Abstract
G protein-coupled receptor (GPCR) biogenesis, trafficking, and function are regulated by post-translational modifications, including N-glycosylation of asparagine residues. α1D-adrenergic receptors (α1D-ARs) – key regulators of central and autonomic nervous system function – contain two putative N-glycosylation sites within the large N-terminal domain at N65 and N82. However, determining the glycosylation state of this receptor has proven challenging. Towards understanding the role of these putative glycosylation sites, site-directed mutagenesis and lectin affinity purification identified N65 and N82 as bona fide acceptors for N-glycans. Surprisingly, we also report that simultaneously mutating N65 and N82 causes early termination of α1D-AR between transmembrane domain 2 and 3. Label-free dynamic mass redistribution and cell surface trafficking assays revealed that single and double glycosylation deficient mutants display limited function with impaired plasma membrane expression. Confocal microscopy imaging analysis and SNAP-tag sucrose density fractionation assays revealed the dual glycosylation mutant α1D-AR is widely distributed throughout the cytosol and nucleus. Based on these novel findings, we propose α1D-AR transmembrane domain 2 acts as an ER localization signal during active protein biogenesis, and that α1D-AR N-terminal glycosylation is required for complete translation of nascent, functional receptor.
Similar content being viewed by others
Introduction
G protein-coupled receptors (GPCRs) are essential membrane proteins that regulate the vast majority of physiological functions in the human body. As a result, GPCRs have been estimated to be targeted by approximately one third of all currently approved medications1. Adrenergic receptors (ARs) are a clinically relevant subfamily of GPCRs. Activated by the endogenous sympathetic neurotransmitters epinephrine and norepinephrine, adrenergic GPCRs consist of three major subtypes: α1, α2, and β. The α1 sub-family – containing α1A, α1B, and α1D subtypes2 – are targets for medications that regulate blood pressure3,4, bladder5,6, prostate7,8, and central nervous system function9,10,11. Thus, understanding the molecular and cellular mechanisms regulating α1-AR function will help spur the development of new medications associated with aberrant α1-AR signaling, such as hypertension, PTSD, schizophrenia, and benign prostatic hypertrophy12,13,14,15.
Among the three α1 subtypes, the α1D-AR remains poorly understood due to technical challenges. Relative to the closely related α1A and α1B-AR subtypes, α1D-AR displays limited functional responses and minimal plasma membrane expression when expressed in heterologous cell culture16,17,18. Although pharmacologically detectable in intact isolated aortae in organ-tissue bath assays19, α1D-AR functional expression rapidly disappears in primary vascular smooth muscle cell cultures within 24–48 hours20. Also, immortalized cell lines that endogenously express α1D-ARs have yet to be discovered. Combined, these experimental clues indicate the molecular and cellular mechanisms governing α1D-AR functional expression in cells are unique amongst the α1-AR subtypes, and thus may be targeted by novel drugs to exogenously regulate α1D-AR signaling in human disease.
Important clues towards solving why α1D-ARs are poorly expressed in cell culture include two structural features distinct to the α1D-AR: (A) a C-terminal PSD-95/Dlg/ZO-1 (PDZ) ligand that ensures the formation of a modular, homodimeric macromolecular complex via binding to PDZ-domain containing proteins Scribble and syntrophin21,22,23,24,25; and (B) an atypical extracellular N-terminal domain (NTD). The average NTD for class A GPCRs is 40 amino acids26, making the 95 amino acid α1D NTD unusually long. We, and others, have previously demonstrated the α1D-AR NTD contains an endoplasmic reticulum (ER) retention signal17,18,27, and that the NTD undergoes an endogenous cleavage event that enhances α1D-AR plasma membrane trafficking and agonist-stimulated functional responses28. Unfortunately, the mechanisms by which the NTD regulates α1D-AR trafficking and function are unknown.
GPCR trafficking is a highly complex process that is regulated in part by multiple factors, including Rab GTPases29,30, TBC domain-containing proteins31, GPCR oligomerization32,33, N-terminal cleavage34,35,36,37, and N-terminal translocation and glycosylation in the ER lumen38,39,40,41. Previous studies employed WGA lectin and deglycosylating enzymes to demonstrate endogenous α1-ARs are glycosylated in rat brain42, but were unable to determine if individual α1-AR subtypes were glycosylated due to technical limitations43. Subsequent studies have produced conflicting results27,44,45,46, and as a result, it remains unclear how NTD glycosylation regulates α1-ARs physiological function. Interestingly, the α1D-AR NTD contains two putative N-glycosylation sites located at N65 and N8247. In this study, we leverage SNAP-epitope tag labeling and label-free dynamic mass redistribution technology to show, for the first time, that the α1D-AR NTD is dual glycosylated, thereby ensuring proper biosynthesis and trafficking of nascent receptors.
Results and Discussion
N-terminal glycosylation is required for complete α1D-AR biogenesis
The α1D-AR N-terminal contains two putative N-glycosylation sites (N65, N82) with both serving as theoretical acceptors for N-glycans within the ER lumen47. N-glycosylation is the covalent attachment of an N-glycan sugar moiety to an asparagine residue within the consensus sequence N-X-S/T, where X is any amino acid except proline48,49. Thus, we sought to examine the glycosylation state of full length α1D-AR using PNGase F deglycosylation assays. To test this possibility, HEK293 cells were transiently transfected with N-terminal SNAP-epitope tagged α1D-AR cDNA constructs (SNAP-α1D). We have previously demonstrated the SNAP epitope-tag facilitates visual analysis of GPCR protein bands directly within polyacrylamide gels, and do not require nitrocellulose paper transfer or antibody staining, thus removing all potential false positive bands22,28,50. Incorporating this powerful technology, HEK293 cell lysates expressing SNAP-α1D were lysed, denatured, and incubated with PNGase F, then subjected to polyacrylamide gel electrophoresis and near-infrared imaging (PAGE NIR). Similar to previous reports27,46, our results were inconclusive, likely due to instability of α1D-AR in the required buffer conditions (Supplementary Fig. S1A). To overcome this technical issue, we utilized lentil lectin affinity purification. Lentil lectin recognizes complex glycans containing α-(1 → 6)-linked fucose on the core GalNAc as well as glucose and/or α-mannose residues, and is active in a variety of buffer conditions51,52. HEK293 cells were transiently transfected with SNAP-α1D and lysates were incubated with lentil lectin sepharose beads. Samples were eluted and subjected to PAGE NIR analysis. Shown in Fig. 1A are the results. In agreement with our previous studies22,28, the input lane demonstrates full length SNAP-α1D is robustly expressed as a monomeric band at ~80 kDa, a larger, more intense band at ~90 kDa (Fig. 1A, arrow, Supplementary Fig. S1B), as higher order oligomers (MW > 180 kDa), and as multiple NTD cleavage products (MW = ~30–35 kDa). Remarkably, both ~90 kDa monomeric and multimeric SNAP-α1D species were detected in the lectin bound lane (Fig. 1A, bound). Although faint, the largest α1D NTD cleavage product28 was also observed in the lectin-bound sample. Thus, this experiment clearly demonstrates, for the first time, that the α1D NTD is N-glycosylated.
