Abstract
Heterodera glycines is one of the most destructive pathogens of soybean. Soybean seeds coated with Bacillus simplex Sneb545 have shown resistance to H. glycines as a result of induced systemic resistance (ISR) in the plants. In this study, we aimed to identify the resistance-inducing determinants from this B. simplex strain. Combining the ISR bioassay, six ISR-active compounds were isolated from a culture of B. simplex Sneb545 using organic solvent gradient extraction, silica gel column chromatography, Sephadex LH-20 column chromatography, and semi-preparative high-performance liquid chromatography (HPLC), and all systems were based on activity tracking. The compounds were determined as cyclic(Pro-Tyr), cyclic(Val-Pro), cyclic(Leu-Pro), uracil, phenylalanine, and tryptophan using 1H NMR and 13C NMR. In plants from seeds coated with Bacillus simplex Sneb545, these six ISR-active compounds delayed the development of H. glycines in soybean roots. Moreover, cyclic(Pro-Tyr), cyclic(Val-Pro), and tryptophan reduced the number of nematodes in soybean roots. The expression levels of defense-related genes with cyclic(Val-Pro), tryptophan and uracil treatment soybean analysed using Quantitative real-time PCR (qRT-PCR). The results indicate cyclic(Val-Pro), tryptophan and uracil induced the expression of defense-related genes involved in the SA- and JA-pathways to against H. glycines. Our research results provide new agents for the control of H. glycines.
Similar content being viewed by others
Introduction
Soybean is an important economic crop, which is widely used in oil and food production. The United States of America, Brazil, and Argentina are the top three producers of soybean, producing approximately 123.7, 117.0, and 55.0 million metric tons annually, respectively, China is ranked fourth globally, with a production of 15.9 million metric tons per year (https://soystats.com/international-world-soybean-production, 2019). Heterodera glycines (soybean cyst nematode, SCN) is one of the most destructive pathogens of soybean1. This nematode is a considerable threat to soybean production and is responsible for annual losses of up to USD 1.5 billion globally2. Chemical nematicides are widely used to control parasitic nematode disease in soybean3. However, their application not only causes widespread environmental pollution but also threatens human health. In addition, drug tolerance of nematodes has been enhanced by the long-term use of chemical nematicides4,5. It is necessary, therefore, to seek safe, effective, and environmentally friendly products to defend soybean plants against SCN6.
The life cycle of SCN consists of four juvenile stages (J1–J4). The embryo in the egg develops into the first stage (J1) of juvenile and then molts in the egg to form the infested second-stage juveniles (J2). The J2 penetrates the root of the host plant, once J2 reach the vascular cylinder, they will establish a feeding site at the root, and then begins to develop into the third (J3) and fourth (J4) juvenile stages in the root system. Finally, matures to an adult female or male and most become adults by 30 days post-infection7,8.
Plants have evolved a great variety of defence mechanisms to fight against pathogens. Plant growth-promoting rhizobacteria (PGPR) play an important role in plant defence by triggering induced systemic resistance (ISR)9,10. The exogenous stimulation of PGPR can induce a plant immune response, which can directly induce salicylic acid (SA)- and/or jasmonic acid (JA)-dependent defence responses11,12. Previous studies have indicated that the expression of SA- and JA-pathway-related genes could be up-regulated in host plants in response to SCN by coating seeds with bacteria13,14,15.
Secondary metabolites produced by PGPR, identified as ISR determinants, play a role in resisting plant pathogens by triggering the ISR response16,17,18. ISR determinants, such as flagellin, pyoverdine, and N-alkylated benzylamine of Pseudomonas spp., affect the level of phytohormones and enhance resistance against disease by regulating the physiological and metabolic responses of plants19,20,21,22,23. In response to ISR, SA in Pseudomonas aeruginosa enhanced plant resistance to Meloidogyne javanica and promoted the colonization of bacteria into the tomato rhizosphere24. In addition, tomato seeds soaked with an SA-chitosan mixture improved phenylalanine ammonia lyase (PAL) activity and SA content, which increased the resistance of tomato plants infected with nematodes25,26. Lipopolysaccharide (LPS) of Rhizobacterium rhizobium protects the potato from Globodera pallida27, and N-acyl-L-homoserine lactone (AHL) plays a key role in the biocontrol activity of rhizobacteria against Alternaria alternata28. Additionally, the phthalic acid methyl ester of Bacillus subtilis acts against Fusarium wilt in tomato29. Therefore, understanding the roles of such secondary metabolites can inform the management of plant-parasitic nematodes.
It is important to explore the basic mode of function for improving the effectiveness of biocontrol agents. Studies have shown that soybean seeds coated with Bacillus simplex Sneb545 showed reduced infection and delayed development of pathogenic nematodes by triggering ISR30,31. This study was aimed to isolate and identify the ISR determinants from Bacillus simplex Sneb545 against H. glycines. Six isolated ISR-active compounds were obtained through organic solvent gradient extraction, silica gel column chromatography, Sephadex LH-20 column chromatography, and semi-preparative high-performance liquid chromatography (HPLC). We also analysed the expression levels of defense-related genes involved in SA and JA pathway of soybean treated with cyclic(Val-Pro), tryptophan and uracil treatment soybean by using qRT-PCR. Our research results provide new agents for the control of H. glycines.
Results
Preliminary screening of ISR determinants from Bacillus simplex Sneb545
Soybean seeds were coated with fermentation supernatant and bacterial suspension for ISR bioassay. The results indicate that the J4 ratio (the numbers of J4 to the total nematodes in per plant root) was 59.8% in the fermentation supernatant treatment, which was significantly lower than the 71.1% in the bacterial suspension treatment and the 72.1% in the control (Fig. 1A). The numbers of nematodes in the root were 51 and 62 in the fermentation supernatant and bacterial suspension, respectively, which were significantly lower than that of the control (Fig. 1B). Thus, the fermentation supernatant was used for further separation and purification that considered by combining the J4 ratio and total number nematodes in the root system.
