous microarray and Chromatin-Immuno-Precipitation (ChIP) followed by microarray (ChIP-chip) analyses proposed that VosA may directly bind to the mcrA promoter and regulate mcrA expression in conidia, that is, McrA is probably a direct target of VosA in conidia30.
McrA is a multifunctional regulator controlling certain gene expression involved in biosynthesis of secondary metabolites34. We revealed that, in conidia, the loss of mcrA increased the mRNA level of aflR, proposing the role of McrA in repressing conidial secondary metabolites. Furthermore, McrA is needed for proper biogenesis of conidial trehalose and downregulation of fksA in conidia, thereby governing the integrity of spores.
Importantly, we have shown McrA’s new role in vegetative growth, autolysis, and cell death in A. nidulans. Autolysis is a naturally occurring phenomenon that is reported as enzymatic self-degradation of the cells, affected by nutrient limitation, aging, and other factors42. Previous studies have noted that FlbA, an RGS protein, negatively regulates vegetative growth by turning off the FadA-mediated G protein to cAMP-dependent protein kinase (PKA) signaling cascade37. The absence of flbA leads to prolonged the activation of FadA-mediated G protein signaling, resulting in the autolytic phenotypes36. Our double deletion analysis has shown that the deletion of mcrA leads to a slight delay in the autolytic phenotypes but could not suppress the elevated cell death caused by ΔflbA. On the contrary, the removal of mcrA restored ST production in the ΔflbA mutant, proposing that McrA represses ST biosynthesis downstream of the FlbA-controlled FadA-PKA signaling route.
Collectively, we suggest a genetic model showing the multiple roles of McrA in A. nidulans (Fig. 6). During vegetative growth, McrA represses secondary metabolites production acting downstream of the FadA (Gα) and SfaD::GpgA (Gβγ) mediated signaling pathway, which is attenuated by the RGS protein FlbA. McrA may have a potential to activate hyphae growth and autolysis in parallel to the FadA-mediated signaling pathway. McrA is also required for proper expression of brlA and thereby conidiation. It is not yet known whether McrA directly binds to the brlA region or it affects other upstream regulators. During spore formation, the VosA-VelB complex activates the expression of mcrA in conidia, thereby controlling trehalose biogenesis and spore viability, and inhibiting ST production and β-glucan synthesis. While this study demonstrated the complex role of McrA in fungal biology, the detailed molecular mechanisms of McrA action need to be investigated. In this regard, we are in the process of identifying the direct targets of McrA via ChIP-seq analysis using the McrA-FLAG fusion protein.
Model for McrA-mediated regulation of cellular and metabolic development in A. nidulans. (see “Discussion” section).
Methods
Fungal strains and culture conditions
A. nidulans strains used in this study are listed in Table 1. Fungal strains were grown on solid or liquid minimal medium (MM) with 1% glucose (MMG) with supplements as described previously43. To determine the numbers of conidia in WT (FGSC4) and mutant strains, 105 spores were spread onto solid MMG and incubated at 37 °C for 2 days. The conidia were then collected from the entire plate and counted with the use of a hemocytometer.
To examine the effects of overexpression of mcrA by expressing an ectopic copy of mcrA under the alcA promoter44,45, all strains were grown in liquid MMG at 37 °C and 220 rpm (Innova 4,330, New Brunswick) for 14 h (designated as time point ‘‘0′’) and then transferred on solid MMG (non-inducing) or solid MM with 100 mM threonine (MMT to induce overexpression of mcrA) and 0.1% yeast extract (w/v).
For Northern blot analysis, samples were collected as described previously26. Briefly, 106 conidia/mL were inoculated in 100 mL liquid MMG in 250 mL flask and cultured at 37 °C and 220 rpm. Samples from liquid submerged culture were collected at designated time points, squeeze-dried and stored at -80 °C until the isolation of RNA. For sexual and asexual developmental induction, 18 h vegetative grown mycelia were filtered and then transferred to solid MMG. The plates were air exposed for asexual developmental induction, or tightly sealed and blocked from light for sexual developmental induction46.