Site-directed mutagenesis and lectin affinity purification analyses reveal α1D-AR is dually glycosylated at N65 and N82. (A) Lysate from HEK293 cells expressing WT SNAP-α1D (input) was incubated with lentil lectin sepharose beads to isolate glycosylated proteins. Bound protein was eluted with methyl-α-D-mannopyranoside (bound), and analyzed using PAGE NIR. SNAP-α1D monomers (►) and higher order oligomers, as well as the previously described N-terminal cleavage product, are present in the elutant. (B) Schematic and (C) PAGE NIR of HEK293 cell lysate expressing WT, single glycosylation mutants (N65Q and N82Q), and double glycosylation mutant (NQQ) SNAP-α1D species. (D) HEK293 cells expressing WT SNAP-α1D were incubated for 16 hr with vehicle (−) or tunicamycin (TUN, +), and analyzed with PAGE NIR. A signal at 43 kDa was observed in the tunicamycin treated samples (○). (E) PAGE NIR of HEK293 cell lysate transfected with N-terminal SNAP-epitope (λ = 800 nm, green) and C-terminal CLIP-epitope (λ = 700 nm, red) dual tagged WT (S-WT-C) and NQQ (S-NQQ-C) α1D-AR constructs. (F,I) PAGE NIR of HEK293 cell lysate expressing S-WT-C or (G,J) S-NQQ-C following 24 hr bortezomib (BTZ) (F,G) or protease inhibitor (PI) treatment (I,J). (H) Quantitation of signals from F and G normalized to vehicle. (K) Quantitation of fluorescent signals from I and J normalized to vehicle. All gels are representative images from n = 3 experiments. For F and G, data are represented as mean ± SEM; Unpaired t tests, ***p < 0.001.
Towards our goal of addressing the importance of each NTD glycosylation site for α1D-AR function, we created single (N65Q or N82Q) and double (NQQ) glycosylation deficient SNAP-α1D mutants using PCR site-directed mutagenesis (see Fig. 1B for schematic). To ensure each α1D-AR NTD mutant was expressed as protein, cDNA constructs were transfected into HEK293 cells and subjected to PAGE NIR analysis. Both the N65Q and N82Q SNAP-α1D NTD mutants display equivalent protein band patterns as SNAP-α1D (Fig. 1C). Unexpectedly, the NQQ SNAP-α1D double mutant did not produce monomeric or higher order oligomeric bands. Instead, NQQ SNAP-α1D was primarily expressed as a single, robust band of ~43 kDa in size. Subtracting the size of the SNAP-epitope tag plus linker (25 kDa) yields a polypeptide of 18 kDa, ‒ roughly equivalent in size to the α1D NTD, transmembrane domain (TM) 1, intracellular loop 1, and TM2. Subsequent lectin-purification assays reveal full length, N65Q and N82Q, but not NQQ, SNAP-α1D species are glycosylated (Supplementary Fig. S1C). To ensure this unexpected NQQ product was due to inhibition of glycosylation, and not a by-product of mutation, cells expressing WT SNAP-α1D were treated with tunicamycin – an inhibitor of N-glycosylation53. 24 hours after transfection with WT SNAP-α1D, HEK293 cells were treated with fresh media supplemented with 5 μg/mL tunicamycin or EtOH vehicle followed by PAGE NIR (Fig. 1D). Interestingly, though faint, the same ~43 kDa species observed in the NQQ SNAP-α1D is also present in the tunicamycin treated samples (Fig. 1D; circle). Thus, this initial round of experiments demonstrates that (A) α1D-AR is glycosylated at N65 and N82; (B) only a single glycosylation site needs to be available for the NTD to become glycosylated and full length α1D protein processing to occur; and (C) removal of both α1D NTD glycosylation sites not only prevents glycosylation, but produces an abnormally short, previously unreported α1D-AR peptide species.
We next tested two potential explanations for this serendipitous, intriguing result: (A) the NQQ double mutation introduces a destabilizing effect, causing the α1D-AR to be targeted for degradation, with the observed 43 kDa band representing the major degradation product; or (B) NQQ is inhibiting proper translation of α1D-AR, causing an early termination after TM2. These hypotheses were tested using a dual epitope-tagging approach. InFusion PCR was used to add C-terminal CLIP-epitope tags to WT SNAP-α1D (S-WT-C) and NQQ SNAP-α1D (S-NQQ-C). CLIP is a homolog of SNAP that covalently interacts with benzylcytosine conjugates, displaying no cross-reactivity for the SNAP substrate, benzylguanine54. We reasoned that if A is true, CLIP substrate fluorescence in the 700 channel (red) would be observed in both the S-WT-C and S-NQQ-C PAGE NIR lanes. Conversely, we would expect to detect no 700 signal in the S-NQQ-C lane if B were true, as the CLIP tag would not be transcribed if α1D-AR translation was halted at TM2. Thus, S-WT-C and S-NQQ-C α1D-AR cDNA constructs were expressed in HEK293 cells and subjected to PAGE NIR analysis. Fig. 1E shows that overlapping CLIP (red) and SNAP (green) substrate signals are detectable in the S-WT-C lane (left). Contrarily, no CLIP signal is observed in the S-NQQ-C lane, and only the previously observed 43 kDa SNAP-α1D species (Supplementary Fig. S1D).
As an orthogonal approach, HEK293 cells expressing either S-WT-C or S-NQQ-C were incubated with bortezomib (BTZ) ‒ a proteasomal inhibitor (Fig. 1F,G) ‒ or protease inhibitor (PI) cocktail (Fig. 1I,J) for 24 hours followed by PAGE NIR analysis. As expected, significant increases of S-WT-C and S-NQQ-C protein bands were observed with BTZ treatment (Fig. 1H; S-WT-C = 166.3 ± 4.3%, mean ± SEM of vehicle; S-NQQ-C = 186.3 ± 6.0% mean ± SEM of vehicle; Unpaired t test; p < 0.001), but not with PI cocktail treatment (Fig. 1K; S-WT-C = 96.7 ± 0.3%, mean ± SEM of vehicle; S-NQQ-C = 97.0 ± 2.5%, mean ± SEM of vehicle; Unpaired t test, p > 0.05). However, neither BTZ nor PI cocktail had any discernable effect on the molecular weight of the NQQ band; nor were CLIP signals observed in either condition. Taken together, these findings indicate that the NQQ α1D-AR species is not created by proteolytic cleavage and/or degradation of full-length α1D-AR.
