Abstract
Chromium is one of the heavy metal pollutants that causing risky health issues when discharged into the aquatic ecosystems. The current investigation focused on the bioremoval of Cr6+ depending on the bacterial sorption process by using Pseudomonas sp. NEWG-2 which was identified on the basis of morphological, cultural characteristics, 16S rRNA sequencing and phylogenetic analysis as Pseudomonas alcaliphila strain NEWG-2. It is clear from the FCCD experiments that the bacterium can grow normally and remove 96.60% of 200 mg/l of Cr6+ using yeast extract (5.6 g/l), glucose (4.9 g/l), pH (7) for 48 h incubation period. SEM and EDS analyses proved that the Cr6+ was biosorbed by P. alcaliphila NEWG-2. FTIR spectra indicated that the phenolic, carbonyl ester, acetyl, carboxylate, alkanes and carbonyl were the main groups involved in the chromium biosorption. Of the equilibrium isotherms models, the Langmuir model was more obedient, with a maximum uptake (qmax) of 10 mg/g (bacterial-alginate beads), than the Freundlich one. The findings reveal the efficiency of P. alcaliphila NEWG-2 in Cr6+ biosorption, with feasibility in the treatment of chromium-contaminated water as a green-technology tool. Interestingly, to the best of our knowledge, this is the first report on Cr6+ biosorption process by P. alcaliphila.
Similar content being viewed by others
Introduction
The emerging threat of heavy metals pollution has been recognized with an adverse impact on the environment, especially in developing countries. Due to the large-scale of industrial applications, heavy metals could be considered a major health risk with biomagnified toxicity in humans, especially at the higher concentrations. Heavy metals dispersed, in free forms, in both terrestrial and aquatic ecosystems, could be extended to the human food chain, that is why, the discharge of heavy metals into the aquatic ecosystem is a concern over the last few decades1,2,3. Moreover, the non-biodegradable nature and accumulation in living organisms ensure the prolonged presence of heavy metals in the environment, and therefore great interests have been made to eradicate such ecotoxicological hazards2,4. On the other side, some of the heavy metals, such as Cu2+, Zn2+, Fe2+/Fe3+, in trace amounts are essential for numerous vital biological process in living organisms, like enzymatic reaction process, but at higher levels may cause extreme toxicity, owing to the inhibition of metabolic reactions of the organism5,6.
Owing to the rapid growth of industries (e.g. leather, textile, mining and electric manufacturing), the problem of chromium-contaminated areas has emerged7. Depending on the oxidation state and concentration, chromium is one of the metals that could be either constructive or lethal to the biological systems. Chromium below 100 ppm is an essential and non-toxic mineral and, further, plays a functional role in both the nucleic acid synthesis and the metabolism of lipids, glucose and amino acids8,9. Contrarily, another category classified chromium, especially hexavalent chromium (Cr6+), as toxigenic due to having a high oxidation state with exerting mutagenic and carcinogenic potential on various biological systems10. As stated by the WHO and Indian Standard Institution, 0.05 mg/l is the allowed level of Cr6+ in drinking water, while the allowable limit of Cr6+ in industrial effluents ranges from 2.0 to 5.0 mg/l11.
In the biological systems, however, most of the cellular chromium exists in the trivalent (Cr3+) state, compared to Cr6+, the Cr3+ form is more soluble and has stable oxidation property, and thus shows less biotoxicity. Several symptoms appear on a person contaminated by chromium, of such symptoms, nasal irritation, ulceration, skin irritation, eardrum perforation and necrosis are the most common12. In the environmental systems, the harmful effects extended to the number and balance of the microbial communities in various ecosystems elements, including water and soil13.
Environmentally, the efficient removal of heavy metals from industrial wastes, particularly from the aqueous wastes, is one of the most important concerns of the world14. The previously common and traditional methods applied the oxidation–reduction reactions, solvent extraction, ion exchange, chemical precipitation and adsorption for the elimination of heavy metals. These techniques are relatively expensive and ineffective, especially when the concentration of heavy metals exceeds 1 mg/l15.
Otherwise, biosorption is emerging as an effective eradication process of heavy metals from aqueous solutions. In which, various plants (including aquatic plants and seaweeds) and microorganisms (bacteria, fungi, yeast and algae) have been recognized as operative bio-sorbent agents3,16,17,18. The bio-removal process by microorganisms is an innovative, low cost and eco-friendly19. Diverse mechanisms were proposed for the microbial biosorption of heavy metals, e.g. transport across the cell membrane, biosorption by cell walls and entrapment in extracellular capsules, precipitation, complexation and oxidation–reduction reactions5.
The employment of bacterial biomass for metal removal treatment of wastewater is perceptively suggested by many investigators. During metal-bacteria interaction, the polarizable groups (e.g. phosphate, carboxyl, hydroxyl and amino groups) are responsible for metal binding capacity and capable of interacting with cations20,21. Through the removal processes by microorganisms, chromium can be eradicated during three main processes, i.e. biotransformation, bioaccumulation and biosorption, the latter is superior to other ones, with highly selective depending upon the binding capacity of biological materials used as bio-sorbents19,22. Interestingly, the biosorption process can take place by living and dead biomass, but the living biomass has advantages, i.e. the metal can be removed during the growth, hence eliminating the processes of microbial reproduction, drying and storage23,24.
Wherein, this study aimed to apply the face-centered central composite design to optimize the biosorption process of hexavalent chromium ions by varying the nutritional (yeast extract, glucose) and physical (pH and interval periods) conditions, using P. alcaliphila NEWG-2. The aptness of the sorption process by immobilized bacterium cells using Freundlich and Langmuir isotherms models was evaluated.
Results and discussion
The ecosystem's healthiness and balance are greatly governed by heavy metals pollution. Chromium discharged from the effluent of the tannery and/or metal industries is a major risk and disturbance factor, in this respect25. That is because of the high toxicity, e.g. carcinogenic and teratogenic features, especially in the form of the hexavalent state. The annual discharged quantity of chromium into the environment is approximately 1,70,000 tonnes26. The relatively expensive and ineffective traditional disposal procedures motivate interests towards safe and effective alternative strategies, mainly biological-based strategy. One of such profoundly driven approach in recent times is biosorption through microbial biomass and their products2. The biosorption of this metal could occur during the metabolic pathways of the microorganism, which are known to survive and tolerate heavy metals stress in the contaminated environment.
Comparing to the other microorganisms, bacteria have many merits, such as the small in size, the abundant distribution, the more resistant to harsh environmental conditions, the ability to be cultured under controlled conditions, and further, possess various mechanisms of bioleaching, including bioaccumulation, biomineralization, biotransformation and biosorption22,27.