Generation of A. nidulans strains
Oligonucleotides used in this study are listed in Table 2. The double joint PCR (DJ-PCR) method47 was used to generate the ΔmcrA and ΔflbA mutants. Both 5′ and 3′ flanking regions of each gene were amplified from genomic DNA of A. nidulans FGSC4 using OHS767;OHS769 and OHS768;OHS770 (for mcrA), and OMK607;OMK613 and OMK614;OMK610 (for flbA). The A. fumigatus pyrG+ marker was amplified from A. fumigatus AF293 genomic DNA with the primer pair OMK589;OMK590. The final mcrA or flbA deletion construct was amplified with OHS771;OHS772 or OMK611;OMK612, respectively. To generate the ΔflbA ΔmcrA double mutant, 5′ and 3′ flanking regions of flbA (OMK607;OMK608 and OMK609;OMK610) were amplified. The A. nidulans pyroA+ marker was amplified from FGSC4 genomic DNA with the primer pair ONK395;ONK396. The flbA deletion cassette was introduced into TMK19. Protoplasts were generated using the Vinoflow FCE lysing enzyme (Novozymes)48. At least three independent deletion mutant strains were isolated and confirmed by PCR analysis.
To complement the deletion of mcrA, the mcrA locus such as its 2 kb 5′ UTR and coding region was amplified with the primer pair OMK657;OHS878, digested with EcoRI and NotI, and cloned into pHS1326, which contains 3/4 pyroA, a 3xFLAG tag, and the trpC terminator. The resulting plasmid pMK23 was then introduced into the recipient ΔmcrA mutant TMK19, in which a single copy mcrA+ is confirmed to be inserted into the pyroA4 locus, to give rise to TMK20.
To generate the alcA(p)::mcrA fusion construct, the mcrA ORF derived from FGSC4 genomic DNA was amplified using the primer pair OHS875;OHS878. The amplicon was double digested with EcoRI and NotI and cloned into pHS3, which has the alcA promoter and the trpC terminator49. The resulting plasmids pHSN74 was then introduced into TNJ36. The mcrA overexpression (OEmcrA) mutant, THS41, was screened by Western blot analysis using monoclonal anti-Flag antibody (M2 clone, Sigma-Aldrich).
Nucleic acid manipulation
Genomic DNA isolation was performed as previously described48. Total RNA for Northern blot was isolated from each sample using Trizol reagent (Thermo Fisher Scientific) following the protocol provided by the manufacturer’s instructions. For Northern blot analysis, DNA probes were prepared by PCR amplification of the coding region of individual genes with suitable oligonucleotide pairs using WT genomic DNA as a template. Probes were labeled with 32P-dCTP (PerkinElmer) using the Random Primer DNA Labeling Kit (Takara Bio) and purified by Illustra MicroSpin G-25 columns (GE Healthcare).
For quantitative real-time PCR, complementary DNA was synthesized using the GoScript Reverse Transcription system (Promega) using the total RNA was isolated from each sample using Trizol reagent. qRT-PCR was performed with each gene-specific primer set and iTaq universal SYBR Green supermix (Bio-Rad) and using a CFX96 Touch Real-Time PCR system (Bio-Rad).
Determination of cell viability
For spore viability, WT and mutant strains were inoculated onto MMG and cultured for 2, 5, 8, 10, and 20 days15. Conidia were collected from the cultured plates. After then, about 100 conidia were spread onto MMG plates and the plates were then incubated for 48 h. Survival rates were calculated as the ratio of the number of colony forming unit to the number of spores inoculated.
Fungal cell viability was determined by the percent reduction of alamarBlue (Bio-Rad). The alamarBlue assay reagent was placed into each well of a 24-well plate, which has 1 mL of fresh liquid MMG with 0.5% YE and 0.5 mL of individual cultures with an equal amount of the mycelium, at a final concentration of 10% of the reaction volume. After the plate was incubated at 37 °C for 6 h in the dark38, the absorbance of each well was detected by A570 and A600 nm wavelength. The reduction percent of alamarBlue was calculated as described previously50,51. The values are designated as the mean standard deviation for triplicates of individual cultures.
Sterigmatocystin extraction and thin-layer chromatography (TLC) analysis
Sterigmatocystin (ST) was extracted from fresh conidia and examined as described31. Briefly, 105 conidia were inoculated into 2 mL liquid complete medium (CM) and slant cultured at 37 °C for 3 days. ST was extracted by adding 2 mL of CHCl3, and the organic phase was transferred into 1.5 mL tubes and then centrifuged at 10,000 rpm for 2 min. The CHCl3 layer was collected, dried, and then resuspended in 100 μL of CHCl3. Approximately 10 μL of each sample was applied onto a TLC silica plate including a fluorescence indicator (Kiesel gel 60, 0.25 mm thick, Merck). ST standard (Sigma-Aldrich) was loaded onto the TLC plate. The TLC plate was then developed with toluene:ethyl acetate:acetic acid (80:10:10, v/v/v), where the Rf value of ST was 0.65. Aluminum chloride (20% w/v in 95% ethanol) was sprayed onto the TLC plate and the plate was baked at 70 °C for 5 min to enhance the detection of ST. The TLC plate was exposed to UV of A320 nm, and ST levels were measured. This experiment was performed in triplicate.