To further confirm the identity of this unexpected NQQ species, HEK293 cells were transiently transfected with either WT SNAP-α1D or NQQ SNAP-α1D, lysed, and SNAP-fusion proteins were isolated with SNAP-Capture beads. Due to the covalent nature of the SNAP-Capture:SNAP-tag, an on-bead digest was performed using Trypsin and Glu-C proteases. Samples were subjected to MS/MS analysis (SNAP MS/MS). As shown in Fig. 2A, identified peptides spanned the entirety of the WT SNAP-α1D (Fig. 2A, underlined). Contrarily, only peptides in the N-terminal domain were identified in NQQ SNAP-α1D samples (Fig. 2B, underlined). Furthermore, previously reported α1D-AR interactors syntrophin21,24,25, members of the dystrophin-associated protein complex23, and scribble22,25 were identified in the WT, but not NQQ samples (Supplementary Datas S1, S2). Together, these data provide compelling evidence that glycosylation of both N65 and N82 are necessary for proper biogenesis of full-length α1D-AR, and disruption of these essential glycosylation sites results in early termination of α1D-AR processing after TM2.
SNAP MS/MS analysis identifies distal peptides in WT, but not NQQ SNAP-α1D lysates. (A) WT or (B) NQQ SNAP-α1D was purified from HEK293 lysate using SNAP-Capture pull-down resin, then subjected to on-bead double-enzymatic digestion, and MS/MS analysis. Red text indicates SNAP-epitope tag. Blue text indicates transmembrane domains. Peptides identified in MS/MS analysis are underlined. See Supplementary Datas S1 and S2 for complete data set.
Glycosylation imparts α1D-AR function and plasma membrane insertion
The effects of NTD glycosylation on GPCR function and trafficking are highly divergent. Mutating N-terminal glycosylation sites decreases functional responses of the FSH55, dopamine D256, and neurokinin 1 receptor subtypes57, while loss of glycosylation has no effect on the function of the histamine H2 receptor58. Conversely, blocking N-terminal glycosylation increases binding site density of the human oxytocin receptor59, and signaling efficacy of the vasopressin 1A receptor60. To understand how N-glycosylation impacts α1D-AR function, label-free dynamic mass redistribution (DMR) assays were used to quantify the efficacy of the α1-AR agonist phenylephrine for stimulating α1D NTD glycosylation site mutants. HEK293 cells expressing WT, N65Q, N82Q, or NQQ SNAP-α1D were seeded in 384-well DMR plates and incubated with increasing concentrations of phenylephrine to facilitate concentration-response curve analysis (Fig. 3A). Surprisingly, phenylephrine maximal responses for N65Q (24.99 ± 11.35 pm, mean ± SEM), N82Q (46.64 ± 9.96 pm, mean ± SEM), and NQQ (45.20 ± 8.35 pm, mean ± SEM) were significantly lower than WT SNAP-α1D (112.5 ± 9.27, mean ± SEM; p < 0.01, One-way ANOVA with Tukey’s multiple comparisons post-hoc test).
α1D-AR function and plasma membrane insertion is glycosylation dependent. (A) Dynamic mass redistribution assays quantifying phenylephrine efficacy in HEK293 cells transfected with WT, N65Q, N82Q, or NQQ SNAP-α1D. Data are the mean of 8 replicates ± SEM; **p < 0.01 Emax from WT. (B) Cell surface expression of WT, N65Q, N82Q, or NQQ SNAP-α1D in fixed HEK293 cells labeled with the cell impermeable SNAP substrate, BG782 (top panel, green); nuclear stain, TO-PRO-3 was used to normalize for cell numbers (bottom panel, red). (C) Fluorescence intensity of data from B was normalized to cells expressing SNAP alone. Data are mean of 6 replicates ± SEM; One-way ANOVA with Tukey’s multiple comparisons post-hoc tests, **p < 0.01 from WT SNAP-α1D; ###p < 0.001 from empty SNAP.
Glycosylation has been shown to facilitate plasma membrane trafficking of the angiotensin II type 161, GPR3062, rhodopsin 163, δ-opioid receptor40,64,65, and P2Y2 receptor subtypes66. Therefore, one possible explanation for the reduced function of α1D NTD glycosylation mutants may be aberrant cellular trafficking, leading to a decrease in cell surface expression. This was examined by quantifying WT, N65Q, N82Q and NQQ SNAP-α1D plasma membrane expression levels in fixed HEK293 cells treated with the cell-impermeable SNAP substrate, BG-782 (Fig. 3B,C)22,28,50. Cells were also treated with nuclear stain TO-PRO-3 to normalize for cell number. We observed significant reductions in N65Q (13.40 ± 4.65%, mean ± SEM change from SNAP), N82Q (9.49 ± 5.95%, mean ± SEM change from SNAP), and NQQ (13.64 ± 5.76%, mean ± SEM change from SNAP) cell surface expression in comparison to WT SNAP-α1D (46.13 ± 5.61%, mean ± SEM change from SNAP; p < 0.01, One-way ANOVA with Tukey’s multiple comparisons post-hoc test). Combined, these data strongly indicate both N65 and N82 must be glycosylated to facilitate α1D-AR plasma membrane insertion and agonist-stimulated functional responses in cultured human cells.
TM2 of α1D-AR triggers ER translocation during protein synthesis
TM1 domain is thought to provide the ER localization signal for myriad polytropic integral membrane proteins ‒ including some GPCRs ‒ during protein synthesis38. Though, synthesis of TM2 has also been shown to trigger ribosomal translocation to the ER for some multi-pass transmembrane proteins, such as Cig3067,68 and ProW69. Because the α1D-AR NQQ mutant appears to cause early termination after TM2 (Figs. 1C,E,G,J and 2), we hypothesized that TM2 acts as the ER localization signal for α1D-ARs. To test this, we utilized two orthogonal, but complementary, approaches: sucrose density gradient and confocal imaging.
Previous studies examining α1-AR subcellular localization used cell fractionation/sucrose density gradient to sequester distinct cellular compartments, and then radioligand binding to quantify the number of receptors present in each compartment sample18. Although useful, this method is only able to detect properly folded, functional receptors that are able to bind ligand; and has non-optimal signal-to-noise ratios18. Thus, sucrose density centrifugation protocols were modified to incorporate the sensitivity of SNAP-epitope tag PAGE NIR imaging analysis. This novel experimental approach allows accurate detection of poorly expressing α1D-AR peptide species, regardless of their structural or functional state. Furthermore, the use of the SNAP epitope tag displays increased sensitivity compared to traditional immunoblotting techniques, which can be limited by the inability of antibodies to detect low expression levels of endogenous protein markers70. Thus, HEK293 cells were transfected with SNAP-α1A-AR, which we have previously shown expresses readily at the plasma membrane18, or SNAP-Sec61β, an ER integral membrane protein71. Cells were lysed in detergent free buffer then conjugated to SNAP substrate BG-782. Labelled lysates were then fractionated in a discontinuous gradient (see methods for details), collected, and subjected to PAGE NIR analysis. In each case, the detectable SNAP signal from each isolated fraction was normalized to input. Data were analyzed by area under curve (AUC) to quantify the distribution of each SNAP protein in specific fractions. Figure 4 displays the PAGE NIR band pattern for SNAP-α1A (Fig. 4A, Supplementary Fig. S2A) and SNAP-Sec61β (Fig. 4B, Supplementary Fig. S2B). Subsequent AUC analysis revealed SNAP-Sec61β to be primarily distributed in fractions 1 through 4 with a peak in fraction 2 (91.50% total AUC; Fig. 4C), which is considered to be the ER fraction18. Conversely, SNAP-α1A is significantly concentrated in fractions 6 through 9 with the maximum signal in fraction 7 (100% total AUC).