Primary screening of ISR determinants from B. simplex Sneb545. Soybean seeds treated with fermentation supernatant, bacterial suspension, and a control (sterile water) at 12 dpi inoculation. (A) Percentages of various stage nematodes. The ratio of different developmental stages of SCN to the total number of nematodes within soybean roots. (B) Number of nematodes per plant. The different lowercase letters on the bars represent significant differences (P < 0.05) between treatments. Data are means ± SE. n = 3.
Extraction of ISR determinants from Bacillus simplex Sneb545 supernatant
Soybean seeds were coated with alcohol supernatant and precipitate for ISR bioassay. The results indicate that the J4 ratio was 51.6% in the alcohol supernatant treatments, which was significantly lower than the 74.4% in the control (Fig. 2A). There were 61 nematodes in the roots of the alcohol supernatant treatment plants, which was significantly lower than the number in the control and the ethanol precipitate treatments (Fig. 2B). These findings indicate that the potential ISR determinants were in the alcohol supernatant, therefore, the alcohol supernatant was used for further separation and purification.
ISR bioassay for alcohol precipitate and alcohol supernatant. The soybean seeds treated with alcohol precipitate, alcohol supernatant, and control (2% DMSO) at 12 dpi inoculation. (A) Percentage of various stage nematodes. The ratio of different developmental stages of SCN to the total number of nematodes within soybean roots. (B) Number of nematodes per plant. The different lowercase letters on the bars represent significant differences (P < 0.05) between treatments. The data are means ± SE. n = 3.
The alcohol supernatant solution was extracted using a series of organic solvents, and ISR bioassay was performed using the final aqueous phase (AP) and various organic extracts. The J4 ratio was 65.1%, 58.9%, 60.1%, 39.9%, and 40.4% for the petroleum ether (PET), chloroform (CHCl3), ethyl acetate (ETOAC), n-butanol (nBuOH) and final AP treatments, respectively (Fig. 3A), while the control was 73.8%; the number of nematodes in the roots were 76, 66, 67, 49, and 55, respectively (Fig. 3B). The values for the nBuOH treatments were significantly lower than those of the control, indicating that the ISR-active components were retained in the nBuOH fraction, which was further separated and purified.
ISR bioassay for various organic extracts. The soybean seeds treated with PET, CHCl3, ETOAC, nBuOH, AP, and control (2%DMSO) at 12 dpi inoculation. (A) Percentage of various stage nematodes. The ratio of different developmental stages of SCN to the total number of nematodes within soybean roots. (B) Number of nematodes per plant. The different lowercase letters on the bars represent significant differences (P < 0.05) between treatments. The data are means ± SE. n = 3.
Separation and purification of n-butanol extract
Twelve fractions (S1−S12) were obtained from the n-butanol extract by combining silica gel chromatography and thin layer chromatography and these were then used for ISR bioassays. The J4 ratio for S5 and S11 within the root was 58.7% and 40.9%, respectively (Fig. 4A). The number of nematodes in the roots was 52 and 54 (Fig. 4B) for S5 and S11, respectively, which were significantly lower than the control. Combined with the J4 ratio and number of nematodes in the roots, the S5 and S11 were further separations.
ISR bioassay for various nBuOH extract components. The soybean seeds treated with 12 fractions (S1−S12) extracted from nBuOH and the control (2%DMSO) at 12 dpi inoculation. (A) Percentage of various stage nematodes. The ratio of different developmental stages of SCN to the total number of nematodes within soybean roots. (B) Number of nematodes per plant. The different lowercase letters on the bars represent significant differences (P < 0.05) between treatments. The data are means ± SE. n = 3.
Separation fraction S5 through semi-preparative HPLC
Three main compounds were obtained in fraction S5 by using semi-preparative HPLC. The sub-fractions S5-1, S5-2, and S5-3 were the peak points at 7.540 min, 7.972 min, and 16.006 min, respectively (Fig. 5A,B,C).
Separation of S11 by Sephadex LH-20 column
Fraction S11 was further purified using a Sephadex LH-20 column and the ultraviolet detector (220 nm) determined six sub-fractions named S11-1, S11-2, S11-3, S11-4, S11-5, and S11-6 (Fig. 6). The six sub-fractions were subjected to ISR bioassays. The J4 ratio of S11-6 was 54.5% (Fig. 7A) and the number of nematodes in the roots was 48 (Fig. 7B) for S11-6, which were significantly lower than the control. The results suggest that S11-6 exhibited ISR-activity.
ISR bioassay for the sub-fraction of S11 by Sephadex LH-20 column chromatography. The soybean seeds treated with S11-1, S11-2, S11-3, S11-4, S11-5, S11-6 sub-fraction and control (2%DMSO) at 12 dpi inoculation. (A) Percentage of various stage nematodes. The ratio of different developmental stages of SCN to the total number of nematodes within soybean roots. (B) Number of nematodes per plant. The different lowercase letters on the bars represent significant differences (P < 0.05) between treatments. The data are means ± SE. n = 3.
Separation of S11-6 by semi-preparative HPLC
Three main compounds were found in the fraction S11-6 by using semi-preparative HPLC. The sub-fractions S11-6-1, S11-6-2, and S11-6-3 were the peak points at 3.745 min, 5.242 min, and 7.686 min, respectively (Fig. 8A,B,C).
Identification of active compounds
Compound S5-1: white powder. 1H NMR (600 MHz, DMSO-d6) δ: 9.18 (1H, s, OH), 7.86 (1H, s, NH), 7.05 (2H, m, H-12, H-16), 6.64 (2H, m, H-13, H-15), 4.24 (1H, t, H-9), 4.02 (1H, d, H-6), 3.40 (1H, m, H-3a), 3.26 (1H, m, H-3b), 2.00 (1H, m, H-5a), 1.73 (1H, m, H-5b), 1.84 (2H, m, H-4). 13C NMR (600 MHz, DMSO-d6) δ: 169.25 (C-7), 165.45 (C-1), 156.23 (C-14), 131.16 (C-12, 16), 127.41 (C-11), 115.12 (C-13, 15), 58.74 (C-6), 56.36 (C-9), 44.90 (C-3), 35.05 (C-10), 28.06 (C-5), 22.21 (C-4). According to the literature32, the compound was identified as cyclic(Pro-Tyr) (Fig. 9A).