High-performance liquid chromatography (HPLC) analysis
The HPLC analysis was performed as previous described31. Asexual spores (2 × 108) fungal strains were extracted by adding chloroform into the vials. The samples were vigorously mixed using a vortex mixer. The organic phase was then separated by centrifugation and transferred to new vials. Each sample was evaporated and resuspended with 0.5 mL of HPLC grade acetonitrile:methanol (50:50, v/v). For the control, ST was dissolved using the same solvent and then serially diluted. Samples and the ST standard were filtered using a 0.45 μm pore filter. A linear calibration curve (R2 = 0.998) was constructed with a ST dilution series, 10 μg/mL, 1 μg/mL, 0.5 μg/mL, 0.1 μg/mL, and 0.005 μg/mL. HPLC-diode array detection (DAD) analysis was carried out using a Series 1,100 binary pump with an auto sampler and Nova-Pak C-18 column (Agilent Technologies). The mobile phase was consisted of acetonitrile:water (60:40, v/v). The flow rate was 0.8 mL/min and ST was detected at a wavelength of 246 nm. Retention time for ST was approximately 5.6 min. Samples (10 μL) were auto-injected and run in triplicate.
β-(1,3)-glucan analysis
The β-(1,3)-glucan concentration in conidia was determined using the Glucatell assay (Associates of Cape Cod). All samples were tested according to the manufacturer’s instructions52. Briefly, 2-day old conidia were collected with ddH2O and resuspended in 25 mL of ddH2O. Each sample was mixed with 25 μL of Glucatell reagent and incubated at 37 °C for 30 min. To stop the reaction, diazo-reagents were added and optical density was determined at 540 nm. The mean rate of optical density change was determined for each well, and the β-(1,3)-glucan concentration was determined by comparison to a standard curve. This assay was performed in triplicate.
Conidial trehalose analysis
The amount of conidial trehalose was measured as previously described31. Fungal strains were inoculated onto MMG solid and then cultured for 2 days. After culture, 2 × 108 conidia were collected, resuspended in ddH2O, and incubated at 95 °C for 20 min. The supernatant was separated by centrifugation, transferred into a new tube, mixed with equal volume of 0.2 M sodium citrate (pH5.5), and incubated with trehalase (3 mU, Sigma-Aldrich), which hydrolyzes trehalose to glucose. The amount of glucose produced from trehalose was assayed with a glucose assay kit (Sigma-Aldrich). Samples untreated with trehalase served as negative controls.
Microscopy
The colony photographs were taken by using a Sony digital camera (DSC-F28). Photomicrographs were taken using a Zeiss M2 Bio microscope equipped with AxioCam and AxioVision (Rel. 4.8) digital imaging software.
Statistical analysis
Statistical differences between WT (or control) and mutant strains were assessed with the use of Student’s unpaired t-test. Data are reported as mean ± standard deviation (SD). P values < 0.05 were considered significant.
References
Ebbole, D. J. The conidium. In Cellular and Molecular Biology of Filamentous Fungi (eds Borkovich, K. A. & Ebbole, D. J.) 577–590 (American Society for Microbiology Press, Washington, 2010).
Latge, J. P. The pathobiology of Aspergillus fumigatus. Trends Microbiol 9, 382–389 (2001).
Bennett, J. W. An overview of the Genus Aspergillus. In Aspergillus: Molecular Biology and Genomics (eds Goldman, G. H. & Osmani, S. A.) 1–17 (CRC Press, Boca Raton, 2010).
Calvo, A. M., Wilson, R. A., Bok, J. W. & Keller, N. P. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66, 447–459 (2002) (table of contents).
Yu, J. H. & Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol 43, 437–458. https://doi.org/10.1146/annurev.phyto.43.040204.140214 (2005).
de Vries, R. P. et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol 18, 28. https://doi.org/10.1186/s13059-017-1151-0 (2017).
Park, H.-S. & Yu, J.-H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. https://doi.org/10.1016/j.mib.2012.09.006 (2012).
Adams, T. H., Wieser, J. K. & Yu, J.-H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54 (1998).
Timberlake, W. E. Molecular genetics of Aspergillus development. Annu. Rev. Genet. 24, 5–36. https://doi.org/10.1146/annurev.ge.24.120190.000253 (1990).