Sucrose density centrifugation of SNAP-epitope tagged plasma membrane and endoplasmic reticulum markers. HEK293 cell lysates expressing (A) SNAP-α1A-AR (plasma membrane) or (B) SNAP-Sec61β (endoplasmic reticulum) were labeled with SNAP substrate, BG-782, fractionated by discontinuous sucrose density gradient centrifugation, and analyzed using PAGE NIR. I = input. (C) Quantification of fluorescence signal from A and B normalized to respective inputs. Data are mean of 2 experiments ± SEM.
Next, WT, N65Q, N82Q and NQQ SNAP-α1D cDNA constructs were examined (Fig. 5A–D). As expected based on the findings of previous studies performed by us and others16,17,18, WT SNAP-α1D (Fig. 5A, Supplementary Fig. S3A) displayed a similar distribution pattern as SNAP-Sec61β, with a major peak spanning from fractions 1 to 4 (92.47% total AUC; Fig. 5E) and a minor peak in fractions 6 to 8 (7.53% total AUC). Similarly, N65Q SNAP-α1D (Fig. 5B, Supplementary Fig. S3B) was bi-modally distributed, with peaks in fractions 1 to 3 (46.33% total AUC) and fractions 5 to 7 (53.67% total AUC; Fig. 5E). N82Q SNAP-α1D (Fig. 5C, Supplementary Fig. S3C) was largely concentrated in fractions 1 through 4 with the maximum signal in fraction 2 (89.13% total AUC). A minor peak was also observed in fractions 6 to 7 (10.87% AUC; Fig. 5E). Remarkably, NQQ SNAP-α1D (Fig. 5D, Supplementary Fig. S3D) formed a single, strong peak spanning fractions 1 to 3, with the majority of the protein concentrated to the first fraction (100% total AUC; Fig. 5E), which corresponds with a primarily cytosolic localization.
Sucrose density centrifugation of WT and glycosylation deficient SNAP-α1D constructs. Representative PAGE NIR of HEK293 cell lysates transfected with (A) WT, (B) N65Q, (C) N82Q, or (D) NQQ SNAP-α1D following sucrose density centrifugation. I = input. (E) Quantification of band intensity from A-D normalized to respective inputs. Data are presented as mean of 3 experiments ± SEM.
These findings were subsequently corroborated with confocal microscopy imaging analysis. HEK293 cells were transiently transfected with WT (Fig. 6A–D), N65Q (Fig. 6E–H), N82Q (Fig. 6I–L), NQQ (Fig. 6M–P) SNAP-α1D, or empty pSNAP vector (SNAP; Fig. 6Q–T). Cells were fixed with paraformaldehyde and detergent-permeabilized, then incubated with SNAP substrate Alexa Fluor 488 to label SNAP proteins and red fluorescence-Cytopainter stain to label the ER. Visual analysis reveals robust puncta of WT (Fig. 6D), N65Q (Fig. 6H), and N82Q (Fig. 6L) SNAP-α1D within the ER. Contrarily, both NQQ SNAP-α1D (Fig. 6P) and SNAP (Fig. 6T) are diffuse throughout the cell cytosol and nucleus. Colocalization quantification analysis (Fig. 6U) revealed a significant correlation between WT (Pearson’s coefficient = 0.72 ± 0.03, mean ± SEM, p < 0.001 from SNAP, p < 0.01 from NQQ), N65Q (Pearson’s coefficient = 0.70 ± 0.04, mean ± SEM, p < 0.001 from SNAP, p < 0.01 from NQQ), and N82Q (Pearson’s coefficient = 0.53 ± 0.07, mean ± SEM, p < 0.001 from SNAP, p < 0.01 from NQQ) SNAP-α1D constructs with ER stain CytoPainter, when compared with NQQ SNAP-α1D (Pearson’s coefficient = 0.19 ± 0.03, mean ± SEM, p = 0.12 from SNAP) and SNAP (Pearson’s coefficient = 0.03 ± 0.05, mean ± SEM). Together, our cell fractionation and confocal microscopy data support the hypothesis that α1D-AR requires the synthesis of TM2 prior to ER localization, where the N-terminal translocates into lumen, becomes glycosylated, and translation continues. If this does not occur, translation is prematurely terminated and the non-functional polypeptide is likely degraded via the ER-associated degradation (ERAD) pathway71,72,73,74. Further studies are necessary to determine the extent to which this mechanism is involved in the turnover of nascent α1D-AR and other GPCRs.
Confocal imaging reveals NQQ SNAP-α1D is localized to cytosol and nucleus in HEK293 cells. HEK293 cells transfected with (A–D) WT, (E–H) N65Q, (I–L) N82Q, (M–P) NQQ SNAP-α1D, or (Q–T) empty pSNAP vector were fixed, stained for Hoechst 33342 (blue), fluorescence-Cytopainter stain (red), and SNAP-epitope tag (BG-488; green), and imaged using confocal microscopy. Right panels are merged images of three channels. Scale bar = 10 μm. (U) Pearson’s coefficients of SNAP and ER fluorescence signals were computed to measure the extent of colocalization. Data are mean of 5–6 cells ± SEM; One-way ANOVA with Tukey’s multiple comparisons post-hoc tests, ###p < 0.001 compared to empty SNAP; **p < 0.01 compared to NQQ SNAP-α1D, n.s = p > 0.05.
Conclusion
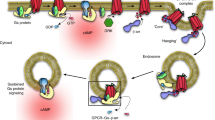
Our findings support a model in which TM2, not TM1, triggers ribosomal translocation to the ER during α1D-AR synthesis67,68,69,75. Upon docking with the ER, the N-terminus is translocated into the ER lumen – possibly via the ER protein complex38,76,77,78 – where glycosylation occurs. This event prevents the N-terminus from retrotranslocating back to the cytosol, which anchors the nascent peptide in the ER membrane in the proper membrane topology, such that the N-terminal will be within the extracellular matrix upon plasma membrane insertion. This event is required before complete translation of the nascent polypeptide continues. However, if glycosylation is prevented, the immature receptor does not anchor in the ER membrane, thus terminating receptor translation after TM2 (see Supplementary Fig. S4 for schematic); and presumably this degenerate polypeptide is degraded via ERAD73 or other cytosolic degradation mechanisms40,79. Furthermore, we show that glycosylation of both N65 and N82 is required for proper function and plasma membrane expression of α1D-ARs.
