Abstract
The brain uptake of biotherapeutics for brain diseases is hindered by the blood–brain barrier (BBB). The BBB selectively regulates the transport of large molecules into the brain and thereby maintains brain homeostasis. Receptor-mediated transcytosis (RMT) is one mechanism to deliver essential proteins into the brain parenchyma. Receptors expressed in the brain endothelial cells have been explored to ferry therapeutic antibodies across the BBB in bifunctional antibody formats. In this study, we generated and characterized monoclonal antibodies (mAbs) binding to the basigin receptor, which recently has been proposed as a target for RMT across the BBB. Antibody binding properties such as affinity have been demonstrated to be important factors for transcytosis capability and efficiency. Nevertheless, studies of basigin mAb properties' effect on RMT are limited. Here we characterize different basigin mAbs for their ability to associate with and subsequently internalize human brain endothelial cells. The mAbs were profiled to determine whether receptor binding epitope and affinity affected receptor-mediated uptake efficiency. By competitive epitope binning studies, basigin mAbs were categorized into five epitope bins. mAbs from three of the epitope bins demonstrated properties required for RMT candidates judged by binding characteristics and their superior level of internalization in human brain endothelial cells.
Similar content being viewed by others
Introduction
Antibodies as therapeutic modalities are highly attractive due to their target specificity, long serum half-life, mechanism of actions, and limited off-target effects compared to small molecules and peptides. During the past decades, huge efforts have been made to optimize the delivery of biotherapeutics across the blood–brain barrier (BBB), which is a major limiting factor for successful antibody treatment of central nervous system-associated disorders. Systemic administration of antibodies results in low brain exposure of < 1% of the injected dose1,2. The low permeability is due to the efficient tight junctions of the brain endothelial cells that form the front line of the BBB, preventing paracellular diffusion of large molecules into the brain. Most of the research effort has been focusing on enhancing transcellular transport as a non-invasive option. The most promising brain delivery strategy of antibodies relies on utilizing endogenous transport mechanisms, such as receptor-mediated transcytosis (RMT), by receptors present at the surface of brain endothelial cells to bypass the BBB. The transferrin receptor 1 (TfR1) has been explored extensively as a target for RMT, and in 1987 Fishman et al.3 succeeded in enhancing brain exposure of antibodies specific for the TfR1. Since then, there has been an increasing interest in developing receptor-mediated drug delivery platforms to enhance brain uptake of central nervous system biotherapeutics. Targeting the TfR1 using antibodies with retained Fc effector led to safety liabilities such as a reduction in circulating reticulocytes, microglial activation, and astrogliosis4,5. Although risk mitigation strategies are under development6,7, alternative drug delivery targets at the BBB are still explored.
Zuchero et al.1 investigated brain uptake mediated by other high abundant receptors and transporters in brain endothelial cells. One of the investigated receptors was the basigin receptor (also known as CD147, neurothelin, 5A11, EMMPRIN, gp42, HT7, OX47, CE9, M6, and HAb18G). They reported similar brain accumulation of anti-basigin and anti-TfR1 antibodies in a mouse model. In addition to enhanced delivery into the brain, the basigin receptor is also highly attractive due to its low expression in neurons8. This is favorable since the mode of action of many brain therapeutic antibodies requires an extracellular target engagement (e.g., amyloid-β, α-synuclein, and tau aggregates), where uptake into neurons is unwarranted. The basigin receptor was first identified in mouse B16 melanoma cells and is part of the immunoglobin (Ig) superfamily9,10. By comparing mouse brain endothelial cells to liver and lung endothelial cells, basigin is 14 fold enriched in brain endothelial cells based on microarray expression analysis11. Moreover, basigin expression was found to be upregulated under ischemic conditions as well as in multiple sclerosis and Alzheimer's disease patients12,13,14,15. Basigin is a highly glycosylated type 1 transmembrane protein and a co-receptor for the lactate transporter, monocarboxylate transporter 1, present at the BBB16. Basigin has four isoforms where the most abundant is the isoform 2 with two extracellular Ig-like domains with intra-domain disulfide bonds17. Basigin has a highly conserved single transmembrane region and a short cytoplasmic tail18,19. The extracellular domain contains three N-glycosylation sites20. Basigin exists in different glycosylated forms – highly glycosylated and lowly glycosylated around 50 and 38 kDa, respectively21,22,23. The glycosylation has been suggested to be essential for basigin's function and localization to the plasma membrane22,24. The subcellular sorting and transcytosis of the basigin receptor in brain endothelial cells have not been well-explored, and most published data is based on trafficking in MDCK cells or non-polarized HeLa cells. After thoroughly studying the TfR1 as a transcytosis-shuttle, it appears likely that for an antibody targeting the basigin receptor via RMT, receptor binding characteristics, such as affinity, valency, pH-dependency, and epitope, would also be crucial for improved brain exposure. It has been reported extensively, that sorting of the antibodies to lysosomes affects the BBB-crossing ability of the antibodies. For TfR1 antibodies, lysosomal localization was suggested to be affected by binding affinity25,26, bivalent target interaction (i.e., avidity binding)27, and pH-dependency of receptor binding28. Hence binding epitope, affinity, and the functional impact of antibody binding may well influence the behavior of the target receptor. Consequently, receptor internalization, subcellular trafficking, and signaling might be affected differently across a diverse panel of antibodies.
In this study, we isolated a number of mouse monoclonal antibodies (mAbs) against the basigin receptor to select a set of candidates for basigin-mediated delivery across the BBB. The first step in our characterization was to define bins of antibodies with overlapping basigin epitopes. Subsequently, representatives from the different bins were subjected to further characterization of affinity, cellular binding and localization, and the ability to internalize. Here we showed a comprehensive characterization of a large panel of basigin antibodies and selection of quality lead candidates potentially to be used in the engineering of BBB crossing antibody constructs.
