Abstract
Seamounts, often rising hundreds of metres above surrounding seafloor, obstruct the flow of deep-ocean water. While the retention of deep-water by seamounts is predicted from ocean circulation models, its empirical validation has been hampered by large scale and slow rate of the interaction. To overcome these limitations we use the growth of planktonic bacteria to assess the retention time of deep-ocean water by a seamount. The selected Tropic Seamount in the North-Eastern Atlantic is representative for the majority of isolated seamounts, which do not affect the surface ocean waters. We prove deep-water is retained by the seamount by measuring 2.4× higher bacterial concentrations in the seamount-associated or ‘sheath’-water than in deep-ocean water unaffected by seamounts. Genomic analyses of flow-sorted, dominant sheath-water bacteria confirm their planktonic origin, whilst proteomic analyses of the sheath-water bacteria, isotopically labelled in situ, indicate their slow growth. According to our radiotracer experiments, it takes the sheath-water bacterioplankton 1.5 years to double their concentration. Therefore, the seamount should retain the deep-ocean water for 1.8 years for the deep-ocean bacterioplankton to grow to the 2.4× higher concentration in the sheath-water. We propose that turbulent mixing of the seamount sheath-water stimulates bacterioplankton growth by increasing cell encounter rate with ambient dissolved organic molecules.
Similar content being viewed by others
Introduction
The 1,000-year-long global thermohaline circulation1 connects the bulk deep water of the modern World Ocean, irrespective of barriers erected by continents, islands and thousands of seamounts2,3,4. While continents and islands shape the circulation, seamounts affect this deep-water flow by creating enclosed circulation cells5, thereby reducing exchange between the so-called ‘sheath-water’ retained by seamounts and the surrounding deep water.
This does not, however, mean that the sheath-water is stagnant. The interaction of seamounts with deep water currents and waves (internal and tidal) causes complex sheath-water dynamics6, specified by the unique geometry of individual seamounts5. The complexity arises from interactions of parallel, rapid, turbulent mixing at centimetre-scales on seamount slopes7,8 and much slower flowing circulations (including Taylor columns) at the seamount-scale9. The complex sheath-water dynamics shapes seamount habitats for resident benthos and plankton5,10,11, causes erosion, controls sedimentation and affects ferromanganese crust formation on seamount slopes12.
A number of seamounts peak close to the ocean surface and mix nutrient-rich deep water with the nutrient-poor surface water enhancing local phytoplankton growth13 and causing a surface seamount effect5,13,14; retention of the produced organic matter in the seamount proximity raises productivity and enriches diversity of the entire seamount-associated ecosystem9,10,15,16,17,18,19. The majority of seamounts do not cause the surface seamount effect because their summits are hundreds or even thousands of meters below the sea surface. Because water retention at seamount-scales is challenging to measure directly, or to assess indirectly through combining observations with high-resolution hydrodynamic modelling9,12,20, the existence of the sheath-water remains a theoretical concept awaiting empirical proof. Measuring the duration of sheath-water retention would help determine the magnitude of interactions between individual seamounts and deep water currents.
Here, we propose a biological approach to test the sheath-water concept and to assess the sheath-water retention time. The most suitable proxy organisms are ubiquitous bacterioplankton, because they grow exclusively on organic molecules dissolved in the deep-ocean water and can be enumerated with high accuracy and precision by flow cytometry21. To ensure that sheath-water bacterioplankton, rather than suspended benthic bacteria, were analysed we compared bacterial abundance in samples collected from more than 5 m above the bottom to those in water samples carefully collected by a remotely operated vehicle (ROV) ~1 m above the bottom. Furthermore we assessed the composition of bacterioplankton for potential “contamination” with re-suspended benthic bacteria using high-throughput sequencing of flow sorted cells22. In the absence of established methods for reliably determining growth of deep-ocean bacterioplankton, we combined two different approaches. First, a custom-built, deep-water incubator was employed to identify those proteins the sheath-water bacterioplankton cells synthesise in situ. Second, we adapted the bioassay approach using amino acid radiotracers at close to ambient concentrations23 for ship-board incubations of deep-water samples.
We combined these techniques in a case study of bacterioplankton growth in the sheath-water of a typical, standalone seamount – Tropic Seamount located in the North-Eastern tropical Atlantic Ocean24. Tropic Seamount was selected because it is sufficiently (~3,000 m) tall to retain deep-ocean water yet its summit is deep enough below the ocean surface (~1,000 m) to cause no pronounced surface seamount effect, which would unnecessary complicate the already challenging assessment.