Materials and Methods
Plasmids and chemicals
Molecular cloning was performed using inFusion HD cloning technology (Clontech/Takara Biotech, Mountain View, CA). The pSNAPf and pCLIP vector, as well as SNAP substrates, BG-782 and Alexa Fluor 488, and CLIP substrate, BC-680 were purchased from New England Biolabs (Ipswich, MA). PageRuler Prestained NIR Protein Ladder was used for all PAGE NIR analyses (Thermo Fisher Scientific, Waltham, MA).
Cell culture
Human Embryonic Kidney (HEK) 293 cells were grown in Dulbecco’s Modified Eagle’s Medium (Corning, Corning, NY) supplemented with 10% fetal bovine serum and 2 mM L-glutamine at 37 °C in 5% CO2. Cells were used ~48 hrs post-transfection with polyethyleneimine unless stated otherwise.
PNGase assay
Cells were lysed in 20 mM Tris-HCl (pH 8), 200 mM NaCl, 5 mM DTT, and 1% NP-40 on ice for 20 min with vortexing every 5 min, followed by 14 K RPM centrifugation at 4 °C for 10 min. 20 μg total protein was incubated with PNGase F (New England Biolabs, Ipswich, MA) for 24 hrs. at 37 °C according to manufacturer’s instructions. After reaction was complete, samples were analyzed by PAGE NIR analysis using a LI-COR Odyssey CLx (LI-COR, Lincoln, NE)22,28,50
Lentil lectin affinity purification
Cells were lysed in 20 mM Tris-HCl (pH 8), 200 mM NaCl, 5 mM DTT, and 1% NP-40 on ice for 20 min. with vortexing every 5 min., followed by 14 K RPM centrifugation at 4 °C for 10 min. The soluble fraction was incubated with Lentil Lectin Sepharose 4B beads (GE Healthcare, Chicago, IL) and 1 μL of 25 μL BG-782 for 1 hr at room temperature. Beads were pelleted and washed 3X in excess lysis buffer. Bound protein was eluted with lysis buffer supplemented with 200 mM methyl-α-D-mannopyranoside at 37 °C shaken at 300 RPM for 10 min. Elutant was collected and subjected to SDS-PAGE electrophoresis, followed by PAGE NIR.
Tunicamycin treatment
HEK293 cells were transfected with WT SNAP-α1D. 24 hr after transfection, cells were washed 3X with PBS, and incubated with media containing 5 ug/mL of tunicamycin or 95% EtOH vehicle for 16 hr. Following incubation, cells were washed 3X with ice cold PBS, lysed, subjected to SDS PAGE, and analyzed by PAGE NIR as described above.
Bortezomib treatment
HEK293 cell were transfected with either S-WT-C or S-NQQ-C. 24 hr after transfections, cells were treated with 1 μM of bortezomib or DMSO vehicle for 24 hr. Following treatment, cells were lysed and lysate analyzed via PAGE NIR.
Protease inhibitor treatment
HEK293 cell were transfected with either S-WT-C or S-NQQ-C. 24 hr after transfections, cells were treated with of Pierce Protease Inhibitor cocktail (Thermo Scientific, Rockford, IL) using a 1:200 dilution or vehicle for 24 hr. Cells were then lysed and lysates were analyzed using PAGE NIR analysis.
SNAP MS/MS
HEK293 cells were transiently transfected with either WT or NQQ SNAP-α1D for 48 hrs. Cells were collected and lysed as above, with the addition of end-over-end rocking for 2 hrs. at 4 °C prior to centrifugation to remove insoluble fraction. Each condition was divided into 4 1.5 mL tubes for 16 hr. incubation with 20 μL of packed SNAP-Capture pull-down resin (New England Biolabs, Ipswich, MA) at 4 °C with end-over-end rocking. Beads were washed 3X with lysis buffer, transferred to new 1.5 mL tubes and washed 3X with 20 mM Tris-HCl pH 8.0 and 2 mM CaCl2. After the final wash the saturated beads were incubated with 20 mM Tris-HCl pH 8.0 supplemented with 5 mM DTT for 30 min at 60 °C with agitation, followed by incubation with 15 mM iodoacetamide for 10 min at RT. Denatured protein was then incubated with 1.5 μg of Trypsin (Sigma, St. Louis, MO) and 1.5 μg of Glu-C endoprotease (Thermo Fisher Scientific, Waltham, MA) for 16 hr at 37 °C with vigorous agitation. Peptides were collected and acidified using formic acid (FA) to a final concentration of 1% FA and desalted using StageTips80. Peptides were eluted from StageTips using elution buffer (40% acetonitrile, 1% FA), dried down and re-suspended in 8% acetonitrile, 1% FA. Samples were then loaded on a self-pulled 360 µm OD x 100 µm ID 15 cm column with a 7 µm tip packed with 3 µm Reprosil C18 resin (Dr. Maisch, Germany). Peptides were analyzed by nanoLC-MS in a 90 minutes linear gradient from 6% to 38% buffer B (buffer A: 0.1% acetic acid; buffer B: 0.1% acetic acid, 80% acetonitrile) on an EASY nLC 1200 (Thermo Scientific, Rockford, IL) and Orbitrap Fusion Lumos Tribrid Mass Spectrometer (FTMS; Thermo Scientific, Rockford, IL). Orbitrap FTMS spectra (R = 60 000 at 200 m/z; m/z 350–1600; 7e5 target; max 20 ms ion injection time) and Top Speed data-dependent acquisition with 3 second cycle time; HCD MS/MS spectra (R = 30 000 at 200 m/z; 31% CE; 5e4 target; max 100 ms injection time) were collected with an intensity filter set at 2.5e4 and dynamic exclusion for 45 second. Mass spectra were searched against the UniProt human reference proteome downloaded on February 20th, 2020 with the addition of SNAP-tag-ADRA1D sequence using MaxQuant v1.6.10.43. Detailed MaxQuant settings: samples were set to fraction 1 and 5 for NQQ mutant and WT, respectively, to allow within-group “match between run”; Trypsin/P and Glu-C were selected in digestion setting. Other settings were kept as default.
Label free dynamic mass redistribution (DMR) assay
DMR assays were performed in 384-well Corning Epic microsensor plates (Corning, Corning, NY) using previously described protocols22,28,50,81. Data were analyzed using GraphPad Prism (La Jolla, CA).
Cell surface assay
Cell surface assay was performed as described previously22,28,50.