Compound S5-2: white powder. 1H NMR (600 MHz, DMSO-d6) δ: 7.96 (1H, s, NH), 4.12 (1H, t, H-6), 3.92 (1H, ddd, H-9), 3.43(1H, m, H-3a), 3.40 (1H, dt, H-3b), 2.34 (1H, m, H-5a), 2.13 (1H, m, H-5b), 1.82 (2H, m, H-4), 1.19 (1H, dq, H-10), 1.01 (3H, d, H-11), 0.85 (3H, d, H-12). 13C NMR (600 MHz, DMSO-d6) δ: 170.65 (C-1), 165.61 (C-7), 59.84 (C-6), 58.62 (C-9), 45.01 (C-3), 28.23 (C-10), 28.19 (C-5), 22.46 (C-4), 18.71 (C-11), 16.76 (C-12). According to the literature32, the compound was identified as cyclic(Val-Pro) (Fig. 9B).
Compound S5-3: white powder. 1H NMR (600 MHz, CD3OD) δ: 7.93(1H, s, NH), 4.28 (1H, t, H-6), 4.15 (1H, m, H-3), 3.53 (2H, m, H-9), 2.32 (1H, m, H-7a), 2.04 to 1.92 (5H, m, H-7b, H-8, H-10), 1.54 (1H, m, H-11), 0.98 (6H, m, H-12, H-13). 13C NMR (600 MHz, CD3OD) δ: 171.83 (C-2), 167.47 (C-5), 8.75 (C-6), 53.14 (C-3), 44.95 (C-9), 37.91 (C -10), 27.58 (C-7), 24.27 (C-11), 22.16 (C-8), 21.80 (C-12 or C-13), 20.71 (C-12 or C-13). According to the literature33, the compound was identified as cyclic(Leu-Pro) (Fig. 9C).
Compound S11-6-1: white powder. 1H NMR (600 MHz, DMSO-d6) δ: 10.93 (1H, s, NH), 10.79 (1H, s, NH), 7.42 (1H, d, H-6), 5.47 (1H, d, H-5). 13C NMR (600 MHz, DMSO-d6) δ: 164.74 (C-4), 154.5 (C-2), 142.2 (C-6), 100.3 (C-5). According to the literature34, the compound was identified as uracil (Fig. 9D).
Compound S11-6-2: white powder. 1H NMR (600 MHz, D2O) δ: 7.42 (2H, m, H-11, 13), 7.37 (1H, t, H-12), 7.32 (2H, m, H-10, 14), 3.99 (1H), dd, H-2), 3.28 (1H, dd, H-3a), 3.12 (1H, dd, H-3b). 13C NMR (600 MHz, D2O) δ: 173.80 (C-1), 134.96 (C-4), 127.57 to 129.24 (C5-9), 55.91 (C-2), 36.22 (C-3). According to the literature35, the compound was identified as phenylalanine (Fig. 9E).
Compound S11-6-3: white powder. 1H NMR (600 MHz, D2O) δ: 10.20 (1H, d, NH-6), 7.73 (1H, d, H-11), 7.53 (1H, d, H-8), 7.31 (1H, s, H-5), 7.28 (1H, ddd, H-10), 7.20 (1H, td, H-9), 4.04 (1H, dd, H-2), 3.48 (1H, dd, H-3a), 3.30 (1H, dd, H-3b). 13C NMR (600 MHz, D2O) δ: 174.27 (C-1), 136.20 (C-7), 126.51 (C-12), 124.85 (C-5), 121.96 to 111.77 (C8-11), 107.33 (C-4), 54.92 (C-2), 26.24 (C-3). According to the literature36, the compound was identified as tryptophan (Fig. 9F).
Active compounds from ISR bioassay
The obtained active compounds, cyclic(Pro-Tyr), cyclic(Val-Pro), cyclic(Leu-Pro), uracil, phenylalanine, and tryptophan, were used for ISR bioassays. The J4 ratio was 64.5%, 58.2%, and 55.2% for cyclic(Leu-Pro), uracil, and phenylalanine, respectively, which were significantly lower than that of the control (85.6%; Fig. 10A). The number of nematodes in the roots was not significantly different from that of the control (Fig. 10B). The results indicated that cyclic(Leu-Pro)-, uracil-, and phenylalanine-coated soybean seeds delayed the development of nematodes in the plant roots.
ISR bioassay for six active compounds. The soybean seeds treated with cyclic(Pro-Tyr), cyclic(Val-Pro), cyclic(Leu-Pro), uracil, phenylalanine, and tryptophan and a control (2%DMSO) at 12 dpi inoculation. (A) Percentage of various stage nematodes. The ratio of different developmental stages of SCN to the total number of nematodes within soybean roots. (B) Number of nematodes per plant. The different lowercase letters on the bars represent significant differences (P < 0.05) between treatments. The data are means ± SE. n = 3.
The numbers of nematodes in the roots were 71, 69, and 76 (Fig. 10B) for cyclic(Pro-Tyr), cyclic(Val-Pro), and tryptophan, respectively, which were significantly lower than that of the control (117). Moreover, the J4 ratio of cyclic(Pro-Tyr), cyclic(Val-Pro), and tryptophan were 73.7%, 68.7%, and 71.9%, which were significantly lower than the 85.6% for the control (Fig. 10A). Therefore, cyclic(Pro-Tyr)-, cyclic(Val-Pro)-, and tryptophan-coated soybean seeds had a significant effect on the number and development of nematodes in roots.