Park, H. S. & Yu, J. H. Biochemistry and Molecular Biology. In The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research) (ed. Hoffmeister, D.) (Springer, Cham, 2016).
Martinelli, S. D. Aspergillus nidulans as an experimental organism. Prog. Ind. Microbiol. 29, 33–58 (1994).
Casselton, L. & Zolan, M. The art and design of genetic screens: Filamentous fungi. Nat. Rev. Genet. 3, 683–697. https://doi.org/10.1038/nrg889 (2002).
Lee, M. K. et al. NsdD is a key repressor of asexual development in Aspergillus nidulans. Genetics 197, 159–173. https://doi.org/10.1534/genetics.114.161430 (2014).
Lee, M. K. et al. Negative regulation and developmental competence in Aspergillus. Sci. Rep. 6, 28874. https://doi.org/10.1038/srep28874 (2016).
Ni, M. & Yu, J. H. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS ONE 2, e970. https://doi.org/10.1371/journal.pone.0000970 (2007).
Seo, J. A., Guan, Y. & Yu, J. H. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172, 1535–1544. https://doi.org/10.1534/genetics.105.052258 (2006).
Adams, T. H., Boylan, M. T. & Timberlake, W. E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362 (1988).
Adams, T. H., Deising, H. & Timberlake, W. E. brlA requires both zinc fingers to induce development. Mol. Cell. Biol. 10, 1815–1817 (1990).
Andrianopoulos, A. & Timberlake, W. E. ATTS, a new and conserved DNA binding domain. Plant Cell 3, 747–748. https://doi.org/10.1105/tpc.3.8.747 (1991).
Andrianopoulos, A. & Timberlake, W. E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14, 2503–2515 (1994).
Mirabito, P. M., Adams, T. H. & Timberlake, W. E. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57, 859–868 (1989).
Park, H. S. & Yu, J. H. Developmental regulators in Aspergillus fumigatus. J. Microbiol. 54, 223–231. https://doi.org/10.1007/s12275-016-5619-5 (2016).
Tao, L. & Yu, J. H. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology 157, 313–326. https://doi.org/10.1099/mic.0.044271-0 (2011).
Wu, M. Y., Mead, M. E., Kim, S. C., Rokas, A. & Yu, J. H. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS ONE 12, e0179571. https://doi.org/10.1371/journal.pone.0179571 (2017).
Wu, M. Y. et al. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. MBio https://doi.org/10.1128/mBio.01130-18 (2018).
Park, H. S., Ni, M., Jeong, K. C., Kim, Y. H. & Yu, J. H. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS ONE 7, e45935. https://doi.org/10.1371/journal.pone.0045935 (2012).
Park, H.-S. & Yu, J.-H. Velvet Regulators in Aspergillus spp. Microbiol. Biotechnol. Lett. 44, 409–419 (2016).
Sarikaya Bayram, O. et al. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6, e1001226. https://doi.org/10.1371/journal.pgen.1001226 (2010).
Park, H.-S. et al. Velvet-mediated repression of beta-glucan synthesis in Aspergillus nidulans spores. Sci. Rep. 5, 10199. https://doi.org/10.1038/srep10199 (2015).
Ahmed, Y. L. et al. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-kappaB. PLoS Biol. 11, e1001750. https://doi.org/10.1371/journal.pbio.1001750 (2013).
Park, H. S., Lee, M. K., Kim, S. C. & Yu, J. H. The role of VosA/VelB-activated developmental gene vadA in Aspergillus nidulans. PLoS ONE 12, e0177099. https://doi.org/10.1371/journal.pone.0177099 (2017).
Ramamoorthy, V. et al. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 8, e74122. https://doi.org/10.1371/journal.pone.0074122 (2013).
Thieme, K. G. et al. Velvet domain protein VosA represses the zinc cluster transcription factor SclB regulatory network for Aspergillus nidulans asexual development, oxidative stress response and secondary metabolism. PLoS Genet. 14, e1007511. https://doi.org/10.1371/journal.pgen.1007511 (2018).
Oakley, C. E. et al. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol. 103, 347–365. https://doi.org/10.1111/mmi.13562 (2017).
Hicks, J. K., Yu, J. H., Keller, N. P. & Adams, T. H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16, 4916–4923. https://doi.org/10.1093/emboj/16.16.4916 (1997).
Lee, B. N. & Adams, T. H. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14, 323–334 (1994).
Yu, J. H. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44, 145–154 (2006).
Shin, K. S. et al. Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot. Cell 8, 738–746. https://doi.org/10.1128/EC.00368-08 (2009).