Results
Generation of novel basigin monoclonal antibodies
Anti-basigin mAbs were generated by standard hybridoma technology. Mice were immunized with recombinant forms of human basigin, corresponding to the extracellular domain of the receptor (basigin-ECD). Hybridomas were screened by enzyme-linked immunosorbent assay (ELISA) for binding to human basigin-ECD and cross-reactivity to rat, mouse, and porcine basigin-ECD. Thirty-two hybridomas with high titers were selected for V-gene recovery and sequencing. A few of the hybridomas were non-clonal, yielding multiple light chain (LC) and heavy chain (HC) sequences. By including all HC/LC combinations from clonal and non-clonal hybridomas, a final set of 54 mAbs were produced recombinantly as human IgG1/κ (G1m3) chimeras for further characterization (see methods in supplementary materials).
Epitope binning of basigin monoclonal antibodies
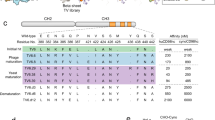
Initially, the 54 basigin mAbs were tested for binding to immobilized human basigin-ECD using biolayer interferometry (BLI), which reduced the number of mAbs to 21 positive binders. Alignment of heavy chain variable domain (VH) sequences was performed to explore the diversity of the basigin antibody panel. The alignment was used to generate a phylogram for the VH sequences (Fig. 1). The individually VH complementarity-determining region 3 (CDR-H3) families, defined by an identical length and more than 80% amino acid sequences identity, should identify antibodies from the same V-D-J recombination lineage. The CDRs were designated according to the IMGT annotation scheme. LC sequences were not included in the phylogenetic tree. However, the LC sequences were almost identical within the CDR-H3 families. The 21 aligned mAbs were representing nine different CDR-H3 families, subsequently grouped into five epitope bins. In-tandem BLI was used to identify the epitope bins (Fig. 2). Antibodies from the same CDR-H3 family are expected to bind the same epitopes, i.e., belong to the same bin. Here antibodies that compete for antigen binding in a cross-blocking matrix are assigned to the same epitope bin. The biosensors were loaded with recombinant basigin-ECD and sequentially introduced to a saturating mAb (mAb1) and a blocking mAb (mAb2) (Fig. 2a). Examples of blocking profiles from a blocking and a non-blocking antibody pair are depicted in Fig. 2b. The epitope binning data was summarized in the matrix showing the Pearson correlation coefficients between the antibodies blocking profiles, which were calculated based on normalized binning data (Fig. 2c). Antibodies with high positive Pearson correlation coefficient (above 0.9) were placed into the same bin (marked in red). The mAbs in light red indicate a positive Pearson correlation coefficient between 0 and 0.9. Green indicates antibody pairs with non-overlapping epitope regions. All antibodies were run in a self-blocking setup as controls (boxed in the matrix with bold lines). The binning experiment resulted in the mapping of five distinct epitope bins (A, B, C, D, and AD) (rightmost column in Fig. 2c). Basigin mAb #85 showed a unique profile with cross-blocking of antibodies in both bin A and D, indicating that bin A and D epitopes are either in structural proximity or neighboring peptide stretches. Basigin mAbs with a baseline drift after loading above 5% of total binding were only used as the blocking antibody (mAb2) to limit the chance for bin misinterpretation. Two bin B representatives were excluded as saturating mAb based on their fast off-rates (#42 and #59). However, basigin mAb#42 and #59 were in the same CDR-H3 family as the other bin B mAbs, indicating that they were correctly assigned to bin B despite its lack of verification in both assay directions. Similarly, only one (#81) of the seven bin C mAbs (#81, #82, #83, #73, #74, #76, and #75) could be confirmed in both assay orientations. As expected, antibodies from the same CDR-H3 families were mapped to the same epitope bind in the BLI assay (last column in Fig. 1). Overall, the alignment showed the diversity of the antibody panel with nine different CDR-H3 families grouped into five different epitope bins by the BLI assay. Bin A is represented by two CDR-H3 families, whereas bin B is comprised of a single CDR-H3 family. Bin C covers three CDR-H3 families, and bind D is represented by two antibodies from different CDR-H3 families. As could be expected, the single candidate representing bin AD belongs to a unique antibody lineage in the panel.
Phylogram of 21 basigin monoclonal antibodies (mAbs). The phylogram was produced with Multiple Sequence Comparison by Log-Expectation (MUSCLE) alignment using heavy chain variable domain (VH) sequences with the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) algorithm, Jukes-Cantor distance measure, and 10,000 Bootstrap replicates. The heavy chain CDR3 (CDR-H3) families are determined based on an identical length and more than 80% amino acid sequence identity. CDRs and mouse VH germline families were annotated according to IMGT. VH family: germline family of the heavy chain variable domain.
Epitope binning assay of the basigin monoclonal antibodies (mAbs). (a) Schematic representation of the epitope binning setup run in-tandem with basigin-ECD immobilized onto streptavidin biosensor followed by binding of the saturating mAb (mAb1) and subsequent the blocking mAb (mAb2). (b) Examples of sensorgrams with basigin mAb#52 as saturating mAb and blocking and non-blocking antibody pairs. (c) Matrix with Pearson correlation coefficients with the saturating mAbs (mAb1) listed in columns, and the blocking mAbs (mAb2) in rows. The highest correlations are marked in red, defined as the Pearson's correlation coefficient above 0.9, and the weak correlation in light red as between 0 and 0.9. Negative Pearson's correlations marked in green, indicating no cross-blocking between antibody pairs. Antibody self-binding pairs are boxed with bold lines. The assigned epitope bins are listed in the rightmost column.