Sucrose density centrifugation
Cells (~6.7 M cells/mL) were suspended in detergent-free lysis buffer (1 mM Tris-HCl pH 7.4, 140 mm NaCl, 10% sucrose) on ice for 20 min with vortexing every 5 min. 19 μL of lysate (~125,000 cells) was labeled with BG-782 at 37 °C. Reacted lysate was gently layered on top of a discontinuous sucrose gradient. Gradient consisted of equal volumes of 65%, 62.5%, 60%, 57.5%, 55%, 52.5%, 50%, and 15% sucrose dissolved in 1 mM Tris-HCl pH 7.4 and 140 mM NaCl. Samples were centrifuged at 134,633 × g at 4 °C for 65 min using a TH-660 rotor (Thermo Fisher Scientific, Waltham, MA). 400 μL fractions were collected and subjected to PAGE NIR analysis. Fluorescence of each fraction was quantified (NIR: λ = 800 nm) using Image Studio software (LI-COR, Lincoln, NE) and analyzed using area under the curve (AUC) analysis with a cut-off of 10% and minimum change in height of 5% from minimum to maximum in GraphPad Prism (La Jolla, CA).
Confocal microscopy
48 hours after transfection, cells were fixed with 4% paraformaldehyde/PBS solution for 10 min. at room temperature, washed with PBS, and permeabilized in 0.1% TritonX-100/PBS for 1 min. Cells were incubated with 1 μM of SNAP Surface Alexa Fluor 488 (New England BioLabs #S9129S, Ipswich, MA) and 1:1000 ER Staining Kit-Red Fluorescence-Cytopainter (Abcam #139482, Cambridge, MA) at 37 °C for 30 min. protected from light. Hoechst 33342 was used for nuclear staining. Cover slips were mounted using ProLong Glass antifade reagent (Thermo Fisher #P36982). Confocal fluorescence microscopy was performed using Leica SP8X laser scanning confocal microscope equipped with a 40x oil immersion objective (Leica Camera, Wetzlar, Germany). The detection pinhole was set to 1 Airy unit, light collection configuration was optimized according to the combination of chosen fluorochromes (Alexa Fluor 488, Texas Red, and Hoechst), and sequential channel acquisition was performed to minimize the risk of bleed-through. The intensity gain was adjusted for each channel before capture in order to avoid saturated pixels. 8 bit, 1024 × 1024 pixel images were collected as Z-stack acquisition. All microscopy was performed in collaboration with the W.M. Keck Microscopy center on the University of Washington School of Medicine campus.
Colocalization analysis
The Alexa Fluor 488 (SNAP) and Texas Red (ER) channels were analyzed for colocalization using Coloc2 plugin for Fiji82. Pearson’s coefficients for a cell were averaged over each slice in a Z-stack. Data were analyzed using GraphPad Prism (La Jolla, CA).
References
Overington, J. P., Al-Lazikani, B. & Hopkins, A. L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Docherty, J. R. Subtypes of functional α1-adrenoceptor. Cell. Mol. Life Sci. 67, 405–417 (2010).
Tanoue, A. et al. The α1D-adrenergic receptor directly regulates atrial blood pressure via vasoconstriction. J. Clin. Invest. 109, 765–775 (2002).
Lyssand, J. S. et al. Blood pressure is regulated by an α1D-adrenergic receptor/dystrophin signalosome. J. Biol. Chem. 283, 18792–18800 (2008).
Hampel, C. et al. Modulation of bladder alpha1-adrenergic receptor subtype expression by bladder outlet obstruction. J. Urol. 167, 1513–1521 (2002).
Bouchelouche, K. et al. Increased contractile response to phenylephrine in detrusor of patients with bladder outlet obstruction: effect of alpha1A and alpha1D-adrenergic receptor antagonist tamsulosin. J. Urol. 173, 657–661 (2005).
Walden, P. D., Gerardi, C. & Lepor, H. Localization and expression of the alpha1A-1, alpha1B and alpha1D-adrenoceptors in hyperplastic and non-hyperplastic human prostate. J. Urol. 161, 635–640 (1991).
Kojima, Y. et al. Up-regulation of α1a and α1d-adrenoceptors in the prostate by administration of subtype selective α1-adrenceptor antagonist tamsulosin in patients with benign prostatic hyperplasia. J. Urol. 186, 1530–1536 (2011).
Olson, V. G. et al. The role of norepinephrine in differential response to stress in an animal model of posttraumatic stress disorder. Biol. Psychiatry 70, 441–448 (2011).
Perez, D. M. & Doze, V. A. Cardiac and neuroprotection regulated by α1-adrenergic receptor subtypes. J. Recept Signal Transduct Res 31, 98–110 (2011).
Raskind, M. A. et al. A trial of prazosin for combat trauma PTSD with nightmares in active-duty soldiers returned from Iraq and Afghanistan. Am. J. Psychiatry 170, 1003–1010 (2013).
Cotecchia, S., Del Vescovo, C. D., Colella, M., Caso, S. & Diviani, D. The alpha1-adrenergic receptors in cardiac hypertrophy: signaling mechanisms and functional implications. Cell Signal. 27, 1984–1993 (2015).
Fusco, F. et al. α1-Blockers improve benign prostatic obstruction in men with lower urinary tract symptoms: a systematic review and meta-analysis of urodynamic studies. Eur. Urol. 69, 1091–1101 (2016).
Hendrickson, R. C. & Raskind, M. A. Noradrenergic dysregulation in the pathophysiology of PTSD. Exp. Neurol. 284, 181–195 (2016).
Akinaga, J., García-Sáinz, J. A. & Pupo, A. S. Updates in the function and regulation of α1-adrenoceptors. Br. J. Pharmacol. 176, 2343–2357 (2019).
Garcia-Cazarin, M. L. et al. The alpha1D-adrenergic receptor is expressed intracellularly and coupled to increases in intracellular calcium and reactive oxygen species in human aortic smooth muscle cells. J. Mol. Signal 3, 6, https://doi.org/10.1186/1750-2187-3-6 (2008).
Petrovska, R. et al. Addition of a signal peptide sequence to the α1D-adrenoceptor gene increases the density of receptors, as determined by [3H]-prazosin binding in the membranes. Br. J. Pharmacol. 144, 651–659 (2005).
Hague, C. et al. The N terminus of the human alpha1D-adrenergic receptor prevents cell surface expression. J. Pharmacol. Exp. Ther. 309, 388–397 (2004).
Piascik, M. T. et al. The specific contribution of the novel alpha-1D adrenoceptor to the contraction of the vascular smooth muscle. J. Pharmacol. Exp. Ther. 275, 1583–1589 (1995).
Fan, L. L. et al. Alpha(1D)-adrenergic receptor insensitivity is associated with alterations in its expression and distribution in cultured vascular myocytes. Acta. Pharmacol. Sin. 30, 1585–1593 (2009).