Cr6+ tolerability of the studied bacterium
From the start point of view, the present investigation was initiated to study the tolerability of Pseudomonas sp. to Cr6+ ions, followed by optimization of the biosorption efficiency by modifying some nutritional and physical culturing conditions. Initially, K2Cr2O7 was used for testing the tolerability of Pseudomonas sp. to different Cr6+ ion concentrations in the batch cultured system, with running over wide ranges of initial Cr6+ concentrations (50–250 mg/l based on Cr6+ ion).
The biosorption capacity of Cr6+, the bacterial growth and the final cultural pH are illustrated in Fig. 1. The bacterial biosorption capacity initiated with 97.2% at 50 mg/l, and kept around the same percent with the increasing of the initial concentration of Cr6+ and reached to saturated value at a concentration of 200 mg/l, with a removal efficiency of 96.1%. Higher concentrations (250 mg/l) decreased the tolerability of the bacterium down to 81%. Although the bacterium was able to grow and reduce Cr6+ ion content in the fermentation medium, the growth and final cultural pH slightly reduced with the increment of Cr6+. There was an inverse relationship between bacterial growth and final cultural pH from one side and Cr6+ concentrations, on the other side, this latter observation indicates the survivability and tolerability of the bacterial strain under the stress of Cr6+. Noticeably, Pseudomonas sp. was found to be able to reduce the yellow color of K2Cr2O7 in the medium (Supplementary Fig. 1), indicating the ability of the bacterium to grow and neutralize Cr6+.
Comparably, previous studies find out that some bacterial strains were tolerant up to 300 mg/l at pH 7.0 and temperature of 37 °C28. Nevertheless, the frequency of changes in metal uptake may be attributed to the metal's characteristics (e.g. the capacity for metal reductions, atomic weight, or ionic size ) and could be due to the characteristics of the bacterium, such as surface area, functional groups and structure20. In this connection, proteins and lipids may play a crucial role in the biosorption process. The capsule and slime layers of bacteria contain polysaccharides, acting as building blocks and/or barriers for heavy metal modulation. The exopolysaccharide is the self-defense of the bacterium against harsh conditions, e.g. pH, temperature, starvation and could play a crucial role in the biosorption process of metals. Likewise, Gupta and Diwan2 reported that the leaching of heavy metal depending on microbial extracellular polymeric substances such as polysaccharides, uronic acid, humic substances and lipids. The species of Pseudomonas contain exopolysaccharides with anionic functional groups, which incorporated in biosorption of heavy metals29,30.
On the other hand, the lower tolerability of some bacteria could be attributed to the mutagenic effect of chromium ion towards bacterial cells31, further, Komori et al.32 reported that mutation in Enterobacter cloacae as a result of DNA damage, leading to disruption of normal cell replication due to interactions with Cr6+. In some cases, the mutated cell may be more resistant to (Cr6+)33. Herein, the decline in the tolerability of Pseudomonas sp. NEWG-2 at the higher concentration of chromium (250 ppm of Cr6+) (Supplementary Fig. 1) could be due to a reduction in biosorption sites on the surface of bacteria34. However, it is worthy to note that the concentration applied in the present study is calculated based on Cr6+ ion content in K2Cr2O7. Consequently, the initial concentration of Cr6+ at 200 mg/l, has been chosen as the tolerable concentration for further investigation during the next trial.
Identification of Pseudomonas sp. strain NEWG-2
The microscopic characterization of the aerobic non-spore-forming, Gram-negative; Pseudomonas strain NEWG-2 showed straight rods of about 1.0 to 5.0 µm long and 0.5 to 1.0 µm wide with polar flagella.
The molecular identification using 16S rRNA gene sequence analysis was employed. The obtained 16S rRNA sequence of Pseudomonas sp. NEWG-2 was determined and the amplified fragment gave sequence with 1,500 bp (Supplementary Fig. 2). The obtained 16S rRNA sequence was subjected to the BLAST search35 on the GenBank database, and the most related sequences were obtained and compared with the present sequence.
The phylogenetic tree, generated by neighbor-joining analysis (Fig. 2), was constructed using MEGA5.0 software36. The isolate showed a high similarity of 99.20% with Pseudomonas alcaliphila. Accordingly, the bacterial isolate was identified as P. alcaliphila strain NEWG-2. The 16S rRNA sequence had been deposited in the DDBJ/EMBL-Bank/Gen Bank database under the accession number of MN025267.
Modeling the optimization of Cr6+ bioremoval process
The medium fermentation conditions of P. alcaliphila NEWG-2 were subjected to a statistical modulating process based on response surface methodology, i.e. the face-centered central composite design (FCCD). The FCCD of the 30-experimental runs were performed to improve the removing efficacy of Cr6+ ions by P. alcaliphila NEWG-2. The design of the matrix includes two nutritional variables, i.e. yeast extract (X1) and glucose (X2) and two physical variables, i.e. pH (X3) and incubation time (X4). The actual and coded levels of the variables as well as the investigated and predicted values of Cr6+ ions removal percent of the 30 runs of the FCCD matrix are displayed in Table 1.
Although the bacterial growth and final culture pH showed marked variation along with the 30-experimental runs of the design matrix (Table 1), their modeling and analysis failed to have significant trend and did not reveal any reasonable relationships (data not shown) and did not have any effect on the bioremoval process, therefore these data were not subjected to additional evaluation or modeling. The subsequent modeling was continued only on Cr6+ removal by the tested bacterium.
The variations in Cr6+ removal ranged from 93.30 to 96.59%. The highest level of Cr6+ removal percent, with a value of 96.59% (run No.8) has occurred at the center values of the tested independent variables. This observation indicates the accuracy of selected independent variables and their levels. In contrast, the minimum Cr6+ removal percent (93.30%) was obtained in the run no. 30, at the level of 3 g/l for both yeast extract and glucose, pH 6 and incubation time for 24 h.
ANOVA and multiple regression analysis
To explore the fitted model, the results were subjected to multiple regression analysis (Tables 2, 3). If the value of the determination coefficient (R2) higher than 0.9, the regression model is described as very significant37. To be an adequate model, the R2 value should not be less than (0.75)38. However, the quadratic model was the best-fitted model, recording the highest values of R2 (0.9860), adjusted R2 (0.9730) and predicted R2 (0.9539) for Cr6+ removal percent using P. alcaliphila NEWG-2. Further, the summary of the fit statistics shows a high significance of the quadratic model (very low P-value < 0.0001), and non-significant lack of fit (F-value = 0.47 and P-value = 0.8541), with a small standard deviation (0.18). So, the quadratic model was selected for fitting the FCCD data of Cr6+ removal by P. alcaliphila NEWG-2 (Table 2).