Material and Methods
Studied areas
This study was conducted in the area of Tropic Seamount (23 °50′N, 20 °40′W) in the tropical North Atlantic on board the Royal Research Ship (RRS) James Cook (cruise number JC142) from 28th October to 6th December 2016 (Fig. 1). This standalone, compact (50 km wide), ferromanganese crust-covered guyot rises steeply from the 4,100 m deep abyssal plane to a 990 m flat summit plateau12. Water samples were collected with a sampling rosette of 20-litre Niskin bottles mounted on a conductivity-temperature-depth (CTD) profiler from the sea surface down to about 5 m above the seafloor (Stations 2–12) and within 1 meter above the seafloor by the ROV, called Isis (Fig. 1a, Supplementary Tables S1 and S2). The two peripheral, south-eastern and southern stations (Stations 1 and 13) served as the local reference of Tropic Seamount. The stations in the middle of the North and South Atlantic subtropical gyres (23 °8.4′N, 36 °21′W and 21 °6′S, 22 °23′W, respectively, Fig. 1b) served as external references. These stations were sampled during the Atlantic Meridional Transect on board the RRS Discovery (cruise number AMT17-D299) from 15th October to 28th November 2005.
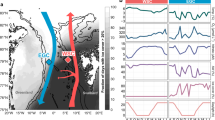
Locations of the sampling sites in the Atlantic Ocean. (a) The sampling sites at Tropic Seamount (St. 2–12) and in the seamount periphery (St. 1 and 13) (Supplementary Table S1): where water samples were collected using bottles mounted on the conductivity-temperature-depth (CTD) profiler at the seamount (green circles) and its periphery (grey-black diamonds); where the samples were incubated with 13C-lysine in situ using the deep-water incubator (DWI, the red circle); where water samples were collected using bottles mounted on the remotely operated vehicle (ROV, yellow stars). The cold colour gradient indicates the total relief and the warm colour gradient indicates the plateau topography on top of the seamount at higher resolution. (b) The sampling sites in the seamount-free middle North and South Atlantic subtropical gyres (NAG and SAG, respectively) relative to Tropic Seamount (Stations 1 and 13).
Bacterioplankton cell counting by flow cytometry and biomass estimation
Seawater samples were fixed with 2% (w/v) paraformaldehyde for 1 h at room temperature (RT). Bacterial cells were stained with SYBR Green I DNA dye25 and counted by flow cytometry (FACSort flow cytometer, Becton Dickinson) using the CellQuest software. Bacterioplankton concentrations were determined using 0.5 μm yellow-green micro-spheres (Fluoresbrite Microparticles, Polyscience) as an internal standard26. To convert bacterioplankton concentrations (N, cells l−1) into biomass (B) we used a mean cellular biomass (Bc) of 11.5 fg carbon or protein cell−1 27:
under the assumption of equal cell carbon and protein contents28.
Flow cytometric sorting of dominant bacterioplankton population
Deep-ocean bacterioplankton cells were concentrated from approximately 3 litre volumes directly from Niskin bottles using sterile 0.2 μm Sterivex filter units (Millipore, Watford). Concentrated samples were fixed with Lugol iodine solution and discoloured with thiosulfate solution29 before being stained with the DNA-specific Hoechst 33342 dye at a final concentration of 0.1 μg ml−1. Stained samples were analysed and target populations flow sorted with the custom-configured MoFlo XDP instrument (Beckman-Coulter, High Wycombe, UK) using the Summit 4.0 software as described previously22. Briefly, bacterial populations were visualised using a combination of the forward angle light scatter [FSC] of a UV diode laser (355 nm, 100 mW; JDSU, CY355-100, Thailand) and the dye fluorescence at 457 ± 25 nm. The flow cytometric plot was drawn using the FlowJo® v10 flow cytometric analysis software (Tree Star).