Chen, Z., Hague, C., Hall, R. A. & Minneman, K. P. Syntrophins regulate alpha1D-adrenergic receptors through a PDZ domain-mediated interaction. J. Biol. Chem 281, 12414–12420 (2006).
Janezic, E. M. et al. Scribble co-operatively binds multiple α1D-adrenergic receptor C-terminal PDZ ligands. Sci. Rep. 9, 14073, https://doi.org/10.1038/s41598-019-50671-6 (2019).
Lyssand, J. S. et al. Alpha-dystrobrevin-1 recruits alpha-catulin to the alpha1D-adrenergic receptor/dystrophin-associated protein complex signalosome. Proc. Natl. Acad. Sci. USA 107, 21854–21859 (2010).
Lyssand, J. S. et al. Syntrophin isoforms play specific functional roles in the α1D-adrenergic receptor/DAPC signalosome. Biochem. Biophys. Res. Commun. 412, 596–601 (2011).
Camp, N. D. et al. Individual protomers of a G protein-coupled receptor dimer integrate distinct functional modules. Cell Discov. 1, 15011, https://doi.org/10.1038/celldisc.2015.11 (2015).
Wallin, E. & von Heijne, G. Properties of N-terminal tail in G-protein coupled receptors: a statistical study. Protein Eng. 8, 693–698 (1995).
Pupo, A. S., Uberti, M. A. & Minneman, K. P. N-terminal truncation of human α1D-adrenoceptors increases expression of binding sites but not protein. Eur. J. Pharmacol. 462, 1–8 (2003).
Kountz, T. S. et al. Endogenous N-terminal domain cleavage modulates α1D-adrenergic receptor pharmacodynamics. J. Biol. Chem. 291, 18219–18221 (2016).
Li, C. et al. The GTPase Rab43 controls the anterograde ER-Golgi trafficking and sorting of GPCRs. Cell Rep. 21, 1089–1101 (2017).
Wu, G. Regulation of post-Golgi traffic of G protein-coupled receptors. Subcell. Biochem. 63, 83–95 (2012).
Wei, Z. et al. Specific TBC domain-containing proteins control the ER-Golgi plasma membrane trafficking of GPCRs. Cell Rep. 28, 554–566 (2019).
Balasubramanian, S., Teissére, J. A., Raju, D. V. & Hall, R. A. Hetero-oligomerization between GABAA and GABAB receptors regulates GABAB receptor trafficking. J. Biol. Chem. 279, 18840–18850 (2004).
Uberti, M. A., Hague, C., Oller, H., Minneman, K. P. & Hall, R. A. Heterodimerization with beta2-adrenergic receptors promotes surface expression and functional activity of alpha1D-adrenergic receptors. J. Pharmacol. Exp. Ther. 313, 16–23 (2005).
Nordström, R. & Andersson, H. Amino-terminal processing of the human cannabinoid receptor 1. J. Recept. Signal Transduct. Res. 26, 259–267 (2006).
Dunham, J. H., Meyer, R. C., Garcia, E. L. & Hall, R. A. GPR37 surface expression enhancement via N-terminal truncation or protein-protein interactions. Biochem. 48, 10286–10297 (2009).
Mattila, S. O., Tuusa, J. T. & Petäjä-Repo, U. E. The Parkinson’s-disease-associated receptor GPR37 undergoes metalloproteinase-mediated N-terminal cleavage and ectodomain shedding. J. Cell Sci. 129, 1366–1377 (2016).
Hakalahti, A. E. et al. Human beta1-adrenergic receptor is subject to constitutive and regulated N-terminal cleavage. J. Biol. Chem. 285, 28850–28861 (2010).
Chitwood, P. J., Juszkiewicz, S., Guna, A., Shao, S. & Hegde, R. S. EMC is required to initiate accurate membrane protein topogenesis. Cell 175, 1507–1519 (2018).
Andersson, H., D’Antona, A. M., Kendall, D. A., Von Heijne, G. & Chin, C. N. Membrane assembly of the cannabinoid receptor 1: impact of a long N-terminal tail. Mol. Pharmacol. 64, 570–577 (2003).
Petäjä-Repo, U. E., Hogue, M., Laperrière, A., Walker, P. & Bouvier, M. Export from the endoplasmic reticulum represents the limiting step in the maturation and cell surface expression of the human δ opioid receptor. J. Biol. Chem. 278, 13727–13736 (2000).
Van Craenenbroeck, K. et al. Folding efficiency is rate-limiting in dopamine D4 receptor biogenesis. J. Biol. Chem. 280, 19350–19357 (2005).
Sawutz, D. G., Lanier, S. M., Warren, C. D. & Graham, R. M. Glycosylation of the mammalian α1-adrenergic receptor by complex type N-linked oligosaccharides. Mol. Pharmacol. 32, 565–571 (1987).
Jensen, B. C., Swigart, P. M. & Simpson, P. C. Ten commercial antibodies for alpha-1-adrenergic receptor subtypes are nonspecific. Naunyn Schmiedebergs Arch Pharmacol. 379, 409–412 (2009).
Lopez-Gimenez, J. F., Canals, M., Pediani, J. D. & Milligan, G. The α1b-adrenoceptor exists as a higher-order oligomer: Effective oligomerization is required for receptor maturation, surface delivery, and function. Mol. Pharmacol. 71, 1015–1029 (2007).
Björklöf, K. et al. Co- and posttranslational modification of the α1B-adrenergic receptor: Effects on receptor expression and function. Biochem. 41, 4281–4291 (2002).
Vicentic, A., Robeva, A., Rogge, G., Uberti, M. & Minneman, K. P. Biochemistry and pharmacology of epitope-tagged alpha(1)-adrenergic receptor subtypes. J. Pharmacol. Exp. Ther. 302, 58–65 (2002).
Perez, D. M., Piascik, M. T. & Graham, R. M. Solution-phase library screening for the identification of rare clones: isolation of an α1D-adrenergic receptor cDNA. Mol. Pharmacol. 40, 876–883 (1991).
Helenius, A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell. 5, 253–265 (1994).
Imperiali, B. & O’Connor, S. E. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 3, 643–649 (1999).
Camp, N. D. et al. Dynamic mass redistribution reveals diverging importance of PDZ-ligands for G protein-coupled receptor pharmacodynamics. Pharmacol. Res. 105, 13–21 (2016).
Kornfeld, K., Reitman, M. L. & Kornfeld, R. The carbohydrate-binding specificity of pea and lentil lectins: Fucose is an important determinant. J. Biol. Chem. 256, 6633–6640 (1981).
Debray, H. et al. Specificity of twelve lectins towards oligosaccharides and glycopeptides related to N-glycosylproteins. Eur. J. Biochem. 117, 41–55 (1981).
Guillemette, T. et al. Methods for investigating the UPR in filamentous fungi. Methods Enzymol. 490, 1–29 (2011).