The data were subjected to ANOVA for further exploring the aptness of the various interaction effects of the quadratic model (Table 3). However, the experimental and predicted values of chromium removal are in a decent agreement, and the value of adjusted R2 and predicted R2 are high enough to indicate a high model significance. Predicted R2 measures the model efficacy and significance in the prediction of new response values. The values of both predicted and adjusted R2 should not be ˃ 20% of each other to be in decent agreement39. In the present study, the predicted R2 is, realistically, in line with the adjusted R2 value, explaining the high agreement between the experimental and the predicted values of Cr6+ removal. The model variability (95.39%) is, thus, satisfactory enough for predicting the values of experimental variables, within the tested range.
Linear, mutual interactions or quadratic coefficient estimate with a negative value indicates that the effects of such variables are negative (antagonistic effect) on Cr6+ removal percent by P. alcaliphila NEWG-2, that is to say, an opposite association between the investigated variable (s) and the removal of Cr6+. While positive coefficient value means a synergistic effect and the variable(s) increase chromium removal percent by P. alcaliphila NEWG-2 in the investigated region of the experiment. The small value of the coefficient of variation (C.V. = 0.19%) reveals an improved accuracy and trustiness of the experiments. The value of adequate precision (24.38) is greater than 4, which is desirable and indicates the reliability of the model. The value of the sum of squares of prediction error (PRESS) is 1.53. The general mean and standard deviation values of the experimental runs are 94.95 and 1.53, correspondingly (Table 3).
The Fisher’s test (F-value = 75.73) and the very small value of probability (P-value less than 0.0001) obtained from ANOVA demonstrate that the model is very significant. Values of P ˃ 0.05 designate that the model term is significant40. When evaluating the prediction ability of the model in the Cr6+ removal by P. alcaliphila NEWG-2, the model is considered accurate with a high cogency since the statics recorded high values of adjusted R2 (0.973), F-value (75.73) and adequate precision (24.38), low values of standard deviation (0.18), coefficient of determination (0.19) and PRESS (1.53) and non-significance lack-of-fit (P-value = 0.8541 and F-value = 0.47) (Table 3).
Moreover, the significance of each parameter’s coefficient was determined by the P-values and F-value, as a rule, the parameter is significant if the P-value < 0.05. Based on such rule, the significant coefficients are yeast extract (X1), pH (X3) and incubation time (X4) as linear coefficients, X1X3, X2X3 and X3X4 as interaction coefficients and finally all the quadratic effect of the factors. On the other hand, the interactions between X1X2, X1X4 and X2X4 are not significant and not significantly contributed to the Cr6+ removal by P. alcaliphila NEWG-2.
The coefficients of regression were used to generate the equation model. The value of Cr6+ removal by P. alcaliphila strain NEWG-2 could be predicted by fitting data with the equation of the second-order polynomial in terms of the next regression model:
where Y is the value of predicted chromium removal percent, X1 = yeast extract concentration, X2 = glucose concentration, X3 = pH and X4 = incubation time.
Model adequacy checking
To confirm the adequacy of the model, some analytical statics were checked and depicted in Fig. 3. Plotting the normal probability of the data of the experimental residuals (Fig. 3A) shows that data points concentrated closely along the straight line, meaning that the residuals follow the normal distribution without linearity41. Where most of the observations gathered around the center and the values located away from the general mean and dwindled equally on both sides of the central peak. Extreme residual values on both sides are not desired. The residuals versus predicted values (Fig. 3B) was plotted, the residuals were found to be scattered randomly around the centerline and no specific patterns could be drawn, this, in turn, indicates that the residuals not correlated and distributed independently, consequently have constant variance, therefore, the model is adequately meeting the postulations of the study. Plotting the values of predicted versus actual data points (Fig. 3C) showed that the data points split equally along the 45° line. This can help detect if any value(s) cannot be easily predicted by the model. However, all data points are detectable, assuring the aptness of the model.
Generating of the three-dimensional (3D) surface plot
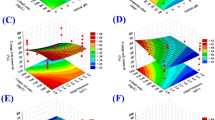
Following the previous assurance of the aptness of the model, the relationship between the interaction of each couple of the tested variables and the Cr6+ removal was explored. For such purpose, the 3D-plots were created (Fig. 4) by drawing the two independent factors on X- and Y-axes against Z-axis (Cr6+ removal percent), while the other two factors are kept at the central points.
The 3D-surface plot of the simultaneous effect of yeast extract and glucose on Cr6+ removal was constructed (Fig. 4A). The maximum Cr6+ removal was located around the central points, out of this range, a rather small percentage of chromium removal was noticed. The removal percent increased with the increment of glucose concentration, but the higher-level supports a fairly low percentage of Cr6+ removal percent. The 3D-surface plot of Fig. 4B shows Cr6+ removal efficacy as a function of yeast extract concentration and pH. The Cr6+ removal percent maximized at pH and yeast extract around the midpoints of both. The same trend of Cr6+ removal was observed for the other pair-wise combinations, i.e. yeast extract and incubation time (Fig. 4C), glucose and pH (Fig. 4D), glucose and incubation time (Fig. 4E) and pH and incubation time (Fig. 4F).
Experimental validation of the model
According to the model's Eq. (1), the highest theoretical value of Cr6+ removal was calculated to be 96.33% and the predicted values of the tested variable were 5.6 g/l (yeast extract), 4.9 g/l (glucose), 7 (pH) and 48 h incubation period. Under these conditions, the Cr6+ removal was experimentally verified and reached up to 96.60%. The experimental data verified a high grade of model precision, confirming the validation of the model under the levels of the factors used. It’s obvious that the biosorption process of chromium has been influenced by tested variables levels.
These results are comparable with the previous findings on Bacillus REP02, grown on various yeast extract and dextrose levels using Box-Behnken design, the predicted (98.86%) and experimentally (99.0%) values of Cr2+ removal were closely related42. Other findings reported that the maximum removal of Cr2+ being 94%, was occurred at pH 6.0 by coated bacteria6. Whereas Abhirami et al.10 reported 87.19% removal of Cr6+ at pH 4.0.
Ranjithkumar and Mahalingam28 stated that at pH values of 5.7 and 9, the Cr6+ tolerability of Pseudomonas sp. was up to 400 mg/l. Another data stated the significant impact of pH on the Cr6+ biosorption among the various variables tested using Box-Behnken design43. Whereas, the maximum removal of Cr6+ by Cyanobacteria was 81.72% at pH 11.0 and initial concentration of 15 mg/l21. Additionally, Durga Devi et al.44 investigated the biosorption of Cr6+ by P. fluorescens, the maximum uptake was found to be 800 mg/l. Other factors such as temperature, pH, redox potential and presence of other metals, play a crucial role in the chelation of chromium by Bacillus spp.26.