Induced defense-related gene expression in the root
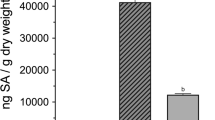
To test the active compounds mediated the ISR defense response against H. glycine, cyclic (Val-Pro), tryptophan and uracil were selected from six active compounds to treat soybeans, then the expression levels of SA-pathway-related genes (PR1, PR2) and JA-pathway-related genes (PR3a, PR3b, PR10) in soybeans were assessed by using qRT-PCR. Soybean was treated with cyclic (Val-Pro) and inoculated with H. glycines J2 (Fig. 11). During the stage of nematode infection, the expression of PR1, PR3b and PR10 were significantly higher than that in control, which increasing 1.9-fold, 1.9-fold and 2.3-fold compared with that in control at 1 dpi. The transcript level of PR1 and PR3b were 2.6-fold and 2.1-fold significantly higher than that in control at 5 dpi. A significantly enhanced transcription of PR2 was 1.9-fold compared with that in control at 10 dpi.
The expression of defense genes in soybean roots with cyclic(Val-Pro) treatment. Soybean root samples were taken at 1, 5, and 10 d after H. glycines inoculation, and the expression levels of five defense genes were analyzed by qRT-PCR: (A) PR1, (B) PR2, (C) PR3a, (D) PR3b and (E) PR10. Error bars represent standard errors; a t-test was used to determine significant differences between the cyclic(Val-Pro)-treated sample and the control (∗ P < 0.05).
Soybean was treated by tryptophan and inoculated with H. glycines (Fig. 12). At 1 dpi, the expression of PR1 and PR10 were significantly higher than that in control, which increasing 1.8-fold and 2.4-fold compared with that in control. The transcript level of PR3a and PR10 were 2.0-fold and 1.9-fold significantly higher than that in control at 10 dpi. Soybean was treated by uracil and inoculated with H. glycines (Fig. 13). A significantly enhanced transcription of PR3b was 1.5-fold compared with that in control at 5 dpi. The transcript level of PR1 and PR10 were 2.9-fold and 2.4-fold significantly higher than that in control at 10 dpi.
The expression of defense genes in soybean roots with tryptophan treatment. Soybean root samples were taken at 1, 5, and 10 d after H. glycines inoculation, and the expression levels of five defense genes were analyzed by qRT-PCR: (A) PR1, (B) PR2, (C) PR3a, (D) PR3b and (E) PR10. Error bars represent standard errors; a t-test was used to determine significant differences between the tryptophan-treated sample and the control (∗P < 0.05).
The expression of defense genes in soybean roots with uracil treatment. Soybean root samples were taken at 1, 5, and 10 d after H. glycines inoculation, and the expression levels of five defense genes were analyzed by qRT-PCR: (A) PR1, (B) PR2, (C) PR3a, (D) PR3b and (E) PR10. Error bars represent standard errors; a t-test was used to determine significant differences between the uracil-treated sample and the control (∗ P < 0.05).
Discussion
Previous studies revealed that soybean seeds coasted with a fermentation broth of B. simplex Sneb545 showed reduced infestation and delayed development of nematodes as a result of ISR30,31. In this study, we assessed the ISR determinants by coating the soybean seeds with this bacterial strain, and all systems were based on activity tracking. Our results identified ISR determinants in the supernatant of the fermentation broth, which is consistent with the results of other studies29,37. Combining ISR bioassays, six ISR-activity compounds (S5-1, S5-2, S5-3, S11-6-1, S11-6-2, and S11-6-3) were obtained from the supernatant using progressive separation and purification steps. The compounds were finally identified as cyclic(Pro-Tyr), cyclic(Val-Pro), cyclic(Leu-Pro), uracil, phenylalanine, and tryptophan using the methods of 1H NMR and 13C NMR.
Cyclic(Pro-Tyr), cyclic(Val-Pro), and cyclic(Leu-Pro) are cyclic dipeptides that belong to 2,5-diketopiperazine (DKP) derivatives, which have a stable six-membered ring structure with two hydrogen bond donors and two hydrogen bond acceptors. Hydrogen bonds have an important effect on interactions between drugs and receptors, hence cyclic dipeptides play a key role in medicinal chemistry38. Cyclic(Ala-Val), cyclic(Pro-Tyr), and cyclic(Phe-Pro) are signal molecules that activate the biosensors of N-acyl-L-homoserine lactone (AHL), consequently regulating group behaviour39. In addition, natural cyclic dipeptides have a broad range of biological activities, such as antibacterial40, antifungal41, antiviral42, and antitumor43. Cyclic dipeptides from B. vallismortis induced resistance in Arabidopsis against Pseudomonas infection44. In this study, cyclic(Pro-Tyr), cyclic(Val-Pro), and cyclic(Leu-Pro) isolated from B. simplex against to H. glycines through ISR.
Uracil is a pyrimidine organic compound and exists in all living species. Pyrimidines play a substantial role in numerous aspects of plant metabolism and development45. Tomato seedlings were treated with uracil significantly increased the plant height and the fresh and dry weights of the shoot, and also resulted in the formation of more flowers46. Tobacco BY-2 cells regulate salvage activity of uracil to establishes proper cellular conditions for the induction of programmed cell death when increase s levels of nitric oxide and hydrogen peroxide47. In this study, uracil isolated from the B. simplex fermentation broth delayed the development of H. glycines in soybean root. Phenylalanine and tryptophan are amino acids that play a key role in plant growth and disease resistance48. Previous studies demonstrated that the contents of free amino acids increased in cotton seedlings inoculated with root-knot nematodes49. Phenylalanine has also been shown to suppress the development of nematodes in plants, and influences the survival of M. javanica J250. Furthermore, phenylalanine is a precursor of SA biosynthesis. SA is a systemic signal for directing protein synthesis related to plant resistance to pathogens, which activates the defence responses of plant51,52. Tryptophan forms tryptamine through biosynthetic pathways, providing a common backbone for many secondary metabolites that may have different biological activities53, such as auxins (IAA), phytoalexins (camalexin), and alkaloids, which promote plant development and resist pathogen infection54,55,56. These results showed that the amino acids in plants play an important role in resistance to nematodes. In this study, phenylalanine and tryptophan isolated from the B. simplex fermentation broth delayed the development of H. glycines, which may have affected the amino acid content or regulated the metabolism level of phytoalexin in soybean.