Affinity determination by surface plasmon resonance
The 21 candidates identified in the binning experiments as positive binders were selected for kinetic profiling of their binding to recombinant basigin-ECD by surface plasmon resonance (SPR) (Fig. 3). A pattern between epitope bin and target affinity was observed by comparing the binding kinetics measured by SPR (Table 1). Bin A mAbs have the highest affinities, followed by bin B, whereas most of bin C mAbs were low in affinities. Basigin mAb#85 in bin AD showed strong binding with a KD of 0.4 nM together with bin A antibodies that all have a high affinity in the low nanomolar range (0.3–4 nM). The basigin antibodies in bin B covered KD values ranging from 7 to 40 nM. The bin B mAbs originate from the same CDR-H3 family, but the CDR-H3 sequences did not contribute to the differences in affinity observed within epitope bin B. Bin C mAb#81 and #82 had binding affinities ranging from KD values of 10 to 20 nM, whereas mAb#73–76 and #83 had poor affinities with KD values above > 100 nM. The KD values of antibodies in bin D were calculated as 6 nM (mAb#79) and 20 nM (mAb#80). A slower dissociation of mAb#79 caused this difference. All bin A mAbs, mAb#85 (bin AD), and mAb#79 (bin D) showed fast association rates and slow dissociation rates. The dissociation rates were faster for bin B but showed similar association rates as the other epitope bins. Some of the kinetic constants were difficult to determine due to a poor data fit (Chi2 values in the last column in Table 1). The candidate set was dominated by antibodies with fast association kinetics, which posed a general challenge for proper kinetics analysis. The sensorgrams for basigin mAb#73–76 illustrated the combined fast association and dissociation rates observed for these antibodies, which made determining proper kinetic constants technically difficult (Fig. 3e). The fast on-rate kinetics observed for several candidates may contribute to the unspecific binding phase observed as an association "hump" in the sensorgrams (e.g., mAb#83 and mAb#73–76) and resulting in a corresponding negative response in the dissociation phase (Fig. 3e). The effect was particularly pronounced for mAb#75. For candidates with KD values higher than 40 nM, ligand saturation was not reached as expected in the analyte titration from 0–600 nM. The binding stoichiometry, when calculated based on the observed maximal response (Rmax) and the theoretical Rmax for a 2-binding site ligand, was 0.7 or less for the higher KD antibodies.
Surface plasmon resonance sensorgrams representing the binding of basigin extracellular domain to captured anti-basigin monoclonal antibodies. The fitted curves used for equilibrium dissociation constant (KD) calculations were obtained using the Langmuir 1:1 binding model in the Biacore S200 evaluation software and plotted in GraphPad Prism (colored in black). (a) bin A, (b) bin D, (c) bin AD, (d) bin B, and (e) bin C. mAb monoclonal antibody, RU response unit.
Screening of cell association to human brain endothelial cells using flow cytometry
Since the BLI and SPR assays were done with a recombinant basigin-ECD, it was also important to screen all the basigin mAbs for binding to the cell surface-expressed basigin receptor. The evaluation of cell association was by flow cytometry using the human brain endothelial cell line, hCMEC/D3, which has endogenous expression of the basigin receptor. All the bin A, B, D, and AD mAbs were positive for binding the hCMEC/D3 cells with up to a 600-fold change in median fluorescence intensity compared to controls cells (2nd ctrl, stained only with the secondary antibody) (p-value < 0.001) (Fig. 4a). Histograms of selected mAbs representing each epitope bin visualize the shift in populations based on binding of the anti-basigin mAb to the receptor on hCMEC/D3 cells compared to control cells (Fig. 4b). The remaining flow cytometry histograms and the gating strategy can be inspected in supplementary Figure S1 and S2. No gating of negative and positive populations was applied due to the clean division of positive and negative cell populations. None of the bin C mAbs was capable of binding the native basigin receptor, as shown by the complete lack of a positive cell population and overlap with negative control cells (mAb#73 in Fig. 4b). Based on the kinetic profiling and cell association, the lowest affinity basigin mAbs (KD values > 100 nM) were excluded from further characterization reducing the number evaluated from 21 to 16 mAbs.
Basigin monoclonal antibody (mAb) association with hCMEC/D3 cells analyzed by flow cytometry. (a) The data was log-transformed and plotted with ± standard error of the mean (SEM). Multiple comparisons of hCMEC/D3 cell-association with the different basigin mAbs with controls cells stained only with the secondary antibody (2nd ctrl) were performed by one-way ANOVA with Dunnett's multiple comparison post hoc test. ***p < 0.001. (b) Selected histograms with the secondary control in blue and basigin mAbs in red. See also supplementary Figures S1 and S2 for gating strategy and the remaining flow cytometry histograms.
Immunofluorescence staining of hCMEC/D3 cells with different basigin monoclonal antibodies
Sixteen representative mAbs from the different epitope bins were further characterized by immunofluorescence for their ability to interact with the basigin receptor in fixed hCMEC/D3 cells (Fig. 5). Bin A basigin mAbs generally bind the hCMEC/D3 cells with mAb#52 showing the strongest signal. Bin B antibodies yielded both strong, weak, and no binding signal on fixed hCMEC/D3 cells. Surprisingly, bin B mAb#69, #71, #42, and #59 were among the antibodies showing weak or no binding to the fixed cells even though these candidates showed strong binding to the hCMEC/D3 cells by flow cytometry analysis (Fig. 4). For bin C, where none of the antibodies associated with the living cells, mAb#82 showed differentiated, robust signal on fixed hCMEC/D3 cells. Bin D mAb#80 bound strongly to the cells, whereas mAb#79 only resulted in weak staining. The immunofluorescence staining of fixed cells can be used for examination of the subcellular binding pattern of the antibodies. Immunofluorescence staining with mAb#80 (bin D) seemed to be detecting a perinuclear localization of the basigin receptor (XZ projection pictures in Fig. 5). In contrast, bin A mAb#52 seemed to detect basigin receptors closer to the cell surface. Basigin mAb#41 (bin B) behaved differently compared to the rest of the antibodies displaying an actin-like staining pattern. In summary, the immunofluorescence staining revealed that some basigin mAbs were more influenced by fixation of the epitope than others and that different sub-populations of the receptor with different local concentrations and epitope representation may exist.