Importantly, the influence of medium pH values on the biosorption process could be explicated based on that the higher pH values cause the formation of metal hydroxide complexes, which decrease the concentration of chromium ions, thereby, causing a decrease in the equilibrium biosorption capacity45. While, a high electrostatic force of desirability at lower pH values, results in a high removal rate of chromium. Commonly, the influence of pH on the biosorption process could be due to the type of adsorbents and adsorbates20. Another study explored the disturbance of the bioremediation process of metals, where the building of bacterial exopolysaccharides (EPS) could be influenced by pH, in addition to other carbon sources available, temperature and the growth phase of the bacterium during which synthesis occurs46. Further, the strategies of heavy metal remediation through bacterial EPS must be focused on utilizing the non-neutral, negatively charged EPS to be incorporated as a suitable biosorbent. Otherwise, the bioremediation process could be influenced by nutritional factors, e.g. glucose or other carbon sources, whereby the monosaccharides form nucleotide diphosphate or monophosphate sugar, which is a crucial step in the synthesis of biosorbent agent such as EPS47.
In general, the microbial tolerance of heavy metal is attributed to a variety of detoxifying mechanisms, i.e. complexation by exopolysaccharides, binding with bacterial cell envelopes, metal reduction and/or metal efflux5. These mechanisms are sometimes encoded in plasmid genes facilitating the transfer of toxic metal resistance from one cell to another48. Additionally, the microbial metal tolerance can be categorized into (i) specific tolerance, which involves inducible mechanism (ii) nonspecific tolerance, which might be inducible or constitutive, some species of Pseudomonas fall under the nonspecific inducible resistance49. All these mechanisms may be applied as the suggested mode sorption process of Cr6+ by the present P. alcaliphila NEWG-2 strain.
Regarding the carbon source, glucose, d-ribose, d-xylose, d-arabinose, citrate and d-lactate were preferred as an electron donor during the metabolic process by Acinetobacter haemolyticus, while sucrose did not improve Cr6+ reduction despite enhancing the bacterium growth50. Whereas, Bacillus circulans, Pseudomonas aeruginosa and Bacillus coagulans preferred utilization of acetate, succinate, oxalate, citrate and malate as an electron donor during the metabolic process compared to glucose that needs to be catabolized into pyruvate before entering the Krebs cycle pathway51.
Commonly, the oxidation process of the organic compounds in synthetic media of chromium removal process might play a crucial role as an electron donor for the removal process, accumulating various types of functional groups on the surface of bacterial cells to enhance the Cr6+ biosorption50. Furthermore, the accumulation of several anionic organic compounds inside the cell induces the formation of complexes with different metal cations including chromium26.
Another, the reduction process of chromium by bacteria could be occurred in aerobic or anaerobic conditions, in which the aerobic reduction of Cr6+ is associated with a soluble protein fraction utilizing NADH or NADPH as an electron donor52. Whereas, in anaerobic reduction, Cr6+ acts as the terminal electron acceptor through membrane-bound reductase activity53. The species of Pseudomonas aeruginosa, Pseudomonas fluorescens and Enterobacter cloacae showed to be tolerant of Cr6+ under anaerobic conditions33.
Fourier transform infrared spectroscopy (FTIR)
The FTIR spectra of P. alcaliphila NEWG-2 dry biomass were investigated before and after Cr6+ biosorption (Fig. 5). The difference in content could be returned to the interface of the metal ion with the bacterial cell wall functional groups that may include hydroxyl, carboxylate, phosphate and amino groups54. The ion-exchange method is the probable biosorption of metal ions on the surface of the cell. In the range of 400–4,000 cm−1 wave-number, the adsorption spectra were investigated55. The stretching vibrations of O–H at the band of 3,434 cm−1. The peak at 2927 cm−1 indicates the existence of the SP3 C–H bond. Sharp symmetric CH2 stretching band maximum at 2,857 cm−1 CH stretching at 2,844 cm−1. 1748 cm−1 for carbonyl ester group (ester C=O). The Raman band at 1,458 cm−1 was assigned to a scissoring vibration of CH2. The peaks at 1634 and 1655 cm−1 for C–O acetyl at lower wavenumbers. 1550 cm−1 strong N–O stretching. 1,458 and 1,443 cm−1 medium O–H bending. 1,406 cm−1 (COO–) carboxylate group. 1,401 cm−1 (C–O). 1,246 and 1,247 cm−1 were attributed to strong enolic C–O stretching vibration. 1107 cm−1 was assigned to C–O stretching, secondary alcohol. 1,161 cm−1 was attributed to C–O stretching’ ester. The absorption bands at 1,037 cm−1, 1,044 cm−1 assigned to the C–O–C bond. 669,674 cm−1 corresponding to C–Cl. Chromium oxygen (Cr–O) stretching bands have been notable at 433 cm−1. The lower frequency regions of IR spectra of all complexes recorded weak bands around 433–564 cm−1 that is attributed to Cr–N bond56. FTIR spectra confirmed that the phenolic, carbonyl ester, acetyl, carboxylate, alkanes and carbonyl were the main groups involved in chromium biosorption. These results are conceding with the previous investigation, which showed that the presence of different functional groups such as hydroxyl, C–H and C–N was confirmed by FTIR analysis of Cyanobacteria and Azotobacter nigricans NEWG-1 during removal of chromium and copper ions, respectively3,21.
Scanning electron microscopy (SEM)
SEM data that recorded the morphological variation in P. alcaliphila NEWG-2 before and after the biosorption process of Cr6+ are depicted in Fig. 6. The scanning electron micrograph clearly illustrates the surface texture and morphology of P. alcaliphila NEWG-2 with a large magnification. The SEM analysis revealed also a piece of crucial information on the surface morphology of bacterium, with more extension. The graph indicates, obviously, the difference between the micrographs before and after adsorption of Cr6+ ions.
Electron dispersive spectroscopy (EDS)
One useful tool, for analysis of elemental content and/or chemical characterization of sorbents, is the EDS57. Herein, the EDS analysis was studied to investigate how well the Cr6+ attached to the cell wall of P. alcaliphila NEWG-2 biomass. Figure 7 depicts the presence of additional Cr6+peak after the biosorption process by P. alcaliphila NEWG-2 and these data confirmed the ability of this bacteria to remove Cr6+ ions from aqueous effluents.
Isotherms of Cr6+ biosorption by immobilized cells
Biosorption equilibrium isotherm modeling of alginate beads alone or immobilized with P. alcaliphila NEWG-2 was studied for Cr6+. Data of initial and residual concentrations of Cr6+ after 4 h of direct contact at 25 °C, as well as, the summary of constants for both Langmuir and Freundlich's constants are presented in Table 4. Initially, the lower the value of b constant of the Langmuir model and the higher the values of both Kf and n constants of the Freundlich model, indicate directly the higher affinity of the biomass.