SA and JA pathways have been implicated in plant resistance to pathogens. The PR proteins are involved in activating the defense response and are important in plant defenses against nematodes. According to the study reported that tomato plants pre-treated with SA and subsequently infected with root-knot nematodes, PR1 was highly expressed in roots at 1 dpi57. During infection with Phytophthora sojae, the expression of PR3b greatly enhanced following the pre-treatment of soybean seedlings with benzothidiazole (BTH) in 24 h and the transcript abundance of PR10 increased within 24 h following leaf pre-treatment with SA58,59. These results indicate that the SA- and JA-pathways play a role in pathogen infection. In this study, pre-treatment with cyclic(Val-Pro) significantly increased the expression of PR1, PR3b, and PR10 and tryptophan increased the expression of PR1 and PR10 at 1 dpi. The results showed that the SA- and JA-pathways were involved in root resistance to nematode infection at an early stage. Another study reported that pre-treatment with Pochonia chlamydosporia induced the expression of the SA pathway (PR1) and JA pathway (Lox D) at 7 days after nematode inoculation60. In our study, high expression of PR1, PR3b in cyclic(Val-Pro) and PR3b in uracil were observed at 5 dpi. Similarly, the transcript levels of PR2 in cyclic(Val-Pro) and PR3a, PR10 in tryptophan, as well as PR1, PR10 in uracil significantly increased at 10 dpi. This result suggests that the SA- and JA-pathways were involved in root resistance to nematode development. Overall, cyclic(Val-Pro), tryptophan and uracil induced a harmonious interaction between the SA- and JA-pathways to against H. glycines.
In summary, we found six active ISR determinants, cyclic(Pro-Tyr), cyclic(Val-Pro), cyclic(Leu-Pro), uracil, phenylalanine, and tryptophan produced by B. simplex Sneb545 that efficiently controlled H. glycines. In addition, three types of substances cyclic (Val-Pro), tryptophan and uracil were selected to treat soybeans, and the expression levels of defense-related genes were assessed using qRT-PCR. The results indicate cyclic(Val-Pro), tryptophan and uracil induced the expression of defense-related genes involved in the SA- and JA-pathways. In future studies, we will focus on the molecular and metabolic mechanisms of the six active compounds identified, as new biological control agents for the management of high glycine in soybean production.
Materials and methods
Bacteria and nematodes
Bacillus simplex Sneb545 strain was maintained in the Nematode Institute of Northern China (NINC), Shenyang Agriculture University, China30 and grown on lysogeny broth (LB) agar medium at 28 °C for 24 h. A single colony was inoculated to 100 mL LB liquid medium and cultivated at 28 °C for 48 h on a rotary shaker at 175 rpm for further treatments.
Heterodera glycines race 3 was obtained from the experimental field of the NINC. The cysts were obtained by pouring infested soil through a nested 850 μm pore over 250 μm pore sifter followed by centrifugal flotation in a 35% sucrose solution61,62. The cysts were surface-sterilised using 0.5% sodium hypochlorite for 3 min, rinsed with sterile distilled water seven times, and then incubated at 26 °C in the dark14. Second-stage juveniles (J2) were collected for SCN inoculum (1,000 J2 mL-1).
Preliminary screening of ISR determinants from Bacillus simplex Sneb545
The supernatant and bacterial precipitate were collected by centrifuging the fermentation broth at 3,952 × g for 10 min. The bacterial precipitate was washed three times with sterilised water, and then centrifuged at 3,952 × g. The bacterial precipitate was retained and sterile water was added to obtain a bacterial suspension. Soybean seeds were coated with fermentation supernatant and bacterial suspension for ISR bioassay, and seeds coated with sterile water were used as a control.
ISR bioassay
The susceptible soybean seeds, Liaodou 15, were surface sterilised with 0.5% sodium hypochlorite for 10 min and washed seven times with sterilised water63. Sterilised seeds were coated with assay liquid, at a ratio of 1:70 (mL:g), and were then air-dried at room temperature for 12 h and grown in plastic culture tubes (40 × 140 mm) containing sterilised sand15. At the second true leaf stage, the seedlings were inoculated with 2 ml freshly hatched J2 (approximately 1,000) by pipetting into two 1-cm-deep holes around the stem base. The roots were collected 12 d after inoculation and stained with acid fuchsin solution64. The number of different stage nematodes within the root were counted under a microscope (Nikon, Tokyo, Japan) (Fig. 14). ISR bioassay was evaluated using the number of nematodes within the root and the J4 ratio65,66.
Extraction of ISR determinants from Bacillus simplex Sneb545 supernatant
To obtain Sneb545 fermentation broth, the strain was incubated with LB medium (25 L) in the fermenter at 28 °C and 220 rpm for 72 h. The fermentation broth was then separated by centrifugation at 3,952 × g for 10 min, and the supernatant was collected to use as a fermentation broth. The broth was concentrated to 1/10 volume by lyophilisation, and then four times the volume of ethanol was added. The concentrate was kept overnight at 4 °C, and the precipitate and supernatant were obtained by filtering. The concentrated supernatant was placed in a rotary evaporator (BUCHI I-100, Flawil, Switzerland), then the concentrates and precipitates were freeze-dried. The lyophilised powder was dissolved in 2% dimethyl sulfoxide (DMSO) to form a 20 mg/mL solution as the assay liquid for ISR bioassay, and 2%DMSO was used as a control.
The lyophilised supernatant powder was prepared in a 10% aqueous solution, and then extracted three times with PET followed by CHCl3, ETOAC, and nBuOH progressively. The final AP and all extracts were concentrated and then freeze-dried. The lyophilised powders were dissolved in 2% DMSO to form a 20 mg/mL solution as the assay liquid for ISR bioassay, and 2% DMSO was used as a control.