Immunofluorescence staining of basigin in hCMEC/D3 cells with selected basigin monoclonal antibodies. Representative microscopic images of basigin (green) in hCMEC/D3 cells. Nuclei visualized by Hoechst (blue). The pictures are the maximum projection of the z-stack, and the XZ projection is below the pictures. Scale bar 30 µm. The pictures are sorted depending on their epitope bin, as indicated to the left. *indicates that the pictures were processed differently compared to the rest due to avoid over-exposed images.
Internalization of selected basigin monoclonal antibodies
Based on our results, five bin representative basigin antibodies, mAb#52 (bin A, KD 3 nM), mAb#43 (bin B, KD 20 nM), mAb#82 (bind C, KD 20 nM), mAb#80 (bind D, KD 20 nM), and mAb#85 (bin AD, KD 0.4 nM), were selected and examined for their ability to support and trail receptor internalization in the hCMEC/D3 cells (Fig. 6). The antibodies were selected based on robust immunofluorescence staining of the hCMEC/D3 cells. The internalization was determined using high-content screening microscopy with 43 images per condition in three independent experiments and plotted as the percentage increase in spot intensity per cell compared to the control after acid-removal of surface-bound mAbs. The effect of the acid-removal was tested on surface stained cells with and without acid stripping, which resulted in a decrease in spot intensity in acid-treated, indicating that the spots quantified are mainly internalized mAbs (supplementary Figure S3). Additionally, representative confocal z-stack images were manually taken of the different time points (Fig. 6a). A time-dependent increase in basigin mAb internalization was observed for mAb#52, #43, #82, and #80, whereas high-affinity mAb#85 reached the same level of basigin-mediated internalization already at 10 min and did not change from 10 to 30 min (Fig. 6b). Basigin mAb#80 resulted in a much higher spot intensity compared to the other antibodies, which was not surprising since mAb#80 associated strongly with the hCMEC/D3 cells in the immunofluorescence staining (Fig. 5). However, the acid stripping was not efficiently removing mAb#80 from the cell surface, and some of the signal could, therefore, be from surface-bound mAbs. All the tested mAbs appear to allow for receptor internalization, making them relevant candidates as engineering partners to facilitate basigin-mediated transcytosis of therapeutic proteins across the BBB.
Internalization of basigin monoclonal antibodies (mAbs) in hCMEC/D3 cells. (a) Representative confocal images of selected basigin mAbs exposed to hCMEC/D3 cells for 10 and 30 min. Basigin mAbs in green and nuclei visualized by Hoechst in blue. The pictures are the maximum projection of the z-stack, and the XZ projection is below the pictures. Scale bar 30 µm. (b) Quantification of intracellular spots using Cellomics Arrayscan after acid stripping and staining. The intensities are normalized to the negative control and plotted as the percentage increase in spot intensity per cell with ± standard error of the mean (SEM).
Discussion
Zuchero et al.1 showed that targeting basigin increased the uptake of antibodies across the BBB in vivo. However, they did not provide details on their basigin mAb epitopes. Many studies have demonstrated that antibodies can be transported across the BBB, reaching the brain parenchyma using RMT29. However, in most cases, the antibodies used for these experiments are only selected based on their affinity and capability to bind the endogenous receptor in cells. In this study, we successfully generated a broad panel of mAbs targeting the basigin receptor. Here, the antibodies were selected by their epitope, which gave a broader panel of antibodies with possibly more different biological characteristics.
Based on binding to recombinant basigin-ECD by BLI, 21 mAbs out of the 54 were selected for further characterization. The mAbs were initially differentiated based on their epitope binding region, affinity, and ability to bind the basigin receptor on living cells. For comparison, the data for the 21 mAbs are summarized in Table 2. The alignment of the 21 mAb VH sequences revealed nine CDR-H3 families, which were group into five epitope bins. Since it is known that affinity to the receptor can affect the transcytosis ability of the antibodies targeting the BBB25,26, the binding affinity of the selected 21 mAbs was determined.
The flow cytometry analysis of the hCMEC/D3 cells showed that some basigin mAbs that did not associate with the endogenous receptor expressed in the hCMEC/D3 cells still bind basigin-ECD in the BLI analysis (Figs. 2 and 4). The BLI experiment and the flow cytometry analysis of the basigin mAbs allowed for both monovalent and bivalent binding to basigin. The bivalent interaction was dependent on the density of the basigin on the surface of the cells or biosensors, which could cause the differences between the positive binders in the epitope binning and flow cytometry analysis. The bin C mAbs did not bind the cells in the flow cytometry analysis at this concentration, and a higher concentration may be needed for cellular binding of the low-affinity bin C mAbs (KD values > 100 nM).