Molecular identification of flow sorted bacterial populations
For taxonomic identification, ~2 × 103 sorted cells from the main bacterial populations were added to 30 μl of Q5 High Fidelity Master Mix (New England BioLabs) complemented with primers and nuclease-free water (Ambion). The V3-V4 hyper-variable regions (490 bp) of 16S rRNA gene were PCR-amplified using S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 primers30 tailed with the Ion Torrent sequencing adapters. The forward primer also included the PGM barcode adapter (Ion Xpres Barcode Adapters 1–96 Kit, Thermo- Fisher Scientific). PCR products (~490 bp) were gel purified with NucleoSpin Gel and PCR Cleanup kit (Macherey-Nagel, Düren), pooled and used as template for emulsion PCR with the Ion Torrent One-Touch System (ThermoFisher Scientific) at a concentration of 26 pmol l−1. Sequencing of PCR products was done on an Ion Torrent PGM sequencer (ThermoFisher Scientific) using the Hi-Q sequencing chemistry. Sequences were quality trimmed for sequence length (>300 bp), controlled for homopolymers (<2%) and ambiguities (<2%) and separated by barcode using mothur31. Taxonomic affiliation was extracted by sequence comparison to the SSU rRNA SILVA database 119 using the SILVAngs pipeline32.
The in situ incubation experiment for bacterioplankton proteomic analyses
The custom-built deep-water in situ incubator (Fig. 2, depth rated to 6,000 m) was designed for in situ incubation experiments at depth to avoid decompression artefacts. For the bacterioplankton proteomic experiment the incubator was operated using the on deck control unit. The bottles were closed at 1,064 m and the tubing connectors were engaged to add 13C-Lysine tracer at 10 nmol l−1 final concentration. Pumps were switched on to mix the tracer with sampled water, closed in the paired bottles. After 20 hour incubation with the tracer in situ the incubator was recovered on board ship. Bacterioplankton were concentrated using 0.2 μm Sterivex filter units and prepared for sorting on board as described above. For targeted proteomic analysis, 2–5 × 106 cells were flow sorted directly onto 0.2 µm filters and stored at −80 °C until further processing ashore.
The deep-water in situ incubator design. The incubator is a tethered water sampling rosette of twelve 20 L Niskin bottles with lids arranged into six sets of paired bottles. A set (a photo to the left, a scheme to the right) consists of two bottles connected at the top by a tubing line with an in-line propeller pump. The two tubing lines at the bottom are deployed closed and disengaged from the bottles. The bottom tubing line can be simultaneously opened and attached to the paired bottles using the two mechanical closures. Before deployment each tubing line is filled with water solution contained fixative or tracer. The incubator can be either operated via a seabird deck unit or pre-programmed for autonomous operation at a target depth.
Proteomic analyses of bacterioplankton populations
Peptides were extracted from the sorted cells deposited on filters by repeated freeze-thawing to lyse the cells and subsequent tryptic protein digestions with sequencing grade modified Trypsin (Promega, Madison, Wisconsin). Cell debris was removed by centrifugations and peptides were concentrated and desalted on Millipore C18 ZipTip column (Merck Millipore, Darmstadt, Germany). Peptides were separated by EASY-nLCII (Thermo Fisher Scientific, Waltham, Massachussets) with self-packed analytical columns (100 μm × 20 cm) containing C18 RP material (Phenomenex, Aschaffenburg, Germany) and measured on an LTQ Orbitrap Elite (Thermo Fisher Scientific). Based on data dependent MS/MS mode, a Full Scan was done using the Orbitrap analyzer, followed by the analysis of the 10 most intense precursor ions using the LTQ analyzer. Singly charged ions were not taken into account for MS/MS analysis and lock mass option was enabled throughout all analyses. Data processing and protein identification based on this reference database was done with the Sorcerer 2 (SageN research) software. Taxonomic hits from 16S taq-sequencing of sorted populations were used to create a dataset specific protein reference database, containing 3049 entries, with publicly available genomic sequences from environmental samples and cultured representatives. Data analysis was done with the Scaffold 4 (version Scaffold_4.4.8, Proteome Software, Inc., Portland, Oregon) software. Identified proteins of the expressed proteome were manually classified into categories of orthologous groups based on general function categories. To convert leucine uptake rate into biomass production by bacterioplankton leucine content in measured peptides was calculated: on average leucine made up 6.86% of all amino acids by weight or 7.31 mol %.