Separation and purification of n-butanol extract
The n-butanol extract (7.5 g) was dissolved in methanol and then mixed with silica gel in a ratio of 1:1.5. When the solvent was completely evaporated, the homogeneous mixture was chromatographed on a silica gel column (60 × 50 mm ID), then progressively eluted using dichloromethane: acetone at 9:1 and 7:3; dichloromethane: methanol at 95:5, 9:1, 8:2, 7:3 and 5:5; followed by methanol: water at 8:2. Forty millilitres of eluent was collected per tube, with 110 tubes in total. Similar fractions were combined using thin-layer chromatography (TLC), and then freeze-dried. The lyophilised various powders were dissolved in 2% DMSO to form a 5 mg/mL solution as the assay liquid for ISR bioassay, and 2% DMSO was used as a control.
Separation of S5 through semi-preparative HPLC
Fraction S5 was dissolved in methanol and filtered, then further separated and purified by semi-preparative HPLC (Waters 600E, Milford, America). Chromatographic conditions were as follows: semi-preparative AQ-C18 Column (10 × 250 mm, 10 μm, Ultimate); the mobile phase of acetonitrile: water = 15:85 (v: v); flow rate of 1.5 mL/min; temperature of 25 °C; detection wavelength of 213 nm; 100 μL per injection. The eluate was collected corresponding to the absorption peak and the purity was assessed using analytical HPLC (Agilent 1260, Palo Alto, America).
Separation of S11 by Sephadex LH-20 column
The absorbance of fraction S11 was determined by an ultraviolet spectrophotometer (Shimadzu UV2450, Kyoto, Japan). Fraction S11 was chromatographed on a gel Sephadex LH-20 column (100 × 16 mm ID) and using methanol: water at 8:2 as the eluting agent. The flow rate was 0.3 mL/min and the detection wavelength was 220 nm. Similar fractions were combined based on UV detection peaks, and then freeze-dried. The lyophilized various powders were dissolved in 2% DMSO in a 5 mg/mL solution as the assay liquid for ISR bioassay, and 2% DMSO was used as a control.
Separation of S11-6 by semi-preparative HPLC
Fraction S11-6 was dissolved in methanol and filtered, then further separated and purified by semi-preparative HPLC. The chromatographic conditions were similar to those described in “Separation of S5 through semi-preparative HPLC” with some modifications, such as the mobile phase of acetonitrile: water was 10:90 (v: v). The eluate was collected according to the absorption peak and the purity was assessed by analytical HPLC.
Structural identification and ISR bioassay for active compounds
The structures of six active compounds were identified using nuclear magnetic resonance spectroscopy (Bruker 600 MHz, Karlsruhe, Germany). The nuclear magnetic conditions were as follows: the sample was dissolved in deuterated reagent and the scanned range of 1H NMR was from − 2 to 16 ppm and 13C NMR was from 0 to 220 ppm at 25 °C. The active compounds were dissolved in 2% DMSO to 1 mM solution as the assay liquid for ISR bioassay, and 2% DMSO was used as a control.
Real-time PCR analysis of gene expression
Soybeans were treated with cyclic(Val-Pro), tryptophan and uracil, respectively, and sterilised water was used as control. Soybeans were grown as described in ISR bioassay and inoculated with 1,000 J2s of H. glycines. Roots were collected at 1, 5 and 10 dpi, and then were frozen in liquid nitrogen immediately and stored at –80 °C until further use. Total RNA was extracted using the RNA pure Plant Kit (Takara, China) according to the manufacturer’s instructions. Total RNA (1 μg) was converted to cDNA using the RT Reagent Kit (Takara, China). qRT-PCR was performed on the CFX Connect Real-Time PCR Detection System (Bio-Rad, USA). The genes expression of PR genes were detected, and Actin 11 was used as reference gene for normalizing the target genes. The primers for PR167, PR268, PR3a58, PR3b58, PR1059 and Actin 1114 were described in Table 1. The 10-μl reaction mixtures of qRT-PCR contained 5 μL 2 × TB Green premix (Takara), 1 μL cDNA template (1:5 dilution), 3.2 μL sterile distilled water, and 0.8 μL of each forward and reverse primers for selected genes. The reaction conditions were 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. A melting curve analysis was performed after 40 cycles to confirm that a single product was present for each reaction. There were three biological replicates and three technical replicates for each treatment per time-point, and the data were quantified using the 2-ΔΔCt method69.
Statistical analysis
All data were analysed using SPSS Statistics 22 (SPSS Inc., IL, USA). The differences among the means were calculated using ANOVA models. Significant differences between different developmental stages for soybean cysts in roots were determined according to Duncan’s multiple range test (P < 0.05). Significant differences between treatments in the defense gene expression analysis experiments were evaluated using t-tests (P < 0.05).
Data availability
All the data supported the findings of this study are available from the corresponding author by request.
References
Wrather, J. A., Anand, S. C. & Dropkin, V. H. Soybean cyst nematode control. Plant Dis. 68, 829–833 (1984).
Wrather, J. A. et al. Soybean disease loss estimates for the top ten soybean-producing countries in 1998. Can. J. Plant Pathol. 23, 115–121 (2001).
Akhtar, M. Current options in integrated management of plant-parasitic nematodes. Int. Pest Manag. Rev. 2, 187–197 (1997).
Kerry, B. R. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 38, 423–441 (2000).
Wen, Y. et al. Nematotoxicity of drupacine and a Cephalotaxus alkaloid preparation against the plant-parasitic nematodes Meloidogyne incognita and Bursaphelenchus xylophilus. Pest Manag. Sci. 69, 1026–1033 (2013).
Zuckerman, B. M. & Esnard, J. Biological control of plant nematodes: current status and hypotheses. Nematol. Res. (Jpn. J. Nematol.) 24, 1–13 (1994).
Lauritis, J. A., Rebois, R. V. & Graney, L. S. Development of Heterodera glycines Ichinohe on soybean, Glycine-Max (L.) Merr., under gnotobiotic conditions. J. Nematol. 15, 272–281 (1983).