Schultz, C. & Köhn, M. Simultaneous protein tagging in two colors. Chem. Biol. 15, 91–92 (2008).
Davis, D., Liu, X. & Segaloff, D. L. Identification of the sites of N-linked glycosylation on the follicle-stimulating hormone (FSH) receptor and assessment of their role in FSH receptor function. Mol. Endocrinol. 9, 159–170 (1995).
Cho, D. I. et al. The N-terminal region of the dopamine D2 receptor, a rhodopsin-like GPCR, regulates correct integration into the plasma membrane and endocytic routes. Br. J. Pharmacol. 166, 659–675 (2012).
Tansky, M. F., Pothoulakis, C. & Leeman, S. E. Functional consequences of alteration of N-linked glycosylation sites on the neurokinin 1 receptor. Proc. Natl. Acad. Sci. USA 104, 10691–10696 (2007).
Fukushima, Y. et al. Structural and functional analysis of the canine histamine H2 receptor by site-directed mutagenesis: N-glycosylation is not vital for its action. Biochem. J. 310, 553–558 (1995).
Kimura, T. et al. The role of N-terminal glycosylation in the human oxytocin receptor. Mol. Hum Reprod. 3, 957–963 (1997).
Hawtin, S. R., Davies, A. R., Matthews, G. & Wheatley, M. Identification of the glycosylation sites utilized on the V1A vasopressin receptor and assessment of their role in receptor signaling and expression. Biochem. J. 357, 73–81 (2001).
Lanctôt, P. M., Leclerc, P. C., Escher, E., Leduc, R. & Guillemette, G. Role of N-glycosylation in the expression and functional properties of human AT1 receptor. Biochem. 38, 8621–8627 (1999).
Gonzalez de Valdivia, E., Sandén, C., Kahn, R., Olde, B. & Leeb-Lundberg, L. M. F. Human G protein-coupled receptor 30 is N-glycosylated and N-terminal domain asparagine 44 is required for receptor structure and activity. Biosci. Rep. 39, BSR20182436, https://doi.org/10.1042/BSR20182436 (2019).
Webel, R., Menon, I., O’Tousa, J. E. & Colley, N. J. Role of asparagine-linked oligosaccharides in rhodopsin maturation and association with its molecular chaperone, NinaA. J. Biol. Chem. 275, 24752–24759 (2000).
Lackman, J. J., Markkanen, P. M. H., Hogue, M., Bouvier, M. & Petäjä-Repo, U. E. N-glycan-dependent and –independent quality control of human δ opioid receptor N-terminal variants. J. Biol. Chem. 289, 17830–17842 (2014).
Markkanen, P. M. & Petäjä-Repo, U. E. N-glycan-mediated quality control in the endoplasmic reticulum is required for the expression of correctly folded delta-opioid receptors at the cell surface. J. Biol. Chem. 283, 29086–29098 (2008).
Nakagawa, T. et al. N-glycan-dependent cell-surface expression of the P2Y2 receptor and N-glycan-independent distribution of lipid rafts. Biochem. Biophys. Res. Commun. 485, 427–431 (2007).
Monné, M., Hessa, T., Thissen, L. & von Heijne, G. Competition between neighboring topogenic signals during membrane protein insertion into the ER. FEBS J. 272, 28–36 (2005).
Monné, M., Gaffvelin, G., Nilsson, R. & von Heijne, G. N-tail translocation in a eukaryotic polytopic membrane protein. Eur. J. Biochem. 263, 264–269 (1999).
Nilsson, I. M., Witt, S., Kiefer, H., Mingarro, I. & von Heijne, G. Distant downstream sequence determinants can control N-tail translocation during protein insertion into the endoplasmic reticulum membrane. J. Biol. Chem. 275, 6207–6213 (2000).
Cole, N. Site-specific protein labeling with SNAP-tags. Curr. Protoc. Protein Sci. 73, 30.1.1–30.1.16 (2013).
Kopito, R. R. ER quality control: The cytoplasmic connection. Cell 88, 427–430 (1997).
Neal, S. et al. The Dfm1 Derlin is required for ERAD retrotranslocation of integral membrane proteins. Mol. Cell 69, 306–320 (2018).
Brodsky, J. L. Cleaning up: ER-associated degradation to the rescue. Cell 151, 1163–1167 (2012).
Huang, Y., Niwa, J., Sobue, G. & Breitwieser, G. E. Calcium-sensing receptor ubiquitination and degradation mediated by the E3 ubiquitin ligase dorfin. J. Biol. Chem. 281, 11610–11617 (2006).
Öjemalm, K., Halling, K., Nilsson, I. M. & von Heijne, G. Orientational preferences of neighboring helices can drive ER insertion of a marginally hydrophobic transmembrane helix. Mole. Cell 45, 529–540 (2012).
Chitwood, P. J. & Hegde, R. S. The role of the EMC during membrane protein biogenesis. Trends Cell. Biol. 29, 371–384 (2019).
Guna, A. & Hegde, R. S. Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr. Biol. 28, R498–R511 (2018).
Guna, A., Volkmar, N., Christianson, J. C. & Hegde, R. S. The ER membrane protein complex is a transmembrane domain insertase. Science 359, 470–473 (2018).
Petäjä-Repo, U. E. et al. Newly synthesized human δ opioid receptors retained in the endoplasmic reticulum are retrotranslocated to the cytosol, deglycosylated, ubiquitinated, and degraded by the proteasome. J. Biol. Chem. 276, 4416–4423 (2001).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 75, 663–670 (2003).
Harris, D. A. et al. Label-free dynamic mass redistribution reveals low-density, prosurvival α1B-adrenergic receptors in human SW480 colon carcinoma cells. J. Pharmacol. Exp. Ther. 361, 219–228 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
Funding for this research was provided by NIGMS T32GM007750 (E.M.J.), R01GM129090 (S.E.O), and R01GM100893 (C.H.). We would also like to thank the W.M. Keck Microscopy Center, which is partially funded by NIGMS S10OD016240. This work used an EASY-nLC1200 UHPLC and Thermo Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer purchased with funding from a National Institutes of Health SIG grant S10OD021502.
Author information
Authors and Affiliations
Contributions
E.M.J. and C.H. designed experiments and wrote the manuscript. E.M.J., C.H., H.T.L., and S.E.O. designed SNAP MS/MS experiment. E.M.J., S.M.L.L., R.G.W., M.C., E.N., H.T.L., and K.S.L. performed all experiments. All co-authors contributed to editing and reviewing of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Janezic, E.M., Lauer, S.ML., Williams, R.G. et al. N-glycosylation of α1D-adrenergic receptor N-terminal domain is required for correct trafficking, function, and biogenesis. Sci Rep 10, 7209 (2020). https://doi.org/10.1038/s41598-020-64102-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-64102-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.