Out of the 21 basigin mAbs, 16 were selected based on their epitope bin and affinity for the candidate selection step. The 16 mAbs represented all the epitope bins and had KD values below 40 nM. They were tested for immunofluorescence staining of hCMEC/D3 cells to determine whether their epitope and/or affinity was important for their ability to detect the cellular basigin receptors. Overall, the differences in interaction with the basigin within each epitope bin were not explained by their differences in affinity. However, to thoroughly investigate the affinity dependency, affinity engineering of representative basigin mAbs would be needed to exclude the influence of individual and local epitopes. The basigin mAbs detection of the basigin receptor was also independent of their epitope, as an intense immunofluorescence staining was observed across the epitope bins. The contradictory results obtained with mAb#82 in the flow cytometry analysis and immunofluorescence staining could be explained by the fixation step, which could modulate the epitope making it more susceptible. In contrast, basigin mAb#42 did not reveal any immunofluorescence signal in the staining of cells, indicating that the antibody binds a conformational epitope, which cell fixation disrupts.
The basigin receptor exists in different glycosylated forms. It has been reported that the cellular localization of basigin is dependent on its glycosylation state. The lowly glycosylated basigin is suggested to be localized primarily in the endoplasmic reticulum, whereas the highly glycosylated basigin is proposed to be at the cell surface22. The glycosylation sites were conserved across species, indicating a functional or structurally important role of the glycan modifications20. Notably, the N-glycosylation is shown to be different between tissues and species30. The basigin receptors can form dimers, but whether the glycosylation affects the oligomerization is still debated. Glycosylation has been reported to enhance basigin receptor dimerization21,22,23, whereas other studies found that glycosylation was not essential31,32. The different glycosylated forms and dimerization patterns of basigin could explain the different abilities of the anti-basigin mAbs to detect membrane-associated basigin and their subcellular staining pattern. The extracellular domain of human basigin was subjected to enzymatic deglycosylation (Figure S4). The deglycosylation resulted in a shift in molecular weight shown by Western blotting. However, the deglycosylation did not affect the detection of human basigin using basigin mAb#52 (bin A) and mAb#80 (bin D) as these binds both the highly and lowly glycosylated forms.
Internalization is a crucial step in the selection process to identify mAbs capable of crossing the BBB. Five basigin mAbs, one representative for each epitope bin, were selected for internalization as part of the functional characterization. The internalization seemed to be increased from 10 to 30 min for all mAbs but mAb#85. The basigin-mediated internalization of the high-affinity mAb#85 did not change over time, indicating that mAb#85 has reached saturation already at 10 min. Basigin mAb #43, #82, and #80 had all KD values of 20 nM, but their uptake levels were quite different. The difference could be explained by their epitope or receptor oligomer preference. mAb#80 resulted in a high internalization signal compared to the others, but the acid stripping of the hCMEC/D3 cell surface did not efficiently remove the mAb#80 surface binding, suggesting that mAb#80 were sticking to the cell surface. Also, the very strong interaction in the immunofluorescence staining could indicate that mAb#80 interacts with a specific subset of basigin receptors or that it exerts some unspecific interactions. The antibodies interact specifically with basigin shown by a substantial decrease in Western blotting band intensity after siRNA knockdown of basigin compared to samples were scramble siRNA was added (Figure S5). The subcellular localization of the internalized basigin mAbs was hard to interpret due to shrinkage of the cytoplasm after acid treatment.
We have generated a diverse panel of basigin mAbs binding to different epitope regions. In terms of the transcytosis capability of antibodies, a lot of the studies conclude only based on one epitope on a given receptor. It is important to explore the BBB crossing based on both affinity and epitope to be able to select lead candidates for drug delivery. Even though a previous study demonstrated that there was no correlation between epitope and function of basigin mAbs in T cells, this needs to be further analyzed in brain endothelial cells with our repertoire of antibodies33.
It remains to be confirmed if these basigin mAbs can transcytose with the receptor through the brain endothelial cells. The hCMEC/D3 cells are not optimal for testing transport across the cell layer due to the low tightness of the barrier model34. For such experiments, BBB in vitro models based on primary brain endothelial cells or human induced pluripotent stem cells are tighter and more suitable35. The basigin mAbs have cross-reactivity to at least two species (human and pig) out of the four tested (human, pig, rat, and mouse). The cross-reactivity eases test of the mAbs in preclinical models and progression of the lead candidates into clinical studies. The porcine cross-reactivity is convenient for in vitro modeling of the transcytosis using the robust porcine BBB model as well as a good human pharmacokinetic prediction using pigs for the in vivo experiments36,37.
Our work resulted in an excellent and broad panel of anti-basigin mAbs. If mAb#80 (bin D) is primarily binding the basigin receptor retained on the cell surface, it would not be relevant for basigin-mediated BBB delivery. Bin C mAbs cannot be excluded as possible candidates, but based on our studies, this concentration revealed weak cellular association and low uptake of mAb#82. In conclusion, bin A, B, and AD mAbs seemed to be promising candidates for further investigation based on their basigin-mediated internalization. The thorough characterization of the antibodies reported here provides valuable information for the further preclinical in vitro and in vivo characterization of the antibody repertoire and basigin-based BBB transportation.
Methods
Epitope binning using biolayer interferometry
The epitope binning assay was performed at Octet RED384 (Pall Fortebio) in 384-tilted well plates in a tandem setup. All samples were prepared fresh in 1 × PBS-P + buffer-10x (GE Healthcare Life Sciences, cat#88,995,084), which also was used as an assay buffer. First, the Dip and Read streptavidin (SA) biosensors for kinetics (Pall Fortebio, cat#18–5,019) were dipped in assay buffer for 200 s to record a baseline step. Next, 0.5 µg/ml of the biotin-tagged basigin-ECD was loaded on the biosensors for 200 s. After 200 s wash in buffer, the basigin-ECD-coated biosensors were dipped in 100 nM saturating basigin mAb (mAb1) for 1,200 s to reach saturation. The mAb1-basigin coated biosensors were washed for 200 s in buffer and then moved to wells containing an array of blocking basigin mAbs (mAb2) for 1,200 s. Complete self-blocking was ensured with all the basigin mAbs. Basigin mAbs with a baseline drift > 5% were only used as a blocking antibody. The data analysis was performed by using ForteBio Data Analysis software (version 10.0, Fortebio, Fremont, CA, USA), and epitope binning matrices were exported to Microsoft Excel. Data normalization was conducted by division of the mAb2 signal by the reference signal (mAb2 only) and multiplied with 100. The Pearson correlation coefficients were calculated by rows, and columns were ordered according to their correlation. The highest correlations defined as Pearson's correlation coefficient above 0.9 (red), weak correlation as between 0 and 0.9 (light red), and negative correlation below 0 (green).