Niblack, T., Lambert, K. & Tylka, G. A model plant pathogen from the kingdom animalia: Heterodera glycines, the soybean cyst nematode. Annu. Rev. Phytopathol. 44, 283–303 (2006).
Lugtenberg, B. & Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 (2009).
Ryu, C. M., Hu, C. H., Reddy, M. S. & Kloepper, J. W. Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytol. 160, 413–420 (2003).
Pieterse, C. M. J. et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 10, 1571–1580 (1998).
Vryzas, Z. The plant as metaorganism and research on next-generation systemic pesticides: prospects and challenges. Front. Microbiol. https://doi.org/10.3389/fmicb.2016.01968 (2016).
Liu, D. et al. Klebsiella pneumoniae SnebYK mediates resistance against Heterodera glycines and promotes soybean growth. Front. Microbiol. https://doi.org/10.3389/fmicb.2018.01134 (2018).
Zhao, J. et al. Biocontrol potential of Microbacterium maritypicum Sneb159 against Heterodera glycines. Pest Manag. Sci. 75, 3381–3391 (2019).
Zhou, Y. et al. Management of the soybean cyst nematode Heterodera glycines with combinations of different rhizobacterial strains on soybean. PLoS ONE https://doi.org/10.1371/journal.pone.0182654 (2017).
Bakker, P. A. H. M., Ran, L. X., Pieterse, C. M. J. & van Loon, L. C. Understanding the involvement of rhizobacteria-mediated induction of systemic resistance in biocontrol of plant diseases. Can. J. Plant Pathol. 25, 5–9 (2003).
Kuć, J. Concepts and direction of induced systemic resistance in plants and its application. Eur. J. Plant Pathol. 107, 7–12 (2001).
Zamioudis, C. & Pieterse, C. M. Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150 (2012).
De Meyer, G. & Hofte, M. Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology 87, 588–593 (1997).
Meziane, H., Van der Sluis, I., Van Loon, L. C., Hofte, M. & Bakker, P. A. Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6, 177–185 (2005).
Budzikiewicz, H. Bacterial catecholate siderophores. Mini-Rev. Org. Chem. 1, 163–168 (2004).
Ongena, M. et al. Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant Microbe Interact. 18, 562–569 (2005).
Kammerhofer, N. et al. Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol. 207, 778–789 (2015).
Siddiqui, I. A. & Shaukat, S. S. Role of salicylic acid in Pseudomonas aeruginosa strain IE-6S+-mediated induction of systemic resistance against Meloidogyne javanica in tomato. 42, 268–274 (2003).
Zinovieva, S., Vasyukova, N., Udalova, Z., Gerasimova, N. & Ozeretskovskaya, O. Involvement of salicylic acid in induction of nematode resistance in plants. Biol. Bull. Russ. Acad. Sci. 38, 453 (2011).
Vasyukova, N. I. et al. The involvement of phenylalanine ammonia-lyase and salicylic acid in the induction of resistance of tomato plants infested with gall nematode Meloidogyne incognita. Dokl. Biol. Sci. 416, 382–385 (2007).
Reitz, M. et al. Importance of the O-antigen, core-region and lipid A of rhizobial lipopolysaccharides for the induction of systemic resistance in potato to Globodera pallida. Nematology 4, 73–79 (2002).
Schuhegger, R. et al. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918 (2006).
Akram, W., Anjum, T. & Ali, B. Searching ISR determinants from Bacillus subtilis IAGS174 against Fusarium wilt of tomato. Biocontrol 60, 271–280 (2015).
Xiang, P., Chen, L., Zhu, X., Wang, Y. & Duan, Y. Screening and identification of bacterium to induce resistance of soybean against Heterodera glycines. Chin. J. Biol. Control 29, 661–666 (2013).
Kang, W., Zhu, X., Wang, Y., Chen, L. & Duan, Y. Transcriptomic and metabolomic analyses reveal that bacteria promote plant defense during infection of soybean cyst nematode in soybean. BMC Plant Biol. 18, 1. https://doi.org/10.1186/s12870-018-1302-9 (2018).
Qunqun, G., Daosen, G., Boguang, Z., Jie, X. & Ronggui, L. Two cyclic dipeptides from Pseudomonas fluorescens GcM5-1A carried by the pine wood nematode and their toxicities to Japanese black pine suspension cells and seedlings in vitro. J. Nematol. 39, 243–247 (2007).
Yan, P. S. et al. Cyclo(L-leucyl-L-prolyl) produced by Achromobacter xylosoxidans inhibits aflatoxin production by Aspergillus parasiticus. Appl. Environ. Microbiol. 70, 7466–7473 (2004).
Oliveira, D. F. et al. Purification and identification of metabolites produced by Bacillus cereus and B. subtilis active against Meloidogyne exigua, and their in silico interaction with a putative phosphoribosyl transferase from M. incognita. An. Acad. Bras. Ciênc. 86, 525–538 (2014).
Wang, Y.-Z., Zeng, G., Zhang, M. & Liu, Y. Chemical constituents from rhizoma of Anemone altaica. Chin. Trad. Herb. Drugs. 45, 1219–1222 (2014).
Derdau, V., Atzrodt, J., Zimmermann, J., Kroll, C. & Brückner, F. Hydrogen-deuterium exchange reactions of aromatic compounds and heterocycles by NaBD4-activated rhodium, platinum and palladium catalysts. Chem. Eur. J. 15, 10397–10404 (2009).
Gómez-Gómez, L. & Boller, T. Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256 (2002).
Guijie, T. A facile pathway to synthesize diketopiperazine derivatives. Tetrahedron Lett. 43, 865–867 (2002).
Holden, M. et al. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33, 1254–1266 (1999).
Wattana-Amorn, P., Charoenwongsa, W., Williams, C., Crump, M. P. & Apichaisataienchote, B. Antibacterial activity of cyclo(L-Pro-L-Tyr) and cyclo(D-Pro-L-Tyr) from Streptomyces sp. strain 22-4 against phytopathogenic bacteria. Nat. Prod. Res. 30, 1980–1983 (2015).