Microbial amino acid uptake rates assayed using radiotracer dilutions
The ambient bioavailable concentrations of amino acids, leucine and lysine, and microbial uptake rates of these amino acids were estimated using the radiotracer dilution bioassay33,34. Subsamples of 1.6 ml from the surface mixed layer (25 m) were incubated with either 0.1, 0.2, 0.4, 0.6, 0.8 and 1.0 nmol L-[3,5-3H] leucine (103 Ci mmol−1, Hartmann Analytics) or L-[3,5-3H] lysine (32 Ci mmol−1, Hartmann Analytics) at RT for 15, 30, 45 and 60 min. To increase sensitivity of measurements in deep waters (>900 m) 1 litre samples were amended with 3H-leucine or 3H-lysine to final concentrations 0.01, 0.025, 0.05, 0.075, 0.1 nmol l−1 and incubated at in situ temperature (Supplementary Fig. S1). Subsamples of 250 ml were withdrawn after approximately 6, 14, 22 and 30 h. Subsamples were fixed with 2% PFA at RT for 1 h. The particulate material was collected onto 0.2 μm pore size polycarbonate filters (Nucleopore, Whatman) and rinsed twice with deionised water. Filters were placed in plastic 8 ml scintillation vials. Vials were filled with 5 ml of Gold Star scintillation cocktail (Meridian Biotechnologies, Tadworth). Radioactivity retained on the filters was radio-assayed using a liquid scintillation counter (Tri-Carb 3100, Perkin Elmer). Leucine and lysine uptake rates of bacterioplankton were determined using a linear regression model (Supplementary Fig. S1).
Computation of bacterioplankton production using the leucine uptake rates
To convert the leucine uptake rates into bacterioplankton production (P)35 under the same assumption of equal cell carbon and protein contents28 the leucine uptake rates (LUR, mol leucine l−1 d−1, multiplying the hourly rates by 24 h) were multiplied by the molecular weight of leucine (131.2 g) and divided by the fraction of leucine (7.31 mol%/100) in bacterial proteins:
We assume bacterioplankton acquire all leucine exogenously from the leucine-depleted (e.g. 0.05 nmol leucine l−1 concentrations, Supplementary Fig. S1) sheath-waters. The 7.31 mol% value derived from our mass spectrometric analysis of bacterioplankton proteins (see above) is in good agreement with the leucine 7.3 ± 1.91 mol% value determined by high performance liquid chromatography analysis35.
Assessment of the sheath-water retention time using the bacterioplankton growth
The sheath-water retention time equals the time required for bacterioplankton to grow from the concentrations in the seamount periphery (Nperif) or in the deep-ocean (Ndeep) to the concentrations in the seamount sheath-water (Nsheath). Therefore to assess the retention time we need to determine two values: (i) the above concentration difference (∆Nsp = Nsheath − Nperif, ∆Npd = Nperif − Ndeep and ∆Nsd = Nsheath − Ndeep) and (ii) the doubling or generation time (DT) of bacterioplankton in the sheath-water. Because bacterioplankton growth in the sheath-water is restricted by availability of organic nutrients (e.g. Supplementary Fig. S1) we compared the restricted (by both nutrients and mortality), linear growth model with the generally used, unrestricted, exponential model of bacterial growth28 to assess the doubling time extremes.
Linear doubling time (LDT = 1/k, where k is the specific growth rate) of bacterioplankton in days (or years) equals bacterial biomass B [g C or protein l−1] divided by bacterial production P [g C or protein l−1 d−1 or year−1]:
Exponential doubling time (EDT or g) of bacterioplankton in days (or years) equals linear doubling time LDT multiplied by natural logarithm of 2:
In brief the Eq. 4 derived from the following equation: ln(2B0) − ln(B0) = EDT/LDT where B0 is biomass at time zero and 2B0 equated to B0 biomass doubling. The latter equation can be simplified as ln(2B0/B0) = ln2 = EDT/LDT. See the chapter five of the Ingraham, et al.28 for details.
The seamount sheath-water retention time required for bacterioplankton to grow linearly (LRT) or exponentially (ERT) from e.g. Ndeep to Nsheath was calculated according to the following equations:
Results and Discussion
Concentration and phylogenetic characterisation of the sheath-water bacterioplankton
An assessment of bacterioplankton enrichment in the sheath-water of Tropic Seamount requires adequate reference sites (Fig. 1). Because the extent of the sheath-water was unknown, we used a nested approach of double referencing: (i) vertical profiles of bacterioplankton concentrations in the seamount-free areas of the North and South Atlantic gyres served as an external reference (Figs. 1b and 3a), (ii) vertical profiles of bacterioplankton concentrations at the seamount periphery (47 km south-east from the seamount summit centre to Station 1 and 24 km south to Station 13) served as a local reference (Figs. 1a and 3a,b). Bacterial concentrations within 1 m (ROV) and ≥5 m (CTD) above the seamount surface were compared to assess dispersal of re-suspended benthic bacteria in the sheath-water (Figs. 1a and 3b).