Surface plasmon resonance analysis of basigin monoclonal antibodies
Binding affinity analysis of basigin mAbs to basigin-ECD was performed on a Biacore S200 and T200 (GE Healthcare Life Sciences) using a kinetic capture setup on a CM4 chip. The CM4 surface was immobilized with a monoclonal mouse anti-human IgG (Fc) antibody (GE Healthcare Life Sciences, cat#BR100839) using amine coupling (GE Healthcare Life Sciences, cat#BR100050). The chip surface was initially activated with a 1:1 (v/v) mixture of 75 mg/ml 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and 11.5 mg/ml N-hydroxysuccinimide (NHS). The anti-human IgG antibody was diluted in 10 mM sodium acetate to 25 µg/ml and immobilized at a flow rate of 10 µl/min for 7 min. The activated carboxylate groups were blocked by the injection of 1 M ethanolamine hydrochloride-NaOH, pH 8.5. During immobilization, HBS-EP + (GE Healthcare Life Sciences, cat#BR100188) was used as a running buffer. The basigin mAbs were adjusted to a concentration of 1 µg/ml in running buffer and captured on anti-human Fc mAb immobilized sensor followed by injection of basigin-ECD for 200 s and buffer for 300–600 s (association and dissociation, respectively) at a flow rate at 30 µl/min at concentrations ranging from 600–0 nM. No basigin antibody was captured in the reference flow channel (FC1). At the end of each cycle, the sensor surface was regenerated in 3 M MgCl2 for 30 s. 1 × HBS-P + (GE Healthcare Life Sciences, cat#BR-1003–68) with 1 mg/ml bovine serum albumin (BSA) (Rockland, cat#BSA-30) was used as running buffer for the kinetic analyses. Fitting of reference (FC1) subtracted data was performed using the Biacore S200 and T200 evaluation software (version 1.0, GE Healthcare Life Sciences, Uppsala, Sweden) using the pre-defined Langmuir 1:1 interaction model. The association rate constant (ka), dissociation rate constant (kd), and flow rate-independent component (tc) were fitted global, Rmax fitted local, and bulk response (RI) fitted constant. For some of the kinetics analyses, the high concentration of basigin-ECD was excluded due to binding to the reference. The KD values are based on two independent experiments.
Flow cytometry analysis
The cultured hCMEC/D3 cells were detached with Versene (Gibco Life Technologies, cat#15,040–033), resuspended in flow cytometry buffer (phosphate-buffered saline (PBS), 2% normal goat serum (Jackson ImmunoResearch, cat#005–000-121), 2 mM EDTA), and plated in a 96 well round bottom plate with 1–0.5 × 106 cells/well. Cells were stained with Live/dead fixable violet dead cell stain (Thermo Fisher Scientific, cat#L34955) for 15 min and washed twice in flow cytometry buffer before blocking in blocking buffer (PBS, 5% normal goat serum, 2 mM EDTA) for 10 min at 4 °C. The cells were incubated with 1 µg/ml primary basigin antibodies diluted in flow cytometry buffer for 30 min at 4 °C. After being washed twice in flow cytometry buffer, cells were incubated with 2.5 µg/ml secondary goat anti-human IgG Alexa Fluor 647 (Jackson ImmunoResearch, cat#109–605-008) for 30 min at 4 °C. The cells were fixed with BD Cytofix/CytoPerm (BD Biosciences, cat#554,714) for 15 min on ice. As negative controls for the staining procedure, cells were either incubated with secondary goat anti-human IgG Alexa Fluor 647, Live/dead fixable violet dead cell stain, or with buffer only (unstained). Since all the basigin mAbs are the same isotype, the basigin mAbs that did not bind the cells worked as Isotype controls. The samples were acquired using FACSVerse flow cytometry system (BD Biosciences, San Jose, CA, USA) with the BD FACSuite software calibrated with FACSuite CS&T research beads (BD Biosciences, cat#650,622), and analyzed using FlowJo (version 10, Tree Star, Inc., Ashland, OR, USA). The cells were gated based on the forward and side scatters to remove cell debris with an additional viability gate. Pulse geometry gating was used to remove doublets, and the final flow cytometry analysis was based on 50,000 events collected in the single-cell gate for each sample (Supplementary Figure S1). The setup was run three times, and the median fluorescence intensities were log-transformed and plotted in GraphPad Prism 8.0 (GraphPad Software, Inc, CA, USA). The fold change was back-transformed in order to describe the mean differences between samples.