Son, Y. E., Cho, H. J., Lee, M. K. & Park, H. S. Characterizing the role of Zn cluster family transcription factor ZcfA in governing development in two Aspergillus species. PLoS ONE 15, e0228643. https://doi.org/10.1371/journal.pone.0228643 (2020).
Son, Y. E. et al. The role of the VosA-repressed dnjA gene in development and metabolism in Aspergillus species. Curr. Genet. https://doi.org/10.1007/s00294-020-01058-y (2020).
Kim, M. J. et al. The velvet repressed vidA gene plays a key role in governing development in Aspergillus nidulans. J. Microbiol. 57, 893–899. https://doi.org/10.1007/s12275-019-9214-4 (2019).
Emri, T., Molnar, Z., Szilagyi, M. & Pocsi, I. Regulation of autolysis in Aspergillus nidulans. Appl. Biochem. Biotechnol. 151, 211–220. https://doi.org/10.1007/s12010-008-8174-7 (2008).
Kafer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19, 33–131 (1977).
McKnight, G. L. et al. Identification and molecular analysis of a third Aspergillus nidulans alcohol dehydrogenase gene. EMBO J. 4, 2093–2099 (1985).
Waring, R. B., May, G. S. & Morris, N. R. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79, 119–130 (1989).
Park, H. S., Nam, T. Y., Han, K. H., Kim, S. C. & Yu, J. H. VelC positively controls sexual development in Aspergillus nidulans. PLoS ONE 9, e89883. https://doi.org/10.1371/journal.pone.0089883 (2014).
Yu, J. H. et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. https://doi.org/10.1016/j.fgb.2004.08.001 (2004).
Park, H. S. & Yu, J. H. Multi-copy genetic screen in Aspergillus nidulans. Methods Mol. Biol. 944, 183–190. https://doi.org/10.1007/978-1-62703-122-6_13 (2012).
Kwon, N. J., Shin, K. S. & Yu, J. H. Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet. Biol. 47, 981–993. https://doi.org/10.1016/j.fgb.2010.08.009 (2010).
Al-Nasiry, S., Geusens, N., Hanssens, M., Luyten, C. & Pijnenborg, R. The use of Alamar Blue assay for quantitative analysis of viability, migration and invasion of choriocarcinoma cells. Hum. Reprod. 22, 1304–1309. https://doi.org/10.1093/humrep/dem011 (2007).
McBride, J., Ingram, P. R., Henriquez, F. L. & Roberts, C. W. Development of colorimetric microtiter plate assay for assessment of antimicrobials against Acanthamoeba. J. Clin. Microbiol. 43, 629–634. https://doi.org/10.1128/JCM.43.2.629-634.2005 (2005).
Odabasi, Z. et al. Beta-D-glucan as a diagnostic adjunct for invasive fungal infections: validation, cutoff development, and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin. Infect. Dis. 39, 199–205. https://doi.org/10.1086/421944 (2004).
Shaaban, M. I., Bok, J. W., Lauer, C. & Keller, N. P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 9, 1816–1824. https://doi.org/10.1128/EC.00189-10 (2010).
Acknowledgments
We thank all our lab members for helpful discussions. This work was supported by KRIBB Research Initiative Program (KGM5232022) to MKL. The work at WU (KHH) was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea (NRF-2017R1D1A3B06035312) to KHH. The work at UW-Madison (AFA and JHY) was supported by the University of Wisconsin-Madison Office of the Vice Chancellor for Research and Graduate Education (OVCRGE) with funding from the Wisconsin Alumni Research Foundation to J.H. Yu. The work at KNU (HSP) was supported by the National Research Foundation of Korea (NRF) grant to HSP founded by the Korea government (NRF-2016R1C1B2010945).
Author information
Authors and Affiliations
Contributions
MKL, HSP, and JHY conceived and supervised the study; MKL, AFA, HSP, KHH, and JHY designed experiments; MKL, YES, HSP, and AFA performed experiments; MKL, YES, HSP, and JHY analyzed data; MKL, HSP, KHH, and JHY wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, MK., Son, YE., Park, HS. et al. Velvet activated McrA plays a key role in cellular and metabolic development in Aspergillus nidulans. Sci Rep 10, 15075 (2020). https://doi.org/10.1038/s41598-020-72224-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72224-y
This article is cited by
-
srdA mutations suppress the rseA/cpsA deletion mutant conidiation defect in Aspergillus nidulans
Scientific Reports (2023)
-
The function of a conidia specific transcription factor CsgA in Aspergillus nidulans
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.