Bacterioplankton distributions in the water column at Tropic Seamount. (a) Comparison of bacterioplankton concentrations between the water column in Tropic Seamount periphery (St. 1 and 13) and the North and South Atlantic gyres (NAG and SAG, respectively). (b) Comparison of bacterioplankton concentrations between samples collected within 1 m of the seamount surface by a remote operated vehicle (ROV), at Tropic Seamount (St. 2–12) and in its periphery (St. 1). Dotted horizontal lines (a,b) indicate the five water layers defined. (c) Comparison of mean bacterioplankton concentrations in the five water layers between Tropic Seamount and its periphery. Error bars indicate single standard deviations of mean values. The results of the corresponding statistical analyses are presented in Supplementary Table S4. The corresponding sampling locations are shown in Fig. 1.
As would be expected, vertical distribution of bacterioplankton at 300–2,000 m depths in the seamount periphery was more similar to the distribution in the geographically closer North Atlantic subtropical gyre than in the geographically distant South Atlantic subtropical gyre (Fig. 3a). Below 2,500 m bacterioplankton concentrations in the two Atlantic gyres are statistically undistinguishable (Supplementary Table S3), whilst being 1.5× lower than the bacterioplankton concentrations in the seamount periphery (Fig. 3a, Supplementary Table S3). This indicates that the seamount affects bacterioplankton concentration even at the local reference sites: the seamount sheath-water extends beyond 20–40 km from the seamount summit (Fig. 1a). This bias of the local reference is taken into account in the latter analyses.
To assist comparisons, we divided the water column into five layers (Fig. 3), of which the two lowest are punctuated by Tropic Seamount. Bacterioplankton concentrations in the top three layers were similar above the seamount and in its periphery (Supplementary Table S4) confirming the laminar flow of water above the seamount and the minimal seamount effect on overlying waters. Indeed, a Taylor column (that can cause this effect) above Tropic Seamount has been found to be weak12. In the bottom two layers, bacterioplankton concentrations were significantly higher (1.5 and 1.8 times, respectively) at the seamount than in the seamount periphery (Fig. 3a,c), indicating the existence of the bacterioplankton-enriched sheath-water.
To test whether the observed higher bacterial concentrations at the seamount are caused by re-suspended benthic bacteria we compared bacterioplankton concentrations in samples collected ≥5 m (CTD) and ~1 m (ROV) above the slope (Fig. 3a). Because the two datasets were statistically indistinguishable (Supplementary Table S5) the uncertainty remained: the benthic bacteria could be evenly suspended farther than 5 m above the seabed or their presence in bacterioplankton could be insignificant. To resolve the uncertainty we decided to characterise bacterioplankton taxonomically. However, by collecting bacterioplankton cells on filters, we would also collect particles suspended from the seafloor, marine snow particles and other particulate materials, which could bias taxonomic composition of truly bacterioplankton community. To avoid this bias we assessed bacterial diversity of the four main flow sorted bacterioplankton populations (Fig. 4a). To test for homogeneity of the sheath-water bacterioplankton we compared the bacterial diversity within the four populations at three locations above the seamount (2,653, 3,030 and 3,215 m depth). We focused on Bacteria as molecularly and ecologically better characterised group (compared to Archaea or protists) in the deep-ocean. Furthermore, compared to eukaryotes basic metabolic rates and growth of prokaryotes could be directly assessed by the uptake rates of amino acids.