Internalization assay
hCMEC/D3 cells were seeded in 96 well plates (Perkin Elmer, cat#3,904) coated with rat tail collagen type 1 at a cell density of 15,000 cells per well. The cells were incubated with 1 µg/ml of the selected basigin mAbs for 10 and 30 min. The cell surface was acid stripped with 0.2 M acetic acid/0.5 M NaCl in PBS for 4 min on ice to remove surface-bound antibodies (as described in38) and subsequently washed in PBS and fixed in 4% PFA for 10 min at room temperature. After permeabilization with PBS 0.1% Triton X-100 (Sigma-Aldrich, cat#T9284) for 10 min, the internalized basigin antibodies were detected by 1 h incubation at room temperature with Alexa Fluor 568 conjugated goat anti-human secondary antibody (2 µg/ml) (Invitrogen, cat#A21090) diluted in PBS with 2% BSA. Nuclei were stained with Hoechst34580 (2 µg/ml) (Invitrogen, cat#H21486) diluted in H2O for 10 min. After being washed, the cells were analyzed in PBS. Cells only treated with secondary antibody were used as a negative control. All samples were in triplicates, and the experiment was performed three times. Quantification of basigin mAb internalization was done using Cellomics Arrayscan VTI setup (Thermo Fisher Scientific) equipped with an Orca-ER camera (Hamamatsu) using a Zeiss 20 × LD Plan-Neofluar (0.4 numerical aperture (NA)) objective and analyzed with the Spot Detector BioApplication. The algorithm was set to exclude dead cells based on nuclear size and fluorescence intensity. The intensity of the spots with fluorescence intensity greater than the pre-defined background level was quantified and normalized to the cell number per field. No region of interest was applied in this experiment. The three repeated experiments were normalized to the negative control and plotted as internalization signal above control levels in GraphPad Prism 8.0 (GraphPad Software, Inc, CA, USA).
Confocal microscopy
Confocal images were acquired using UltraVIEW VoX Spinning Disk Confocal microscope (Perkin Elmer) with Nikon Eclipse Ti inverted microscope coupled to a CSU-X1 spinning disk head (Yokogawa) and equipped with a Nikon Ti 40x/0.95 NA air objective. Images were acquired with a c13440 Orca-flask 4.0 CMOS camera (Hamamatsu) with settings kept constant between samples unless other indicated. Additional confocal images were acquired with a Leica TCS SP8 confocal laser-scanning microscope (Leica Microsystems) using an HC PL APO CS 40x/0.85 NA air objective, and PMT and HyD detectors. At least three z-stack images were acquired per sample with a z-step size of 0.2 µm (Nikon microscope) or 0.6 µm (Leica microscope). Image processing was done with Volocity (version 6.4, Perkin Elmer, Waltham, MA, USA) and Fiji (NIH)39. Images were exported as raw TIFF files, and only adjustments to contrast and brightness were made. Pictures are shown as maximum XY projection of the entire Z-stack and slices of XZ cross-section. The size of scale bars is indicated in the figure legends.
Statistical analyses
All the raw data or the log-transformed data were normally distributed shown by QQ plots. Multiple comparisons were performed by one-way ANOVA with Dunnett's multiple comparison post hoc test. Results are presented as mean ± standard error of the mean (SEM) with p-values < 0.05 considered as statistically significant, ***p value < 0.001. All the statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc, CA, USA).
References
Zuchero, Y. J. Y. et al. Discovery of novel blood-brain barrier targets to enhance brain uptake of therapeutic antibodies. Neuron 89, 70–82 (2016).
Yu, Y. J. et al. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci. Transl. Med. 3, 84ra44 (2011).
Fishman, J. B., Rubin, J. B., Handrahan, J. V., Connor, J. R. & Fine, R. E. Receptor-mediated transcytosis of transferrin across the blood-brain barrier. J. Neurosci. Res. 18, 299–304 (1987).
Couch, J. A. et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood-brain barrier. Sci. Transl. Med. 5(183ra57), 1–12 (2013).
Pardridge, W. M., Boado, R. J., Patrick, D. J., Ka-Wai Hui, E. & Lu, J. Z. Blood-brain barrier transport, plasma pharmacokinetics, and neuropathology following chronic treatment of the rhesus monkey with a brain penetrating humanized monoclonal antibody against the human transferrin receptor. Mol. Pharm. 15, 5207–5216 (2018).
Weber, F. et al. Brain shuttle antibody for Alzheimer’s disease with attenuated peripheral effector function due to an inverted binding mode. Cell Rep. 22, 149–162 (2018).
Sun, J., Boado, R. J., Pardridge, W. M. & Sumbria, R. K. Plasma pharmacokinetics of high-affinity transferrin receptor antibody-erythropoietin fusion protein is a function of effector attenuation in mice. Mol. Pharm. 16, 3534–3543 (2019).
Sharma, K. et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831 (2015).
Miyauchi, T. et al. Basigin, a new, broadly distributed member of the immunoglobulin superfamily, has strong homology with both the immunoglobulin v domain and the β-chain of major histocompatibility complex class ii antigen. J. Biochem. 107, 316–323 (1990).
Biswas, C. Tumor cell stimulation of collagenase production by fibroblasts. Biochem. Biophys. Res. Commun. 109, 1026–1034 (1982).
Tam, S. J. et al. Death receptors DR6 and TROY regulate brain vascular development. Dev. Cell 22, 403–417 (2012).
Zhang, Y. & Pardridge, W. M. Delivery of β-galactosidase to mouse brain via the blood–brain barrier transferrin receptor. J. Pharmacol. Exp. Ther. 313, 1075–1081 (2005).
Burggraf, D. et al. Matrix metalloproteinase induction by EMMPRIN in experimental focal cerebral ischemia. Eur. J. Neurosci. 22, 273–277 (2005).
Agrawal, S. M., Silva, C., Tourtellotte, W. W. & Yong, V. W. EMMPRIN: a novel regulator of leukocyte transmigration into the CNS in multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neurosci. 31, 669–677 (2011).
Nahalkova, J. et al. CD147, a gamma-secretase associated protein is upregulated in Alzheimer’s disease brain and its cellular trafficking is affected by presenilin-2. Neurochem. Int. 56, 67–76 (2010).
Muramatsu, T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 159, 481–490 (2016).
Yu, X.-L. et al. Crystal structure of HAb18G/CD147. J. Biol. Chem. 283, 18056–18065 (2008).