The amount of metal adsorbed at the equilibrium (qe) versus residual concentration (Ce) sorption isotherm relationship was mathematically expressed and plotted (Fig. 8). The value of qmax of bacterial-alginate beads was 10 mg/g compared with only 7.405 mg/g for alginate beads. This, in turn, reflecting the critical role of the immobilized cells in the biosorption process. The linear regressions of data for the Langmuir and Freundlich isotherms for Cr6+ biosorption using immobilized P. alcaliphila NEWG-2 cells were plotted in Fig. 9. The equilibrium parameter (RL) or the dimensionless constant separation factor is essentially used to express the Langmuir isotherm, which indicates the shape of the isotherm adsorption for the adsorbents and adsorbates. In this respect, RL has various types, i.e. favorable (0 < RL < 1), linear (RL = 1) and, unfavorable (RL > 1) the present value (0.01002) at different initial concentrations of Cr6+ for the Langmuir model indicates a favourable adsorption process. The coefficients of determination (R2) of both Langmuir and Freundlich are close to 1.0, i.e. 0.999 and 0.918, respectively, therefore, adequately, describing the experimental data of Cr6+ biosorption. The higher the R2 the more robustness of the model. Langmuir model was found to be more efficient in this respect.
Linear plots of Langmuir and Freundlich models for the adsorption of Cr6+ by the immobilized P. alcaliphila NEWG-2 in alginate beads and uninoculated-alginate beads. qe is the amount of Cr6+ adsorbed at the equilibrium; ln qe is the natural logharitm of the amount of Cr6+ adsorbed at the equilibrium; Ce is the residual concentration of Cr6+; ln Ce is the natural logharitm of the residual concentration of Cr6+.
Likewise, the biosorption process in alginate beads only (Fig. 9) was apt to Langmuir model than Freundlich, in which the R2 is 0.9225 and 0.7443, respectively. Briefly, the immobilization of the bacterial cells made a difference in the biosorption process and such process is more obey to Langmuir model than Freundlich’s one.
The current data are inconsistent with the findings of Rezaei58, in which the equilibrium isotherm models of the sorption process of Cr6+ by Spirulina sp. preciously followed Freundlich than Langmuir. Various equilibrium models for fitting the experimental data for examining the correlation between solid and liquid phase concentrations of Cr6+ at equilibrium had been investigated21. In common, the two major models of kinetic study, i.e. Langmuir and Freundlich isotherms, were the major models59, in which, these mathematical models can fit the data reasonably well, and hardly reflect the sorption mechanism.
Materials and methods
Bacterium and growth conditions
Pseudomonas sp. was isolated, 2 years ago, from the soil and identified at the genus level by the Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center (Affiliation ID: 60019332), Giza, Egypt. The isolate was kindly provided to the present study.
The fermentation medium consisting of (g/l) glucose (5), yeast extract (5) and MgSO4·7H2O (0.2) and pH 7.2. For the maintenance, the bacterial strain was cultured on slants of the same medium supported with 15 g agar and incubated at 25 ± 1 °C for 48 h, the autoclavation was carried out at 121 °C for 20 min. the bacterium was sub-cultured periodically and preserved at 4 °C.
For inoculum preparation, the bacterium was grown on the previous broth medium under shaking at 100 rpm and 25 ± 1 °C for 48 h, the bacterial count was adjusted to obtain 108 ml−1 CFU, 5% (v/v) inoculum was used to inject 50 ml of fermentation medium Erlenmeyer flasks.
Bacterial tolerance to Cr6+
The potentiality level of Pseudomonas sp. NEWG-2 to survive in Cr6+ was investigated. The preceding fermentation medium was supplemented with various concentrations of K2Cr2O7 to cover a range from 50 to 250 ppm of Cr6+, and incubated for 48 h. Then the bacterial tolerability was evaluated in terms of growth reduction, Cr6+ removal after fermentation and a reduction in the final cultural pH.
Molecular identification
Pseudomonas sp. NEWG-2 was molecularly identified using 16S rRNA sequencing. DNA extraction for the bacterial sample was performed using the Thermo Gene JET Genomic DNA Purification Kit (#K0721), followed by the PCR reaction and sequencing according to the method of El-Naggar et al.60. The universal primers; 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′‐TACGGYTACCTTGTTACGACTT‐3′) were used. The phylogenetic tree was constructed using the Neighbor-Joining method with the software MEGA5. The bar indicates sequence divergence61.
Statistical optimization of Cr6+ biosorption
The matrix of FCCD was performed for statistically modeling of the medium conditions for the maximization of the bioremoval process of Cr6+ by P. alcaliphila NEWG-2. Two nutritional (yeast extract and glucose) and two physical (pH and incubation time) factors were the independent factors used for constructing the design matrix. Each of the four factors was investigated at three levels during the fermentation (Table 1). The FCCD was arranged to include eight axial points, sixteen factorial points, six center points and to estimate the pure error62. Following the laboratory experimentation of the design, the fermented medium was centrifuged (5,000 rpm for 20 min), the biosorption process was evaluated.
Evaluation of the biosorption process
The efficacy of bacterial isolate to survive in the Cr6+-containing medium was evaluated, measuring the bacterial growth (OD at 610 nm) and the ability to modify the fermentation medium pH (using pH-meter with a glass electrode (HI 9,321 microprocessor pH-meter). Finally, the residual Cr6+ was determined in the supernatant after fermentation. The concentration was recorded on a Buck Scientific Accusys 211 series, Atomic Absorption Spectrophotometer, USA by an air/acetylene flame system63.
Data modeling
The obtained data were then fitted to the next second-order polynomial quadratic model:
where; Y is the Cr6+ removal percent, Xi, and Xj are independent variables; β0 model constant, βi, is linear coefficients; βii is the quadratic coefficients and βij, is the interaction coefficients. To evaluate the equation model, laboratory validation was performed to ensure the fitness of the theoretically calculated value of each factor.
FTIR spectroscopy
Before and after Cr6+ removal, the cells of P. alcaliphila NEWG-2 were analyzed using FTIR spectroscopy to detect the functional groups present in the cells. With KBr pellets, the bacterial cells were implemented. The FTIR spectra of P. alcaliphila NEWG-2 were estimated in the range from 400 to 4,000 cm−1 (Thermo Fisher Nicolet IS10, “USA spectrophotometer) at Spectral Analyses Unit, Mansoura University, Egypt3.
SEM investigation
To evaluate the removal of the Cu2+ and to examine the surface of bacterial cells, dry cells of P. alcaliphila NEWG-2 (before and after removal of the Cr6+) were gold-coated and examined at various magnifications using SEM (JEOL TEM-2100) attached to a CCD camera at an accelerating voltage of 200 kV at Central Laboratory, Electron Microscope Unit, Mansoura University, Egypt3.