Nishanth Kumar, S., Nath, V. S., Pratap Chandran, R. & Nambisan, B. Cyclic dipeptides from rhabditid entomopathogenic nematode-associated Bacillus cereus have antimicrobial activities. World J. Microb. Biot. 30, 439–449 (2014).
Kwak, M. et al. Cyclic dipeptides from lactic acid bacteria inhibit proliferation of the influenza A virus. J. Microbiol. 51, 836–843 (2013).
Toske, S. G., Jensen, P. R., Kauffman, C. A. & Fenical, W. Aspergillamides A and B: modified cytotoxic tripeptides produced by a marine fungus of the genus Aspergillus. Tetrahedron 30, 13459–13466 (1998).
Noh, S. W. et al. Cyclic dipeptides from Bacillus vallismortis BS07 require key components of plant immunity to induce disease resistance in Arabidopsis against Pseudomonas infection. Plant Pathol. J. 33, 402–409 (2017).
Mourad, G. S., Snook, B. M., Prabhakar, J. T., Mansfield, T. A. & Schultes, N. P. A fluoroorotic acid-resistant mutant of arabidopsis defective in the uptake of uracil. J. Exp. Bot. 57, 3563–3573 (2006).
Mathur, S. N. & Sharma, R. A. Effect of uracil and 5-Nitrouracil on growth and flowering of Tomato. Physiol. Plant. 21, 911–917 (1968).
Stasolla, C., Loukanina, N., Yeung, E. C. & Thorpe, T. A. Alterations in pyrimidine nucleotide metabolism as an early signal during the execution of programmed cell death in tobacco BY-2 cells. J. Exp. Bot. 55, 2513–2522 (2004).
Rai, V. K. Role of amino acids in plant responses to stresses. Biol. Plant. 45, 481–487 (2002).
Lewis, S. A. & Mcclure, M. A. Free amino acids in roots of infected cotton seedlings resistant and susceptible to Meloidogyne incognita. J. Nematol. 7, 10–15 (1975).
Amdadul Hoque, A. K. M., Rejwan Bhuiyan, M., Ashik Iqbal Khan, M., Mahmud, A. & Uddin Ahmad, M. Effect of amino acids on root-knot nematode (Meloidogyne javanica) infecting tomato plant. Arch. Phytopathol. Plant Protect. 47, 1921–1928 (2013).
Klessig, D. F. et al. Nitric oxide and salicylic acid signaling in plant defense. PNAS 97, 8849–8855 (2000).
Wubben, M. J. E., Jin, J. & Baum, T. J. Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol. Plant Microbe Interact. 21, 424–432 (2008).
Kaminaga, Y. et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 281, 23357–23366 (2006).
Kutchan, T. M. Alkaloid biosynthesis: the basis for metabolic engineering of medicinal plants. Plant Cell. 7, 1059–1070 (1995).
Radwanski, E. R. & Last, R. L. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell. 7, 921–934 (1995).
Last, R. L. & Fink, G. R. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240, 305–310 (1988).
Molinari, S., Fanelli, E. & Leonetti, P. Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in miediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 15, 255–264 (2014).
Han, Q. et al. Effect of a benzothiadiazole on inducing resistance of soybean to Phytophthora sojae. Protoplasma 250, 471–481 (2012).
Xu, P. et al. Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 41, 4899–4909 (2014).
Ghahremani, Z., Escudero, N., Saus, E., Gabaldón, T. & Sorribas, F. J. Pochonia chlamydosporia induces plant-dependent systemic resistance to Meloidogyne incognita. Front. Plant Sci. 10, 945 (2019).
Krusberg, L. R., Sardanelli, S., Meyer, S. L. F. & Crowley, P. A method for recovery and counting of nematode cysts. J. Nematol. 26, 599 (1995).
Jenkins, W. R. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Rep. 48, 692 (1964).
Hosseini, P. & Matthews, B. F. Regulatory interplay between soybean root and soybean cyst nematode during a resistant and susceptible reaction. BMC Plant Biol. 14, 300 (2014).
Bybd, D. W. J., Kirkpatrick, T. & Barker, K. R. An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143 (1983).
Lin, J. et al. Overexpression of a soybean salicylic acid methyltransferase gene confers resistance to soybean cyst nematode. Plant Biotechnol. J. 11, 1135–1145 (2013).
Melito, S. et al. A nematode demographics assay in transgenic roots reveals no significant Impacts of the Rhg1 locus LRR-Kinase on soybean cyst nematode resistance. BMC Plant Biol. 10, 104 (2010).
Hu, Y., You, J., Li, C., Hua, C. & Wang, C. Exogenous application of methyl jasmonate induces defence against Meloidogyne Hapla in soybean. Nematology 19, 293–304 (2017).
Fan, S. et al. GmWRKY31 and GmHDL56 Enhances resistance to Phytophthora sojae by regulating defense-related gene expression in Soybean. Front. Plant Sci. 8, 781 (2017).
Livak, K. & Schmittgen, T. Analysis of relative gene expression data using real–time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgments
We thank Dr. Piao Lei and Dr. Aatika Sikandar for helpful comments on the manuscript. This work was supported by a grant from the National Science & Technology Fundamental Resources Investigation Program of China (2018FY100300), and China Agriculture Research System (CARS-04-PS13), and Foundation of Liaoning Educational Commission (LSNQN201723).
Author information
Authors and Affiliations
Contributions
Z. X. wrote the main manuscript text and conducted the experiments, Z. X. and X. W. worked on data curation and analysis. X. W., J. Z., X. Z. and X. Z. amended the manuscript. Y. W., H. F. and L. C. supervised this work. X. L. provided methodology. Y. D. provided the fund for this study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xing, Z., Wu, X., Zhao, J. et al. Isolation and identification of induced systemic resistance determinants from Bacillus simplex Sneb545 against Heterodera glycines. Sci Rep 10, 11586 (2020). https://doi.org/10.1038/s41598-020-68548-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-68548-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.