Muramatsu, T. & Miyauchi, T. Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol. Histopathol. 18, 981–987 (2003).
Biswas, C. et al. The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 55, 434–439 (1995).
Miyauchi, T., Masuzawa, Y. & Muramatsu, T. The basigin group of the immunoglobulin superfamily: complete conservation of a segment in and around transmembrane domains of human and mouse basigin and chicken HT7 antigen. J. Biochem. 110, 770–774 (1991).
Tang, W. & Hemler, M. E. Caveolin-1 regulates matrix metalloproteinases-1 induction and CD147/EMMPRIN cell surface clustering. J. Biol. Chem. 279, 11112–11118 (2004).
Huang, W. et al. Modulation of CD147-induced matrix metalloproteinase activity: role of CD147 N-glycosylation. Biochem. J. 449, 437–448 (2013).
Tang, W., Chang, S. B. & Hemler, M. E. Links between CD147 function, glycosylation, and caveolin-1. Mol. Biol. Cell 15, 4043–4050 (2004).
Bai, Y., Huang, W., Ma, L.-T., Jiang, J.-L. & Chen, Z.-N. Importance of N-glycosylation on CD147 for its biological functions. Int. J. Mol. Sci. 15, 6356–6377 (2014).
Yu, Y. J. et al. Therapeutic bispecific antibodies cross the blood-brain barrier in nonhuman primates. Sci. Transl. Med. 6, 261ra154 (2014).
Bien-Ly, N. et al. Transferrin receptor (TfR) trafficking determines brain uptake of TfR antibody affinity variants. J. Exp. Med. 211, 233–244 (2014).
Niewoehner, J. et al. Increased brain penetration and potency of a therapeutic antibody using a monovalent molecular shuttle. Neuron 81, 49–60 (2014).
Sade, H. et al. A human blood-brain barrier transcytosis assay reveals antibody transcytosis influenced by pH-dependent receptor binding. PLoS ONE 9, e96340 (2014).
Pulgar, V. M. Transcytosis to cross the blood brain barrier, new advancements and challenges. Front. Neurosci. 12, 1019 (2019).
Kanekura, T., Miyauchi, T., Tashiro, M. & Muramatsu, T. Basigin, a new member of the immunoglobulin superfamily: genes in different mammalian species, glycosylation changes in the molecule from adult organs and possible variation in the N-terminal sequences. Cell Struct. Funct. 16, 23–30 (1991).
Yoshida, S. et al. Homo-oligomer formation by basigin, an immunoglobulin superfamily member, via its N-terminal immunoglobulin domain. Eur. J. Biochem. 267, 4372–4380 (2000).
Cui, H.-Y. et al. Dimerization is essential for HAb18G/CD147 promoting tumor invasion via MAPK pathway. Biochem. Biophys. Res. Commun. 419, 517–522 (2012).
Koch, C. et al. T cell activation-associated epitopes of CD147 in regulation of the T cell response, and their definition by antibody affinity and antigen density. Int. Immunol. 11, 777–786 (1999).
Weksler, B., Romero, I. A. & Couraud, P.-O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS 10, 16 (2013).
Helms, H. C. et al. In vitro models of the blood–brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow Metab. 36, 862–890 (2016).
Nielsen, S. S. E. et al. Improved method for the establishment of an in vitro blood-brain barrier model based on porcine brain endothelial cells. J. Vis. Exp. https://doi.org/10.3791/56277 (2017).
Zheng, Y. et al. Minipig as a potential translatable model for monoclonal antibody pharmacokinetics after intravenous and subcutaneous administration. MAbs 4, 243–255 (2012).
Carroll, R. C. et al. Dynamin-dependent endocytosis of ionotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 96, 14112–14117 (1999).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgments
The authors wish to thank Annemette Boe Marnow, Lina Lykke Ragborg, Lene Skrubbeltrang, Susanne Kristine Lüder, and Kristina Billing Løhmann for their excellent technical assistance. Stinus Lindgreen is thanked for his bioinformatic guidance. The work was financially supported by Lundbeck Foundation under Grant number: 2013-14113 and by H. Lundbeck A/S.
Author information
Authors and Affiliations
Contributions
S.C.C., A.J., S.C., C.B.F.A., and M.S.N. conceived and designed the experiments. S.C.C. and B.O.K. performed the experiments and analyzed the data. S.C.C. drafted the manuscript, and it was edited by B.O.K., A.J., C.B.F.A., S.C., and M.S.N. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.C.C., B.O.K., A.J., and S.C. are employees of H. Lundbeck A/S. H. Lundbeck A/S participated in the interpretation, review, and approval of the publication. C.B.F.A and M.S.N. declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Christensen, S.C., Krogh, B.O., Jensen, A. et al. Characterization of basigin monoclonal antibodies for receptor-mediated drug delivery to the brain. Sci Rep 10, 14582 (2020). https://doi.org/10.1038/s41598-020-71286-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-71286-2
This article is cited by
-
The proteome of the blood–brain barrier in rat and mouse: highly specific identification of proteins on the luminal surface of brain microvessels by in vivo glycocapture
Fluids and Barriers of the CNS (2024)
-
Subcellular trafficking and transcytosis efficacy of different receptor types for therapeutic antibody delivery at the blood‒brain barrier
Fluids and Barriers of the CNS (2023)
-
Towards a translational physiologically-based pharmacokinetic (PBPK) model for receptor-mediated transcytosis of anti-transferrin receptor monoclonal antibodies in the central nervous system
Journal of Pharmacokinetics and Pharmacodynamics (2022)
-
Basigin Antibodies with Capacity for Drug Delivery Across Brain Endothelial Cells
Molecular Neurobiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.