EDS evaluation
Energy-dispersive X-ray analysis was performed, employing JEOL TEM-2100 connected to a CCD camera at an accelerating voltage of 200 kV at Central Laboratory, Electron Microscope Unit, Mansoura University, Egypt3.
Bacterial immobilization
Immobilization of P. alcaliphila NEWG-2 cells was performed in 4% sodium alginate in distilled water, prepared by constant stirring at 60 °C for 30 min64. Following cooling, the sterile sodium alginate gel was supplied with bacterial cells (105 CFU/ml from 48 h old culture) with stirring for 5 min at room temperature. The beads with a diameter of 1.5 ± 0.2 mm were generated by adding drop-wise of the alginate alone or alginate-bacterial biomass combination in a cold sterile solution of CaCl2 (2.5%) through a 3-ml syringe with gentle stirring at room temperature to make spheres beads, then, by washing, three times with distilled sterilized water, the trace of CaCl2 was removed and finally, stored at 4 °C overnight in sterilized distilled water for stabilization and hardness. For biosorption procedure, glass separating funnel (Simax) was packed with the alginate-bacterial beads or alginate beads without bacteria. Initial concentrations ranging from 200 to 400 ppm of Cr6+ ion solution were added and left at 25 °C under shaking (100 rpm). Samples of each concentration from both treatments were collected after 4 h from the separating funnel effluent. The collected fractions were analyzed for the residual ions with the Atomic Absorption Spectrophotometer (Buck scientific 210 VGP, Inc.).
Equilibrium isotherms
The biosorption equilibrium uptake capacity in the absence or presence of the bacterial cells in sodium alginate beads at each concentration of Cr6+ was calculated according to mass balance on the ions expressed using the next equation:
where M is the biomass dry weight (g), Ce is the concentration of Cr6+ (mg/l) at equilibrium, C0 is the original Cr6+ concentration (mg/l), V is the sample volume (liter), and qe is the biomass biosorption equilibrium ions uptake capacity (mg/g).
Freundlich's and Langmuir isotherms were used to characterize the equilibrium between adsorbed ions by the bacterial cells and ions in the solution in this study. The equation of Langmuir isotherm empirical model based on the sorption on the surface is as follows:
Then after arrangement, the next equation is generated;
The b and qmax values (the adsorption equilibrium constants) can be obtained from the intercepts and the slopes of the linear plots; respectively, where experimental data of Ce/qe as the function of Ce.
Freundlich isotherm model
The equation of Freundlich isotherm empirical model based on sorption on a heterogeneous surface is as follows:
K and 1/n: an experimental constant, n represents the concentration effect on the adsorption efficiency and reflects the adsorption strength. K means the adsorption capacity of the adsorbent. The equation can be linearized in the following logarithmic form:
These values Kf and n can be obtained from the intercepts and the slopes of the linear plots; respectively, where experimental data of ln qe as the function of ln Ce.
Experimental design and statistical analysis
The matrix of FCCD and statistical analysis of variance (ANOVA) were performed using the statistical software packages Minitab (version 18, Minitab Inc., U.S.A.) and Design-Expert (version 7, Stat-Ease, Minneapolis, USA). The STATISTICA software (Version 8.0, StatSoft Inc., Tulsa, USA) was employed for plotting the 3D-surface plots. The Origin 2018 software (Version 2018, OriginLab Corporation, Northampton, USA) was used for analysis and graphing of isotherms. Experiments were accomplished in triplicates.
Conclusions
The present investigation concluded the optimization process for biosorption of Cr6+ using statistical modeling; FCCD. The maximum removal of Cr6+ (96.60%) by P. alcaliphila strain NEWG-2 was achieved in a medium containing yeast extract (5.6 g/l) and glucose (4.9 g/l) with an initial pH (7) and fermented for 48 h. The immobilized cells showed also efficacy in the biosorption process of Cr6+ following Langmuir and Freundlich isotherms model. The equilibrium data were compliant to Langmuir isotherm model compared to the other one. Finally, the current new green tool technology; P. alcaliphila NEW-2 is recommended to be the biosorption of chromium from the contaminated water as a safe and cost-effective strategy.
References
Lakherwal, D. Adsorption of heavy metals: A review. Int. J. Environ. Res. Dev. 4(1), 41–48 (2014).
Gupta, P. & Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 13, 58–71 (2017).
Ghoniem, A. A., El-Naggar, N. E., Saber, W. I. A., El-Hersh, M. S. & El-Khateeb, A. Y. Statistical modeling-approaches for optimization of Cu2+ biosorption by Azotobacter nigricans NEWG-2: Characterization and application of the immobilized cells. Sci. Rep. 10, 9491 (2020).
Cay, S., Uyanık, A. & Özaşık, A. Single and binary component adsorption of copper (II) and cadmium (II) from aqueous solutions using tea-industry waste. Sep. Purif. Technol. 38(3), 273–280 (2004).
Singh, V., Chauhan, P. K., Kanta, R., Dhewa, T. & Kumar, V. Isolation and characterization of Pseudomonas resistant to heavy metals contaminants. Int. J. Pharm. Sci. Rev. Res. 3(2), 164 (2010).
Jacob, J. J., Varalakshmi, R., Gargi, S., Jayasri, M. A. & Suthindhiran, K. Removal of Cr (III) and Ni (II) from tannery effluent using calcium carbonate coated bacterial magnetosomes. NPJ Clean Water 1(1), 1–10 (2018).
Wang, J. & Chen, C. Biosorbents for heavy metals removal and their future. Biotechnol. Adv. 27(2), 195–226 (2009).
Singh, D. K., Saksena, D. N. & Tiwari, D. P. Removal of chromium (VI) from aqueous solutions. Indian J. Environ. Health 36(4), 272–277 (1994).
Maduabuchi, J. M. et al. Arsenic and chromium in canned and non-canned beverages in Nigeria: A potential public health concern. Int. J. Environ. Res. Public Health 4(1), 28–33 (2007).
Abhirami, A., Ankita Vijayan, A. T., Purbasha, S. & Bhaskara Rao, K. V. Bioremoval of hexavalent chromium by indigenous bacterial isolates of a tannery effluent. Asian J. Microbiol. Biotechnol. Environ. Sci. 21, 5259–5264 (2019).
Tharannum, S., Murthy, V. K., Nandini, V. & Shruthi, P. T. Toxic heavy metal chromium remediation by processed low cost adsorbent green coconut shell. TOJSAT 5(1), 49–54 (2015).
Gibb, H. J., Lees, P. S., Pinsky, P. F. & Rooney, B. C. Lung cancer among workers in chromium chemical production. Am. J. Ind. Med. 38(2), 115–126 (2000).
Oves, M., Khan, M. S. & Zaidi, A. Biosorption of heavy metals by Bacillus thuringiensis strain OSM29 originating from industrial effluent contaminated north Indian soil. Saudi J. Biol. Sci. 20(2), 121–129 (2013).
Kumar, N. K., Reddy, D. S. R. & Venkateswarlu, P. Application of response surface methodology for optimization of chromium biosorption from an aqueous solution onto Syzigiumcumini (java) seed powder. J. Microbial Biochem. Technol. 2, 020–027 (2010).
Lodeiro, P., Barriada, J. L., Herrero, R. & De Vicente, M. S. The marine macroalga Cystoseirabaccata as biosorbent for cadmium (II) and lead (II) removal: Kinetic and equilibrium studies. Environ. Pollut. 142(2), 264–273 (2006).
El-Naggar, N. E., Hamouda, R. A., Mousa, T. E., Abdel-Hamid, M. S. & Rabei, N. H. Satistical optimization for cadmium removal using Ulva fasciata biomass: Characterization, immobilization and application for almost complete cadmium removal from aqueous solutions. Sci. Rep. 8, 12456 (2018).
El-Naggar, N. E., Hamouda, R. A., Mousa, I. E., Abdel-Hamid, M. S. & Rabei, N. H. Biosorption optimization, characterization and application of Gelidiumamansii biomass for complete Pb2+ removal from aqueous solutions. Sci. Rep. 8, 13456 (2018).
Samuel, M. S. & Chidambaram, R. Hexavalent chromium biosorption studies using Penicillium griseofulvum MSR1 a novel isolate from tannery effluent site: Box-Behnken optimization, equilibrium, kinetics and thermodynamic studies. J. Taiwan Inst. Chem. Eng. 49, 156–164 (2015).
Karthik, C. et al. Evaluation of Cr (VI) reduction mechanism and removal by Cellulosimicrobiumfunkeistrain AR8, a novel haloalkaliphilic bacterium. J. Hazard. Mater. 333, 42–53 (2017).
Ozdemir, G., Ceyhan, N., Ozturk, T., Akirmak, F. & Cosar, T. Biosorption of chromium (VI), cadmium (II) and copper (II) by Pantoea sp. TEM18. Chem. Eng. J. 102(3), 249–253 (2004).
Sen, S., Nandi, S. & Dutta, S. Application of RSM and ANN for optimization and modeling of biosorption of chromium (VI) using cyanobacterial biomass. Appl. Water Sci. 8(5), 148 (2018).
Joutey, N. T., Sayel, H., Bahafid, W. & El Ghachtouli, N. Mechanisms of hexavalent chromium resistance and removal by microorganisms. Rev. Environ. Contam. Toxicol. 233, 45–69 (2015).
Khoo, K. M. & Ting, Y. P. Biosorption of gold by immobilized fungal biomass. Biochem. Eng. J. 8(1), 51–59 (2001).
Chaudhary, P., Chhokar, V., Choudhary, P., Kumar, A. & Beniwal, V. Optimization of chromium and tannic acid bioremediation by Aspergillus niveus using Plackett-Burman design and response surface methodology. AMB Express 7(1), 201 (2017).
Dakiky, M., Khamis, M., Manassra, A. & Mer’Eb, M. Selective adsorption of chromium (VI) in industrial wastewater using low-cost abundantly available adsorbents. Adv. Environ. Res. 6(4), 533–540 (2002).
Singh, A. L. Removal of chromium from waste water with the help of microbes: A review. J. Sci. Technol. 3(3) (2008).
Vieira, R. H. & Volesky, B. Biosorption: a solution to pollution?. Int. Microbiol. 3(1), 17–24 (2000).
Ranjithkumar, M. & Mahalingam, P. U. Screening and characterization of chromium tolerant bacterial strains from electroplating effluent contaminated coil. Int. J. Chem. Mate Environ. Res. 3(4), 68–72 (2016).
Freitas, F., Alves, V. D. & Reis, M. A. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 29(8), 388–398 (2011).
Öner, E. T. Microbial production of extracellular polysaccharides from biomass. In Pretreatment techniques for biofuels and biorefineries 35–56 (Springer, Berlin, 2013).
Wang, Y. T. Microbial reduction of chromate. In Environmental microbe–metal interactions 225–235 (ASM Press, Washington, DC, 2000)
Komori, K., Wang, P. C., Toda, K. & Ohtake, H. Factors affecting chromate reduction in Enterobacter cloacae strain HO1. Appl. Microbiol. Biotechnol. 31(5–6), 567–570 (1989).
Wang, Y. T. & Xiao, C. Factors affecting hexavalent chromium reduction in pure cultures of bacteria. Water Res. 29(11), 2467–2474 (1995).
Saravanan, A., Senthil Kumar, P. & Preetha, B. Optimization of process parameters for the removal of chromium (VI) and nickel (II) from aqueous solutions by mixed biosorbents (custard apple seeds and Aspergillus niger using response surface methodology. Desalin. Water Treat. 57(31), 14530–14543 (2016).
Altschul, S. F. & Koonin, E. V. Iterated profile searches with PSI-BLAST—A tool for discovery in protein databases. Trends Biochem. Sci. 23(11), 444–447 (1998).
Tamura, K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28(10), 2731–2739 (2011).
El-Naggar, N. E., El-Shweihy, N. M. & El-Ewasy, S. M. Identification and statistical optimization of fermentation conditions for a newly isolated extracellular cholesterol oxidase-producing Streptomyces cavourensis strain NEAE-42. BMC Microbiol. 16, 217 (2016).
Le Man, H., Behera, S. K. & Park, H. S. Optimization of operational parameters for ethanol production from Korean food waste leachate. Int. J. Environ. Sci. Technol. 7(1), 157–164 (2010).
Rai, A. & Upadhyay, S. H. A review on signal processing techniques utilized in the fault diagnosis of rolling element bearings. Tribol. Int. 96, 289–306 (2016).
El-Naggar, N. E. Extracellular production of the oncolytic enzyme, l-asparaginase, by newly isolated Streptomyces sp. strain NEAE-95 as potential microbial cell factories: Optimization of culture conditions using response surface methodology. Curr. Pharm. Biotechnol. 16, 162–178 (2015).
El-Naggar, N. E., El-Bindary, A. A. & Nour, N. S. Statistical optimization of process variables for antimicrobial metabolites production by Streptomyces anulatus NEAE-94 against some multidrug-resistant strains. Int. J. Pharmacol. 9, 322–334 (2013).
Venil, C. K., Mohan, V., Lakshmanaperumalsamy, P., &Yerima, M. B. Optimization of chromium removal by the indigenous bacterium Bacillus spp. REP02 using the response surface methodology. ISRN Microbiol. (2011).
Aravind, J., Kanmani, P., Sudha, G. & Balan, R. Optimization of chromium (VI) biosorption using goose berry seeds by response surface methodology. Glob. J. Environ. Sci. Manag. 2(1), 61–68 (2016).
Devi, B. D., Thatheyus, A. J. & Ramya, D. Bioremeoval of hexavalent chromium, using Pseudomonas fluorescens. J. Microbiol. Biotechnol. Res. 2(5), 727–735 (2012).
Jing, X. et al. Biosorption of Cr (VI) from simulated wastewater using a cationic surfactant modified spent mushroom. Desalination 269(1–3), 120–127 (2011).
Czaczyk, K. & Myszka, K. Biosynthesis of extracellular polymeric substances (EPS) and its role in microbial biofilm formation. Polish J. Environ. Stud. 16(6), 799–806 (2007).
Madhuri, K. V. & Prabhakar, K. V. Microbial exopolysaccharides: biosynthesis and potential applications. Orient. J. Chem. 30(3), 1401–1410 (2014).
Silver, S. Bacterial resistances to toxic metal ions-a review. Gene 179(1), 9–19 (1996).
Rajendran, P., Muthukrishnan, J. & Gunasekaran, P. Microbes in heavy metal remediation. Indian J. Exp. Biol. 41, 935–944 (2003).
Zakaria, Z. A., Zakaria, Z., Surif, S. & Ahmad, W. A. Hexavalent chromium reduction by Acinetobacter haemolyticus isolated from heavy-metal contaminated wastewater. J. Hazard. Mater. 146(1–2), 30–38 (2007).
Philip, L., Iyengar, L. & Venkobachar, C. Cr (VI) reduction by Bacillus coagulans isolated from contaminated soils. J. Environ. Eng. 124(12), 1165–1170 (1998).
Camargo, F. A. O., Bento, F. M., Okeke, B. C. & Frankenberger, W. T. Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J. Environ. Qual. 32(4), 1228–1233 (2003).
Shen, H. & Wang, Y. Characterization of enzymatic reduction of hexavalent chromium by Escherichia coli ATCC 33456. Appl. Environ. Microbiol. 59(11), 3771–3777 (1993).
Hussein, H., Krull, R., Abou El-Ela, S. I., & Hempel, D. C. Interaction of the different heavy metal ions with immobilized bacterial culture degrading xenobiotic wastewater compounds. In Proceedings of the Second International Water Association World Water Conference, Berlin, Germany Vol. 1519, 15–19 (2001)
Garg, U., Kaur, M. P., Jawa, G. K., Sud, D. & Garg, V. K. Removal of cadmium (II) from aqueous solutions by adsorption on agricultural waste biomass. J. Hazard. Mater. 154(1–3), 1149–1157 (2008).
Khalil, S. M. Synthesis, spectroscopic and magnetic studies on metal complexes of 5-methyl-3-(2-hydroxyphenyl) pyrazole. J. Coord. Chem. 56(12), 1013–1024 (2003).
Dmytryk, A., Saeid, A., & Chojnacka, K. Biosorption of microelements by Spirulina: towards technology of mineral feed supplements. Sci. World J. 2014, Article ID 356328 (2014).
Rezaei, H. Biosorption of chromium by using Spirulina sp. Arab. J. Chem. 9(6), 846–853 (2016).
Gavrilescu, M. Removal of heavy metals from the environment by biosorption. Eng. Life Sci. 4(3), 219–232 (2004).
El-Naggar, N. E., Mohamedin, A., Hamza, S. S. & Sherief, A.-D. Extracellular biofabrication, characterization, and antimicrobial efficacy of silver nanoparticles loaded on cotton fabrics using newly isolated Streptomyces sp. SSHH-1E. J. Nanomater. (2016).
Gomila, M., Peña, A., Mulet, M., Lalucat, J. & García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 6, 214 (2015).
El-Naggar, N. E., Hussein, M. H. & El-Sawah, A. A. Phycobiliprotein-mediated synthesis of biogenic silver nanoparticles, characterization, in vitro and in vivo assessment of anticancer activities. Sci. Rep. 8, 1–20 (2018).
Allen, S. E., Grimshaw, H. M., Parkinson, J. A. & Quarmby, C. Chemical analysis of ecological materials 521 (Blackwell Scientific Publications, Hoboken, 1974).
Kumar, S. S. & Saramma, A. V. Nitrate and phosphate uptake by immobilized cells of Gloeocapsagelatinosa. J. Mar. Biol. Ass. India 54, 119–122 (2012).
Acknowledgements
The authors are grateful to Dr. Maha Sobhy Elsayed, Central Laboratory for Date Palm Research and Development, Agricultural Research Center, Giza, Egypt, for sincere help in the interpretation and graphing of Equilibrium isotherms.
Author information
Authors and Affiliations
Contributions
N.E.E. provided some necessary tools for experiments, experimental instructions, participated in the statistical analysis, contributed to the interpretation of the results, contributed to the writing and revision of the manuscript. A.Y.E. performed some of the experiments, contributed to the interpretation of the results, contributed substantially to the writing and revision of the manuscript. A.A.G. proposed the research concept, provided some necessary tools for experiments, carried out some of the experiments, collected the data and contributed substantially to the writing of the manuscript. M.S.E. proposed the research concept, performed some of the experiments, experimental instructions, contributed to the interpretation of the results and contributed substantially to the writing, reviewing and revision of the manuscript. W.I.S. proposed the topic, designed the research plan and performed some of the experiments, experimental instructions, participated in the statistical analysis and coordinated and contributed to writing and critical reviewing of the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El-Naggar, N.EA., El-khateeb, A.Y., Ghoniem, A.A. et al. Innovative low-cost biosorption process of Cr6+ by Pseudomonas alcaliphila NEWG-2. Sci Rep 10, 14043 (2020). https://doi.org/10.1038/s41598-020-70473-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70473-5
This article is cited by
-
Green synthesis of collagen nanoparticles by Streptomyces xinghaiensis NEAA-1, statistical optimization, characterization, and evaluation of their anticancer potential
Scientific Reports (2024)
-
Isolation, characterization, identification, genomics and analyses of bioaccumulation and biosorption potential of two arsenic-resistant bacteria obtained from natural environments
Scientific Reports (2024)
-
Bionanofactory for green synthesis of collagen nanoparticles, characterization, optimization, in-vitro and in-vivo anticancer activities
Scientific Reports (2024)
-
Fruit waste-derived cellulose-polyaniline composite for adsorption-coupled reduction of chromium oxyanions
Environmental Science and Pollution Research (2024)
-
Green fabrication of chitosan nanoparticles using Lavendula angustifolia, optimization, characterization and in‑vitro antibiofilm activity
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.