Taxonomic and proteomic characterisation of flow sorted bacterioplankton populations sampled from the seamount sheath-water. (a) Characteristic flow cytometric signatures of Hoechst–DNA stained bacterioplankton, sampled at St. 12. Ellipses on the density plot indicate the four main bacterioplankton populations (P1, P2, P3 and P4 with corresponding relative abundances shown as percentages), from which cells were flow sorted for taxonomic and proteomic analyses. The total cell concentration (TCC) is presented for reference. (b) Comparison of average (n = 2) relative taxonomic composition (% of read abundance) of the four main cytometric populations of bacterioplankton (BPL) sampled at the stations 9 (2,653 m), 10 (3,215 m) and 12 (3,030 m). P1 is dominated by the Rhodospirillales, Sphingomonadales and E01-9C-26 group. P2 is reproducibly dominated by the SAR11 and SAR406 groups. P3 is dominated by more diverse Alphaproteobacteria in addition to the SAR406 group. P4 is dominated by the SAR324 group. (c) Functional versus taxonomic classification of the peptides synthesised by cells from P1, focusing on the representative alpha- and gamma- proteobacteria (PB). The limited peptide data reveal the presence of bifunctional aconitate hydratase, 3-hydroxyacyl-CoA dehydrogenase, succinyl-CoA synthetase, isocitrate dehydrogenase, aspartate aminotransferase, aconitate hydratase and malate dehydrogenase enzymes and proteins responsible for core cellular functions such as protein expression, energy conservation, membrane transport or DNA folding.
Variation in composition and relative abundance of bacterial taxa in each of the four flow cytometric population was low between the three locations (Fig. 4b). The results of taxonomic analyses confirmed the dominance of planktonic bacteria and negligible content of potentially benthic bacteria (Fig. 4b, Supplementary Table S6) independent of sampling depth and location. Specifically, there was no indication that obligate benthic species36,37,38 (e.g. JTB255) were present among the sheath-water bacteria, whilst the dominant identified taxa (e.g. SAR11, Rhodospirillales, SAR324 and SAR406) are commonly found in deep-ocean bacterioplankton39,40 (Supplementary Table S6). Hence, the analysed sheath-water bacteria were indeed planktonic.
Proteins synthesized in situ by dominant cells of the sheath-water bacterioplankton
To ascertain whether the sheath-water bacterioplankton are actively growing, we analysed their protein synthesis (basic metabolism) in situ. We incubated isolated water samples in the deep-water incubator (Fig. 2) with 13C6-lysine label to identify the newly synthesized proteins. Because analyses of metaproteomic libraries remain challenging41, we used flow sorting that guaranteed us targeted proteomic analyses of the two most abundant populations of sheath-water bacterioplankton (the 1st and 2nd population, Fig. 4a) enabling direct metabolic and growth assessment of the bacterioplankton majority.
Low rates of 3H-lysine uptake by the sheath-water bacterioplankton at the ambient lysine concentration (see Supplementary Fig. S1) guided us to add 10 nmol l−1 of 13C6-lysine to overcome potential detection limitation. Even at this artificially high (>2,000 times higher than ambient) concentration we could only detect labelled peptides in cells flow sorted from the 1st population, and the labelled peptides had an isotopic mass shift of merely 5–30%. Approximately ten percent of detected peptides belonged to Gammaproteobacteria related to Alteromonas sp., Marinomonas sp. and Halomonas sp. (Fig. 4c). With Halomonas sp. being among the most abundant (Supplementary Table S6), all the three taxa were represented in the molecular data of the 1st population, validating our proteomic analyses (Fig. 4b,c). The newly synthesised proteins were involved in cell maintenance (ribosomal proteins, chaperones, proteins involved in energy conversion, cold-shock proteins) and active substrate uptake (membrane transporter proteins, flagellar proteins).
Active synthesis of proteins involved in transcription and translation, rather than in replication of nucleic acids (Fig. 4c), indicates preferential cellular maintenance rather than cell division. An isotopic mass shift, present in all detected peptides of cold shock proteins, indicates their constant regeneration. This is likely to reflect the mechanism of cell adaptation to cold (2–7 °C) deep-ocean conditions. Lysine addition predictably induced sheath-water bacteria to synthesise proteins involved in amino acid transport and utilization. Irrespective the artificial nature of this induction we could conclude that multiple taxa of sheath-water bacterioplankton are metabolically active and responsive to nutrient pulses (Fig. 4c), suggesting that availability of organic nutrients could restrict their growth.
Amino acid acquisition and growth of the sheath-water bacterioplankton
To reduce artificial stimulation of bacterial metabolic activity23 in samples incubated on board ship, we traced amino acid (leucine and lysine) uptake at concentrations close to ambient. We found that in the surface waters bacterioplankton uptake of leucine was 3 times faster than lysine uptake, whilst in the sheath-water this difference rose to >100 (Supplementary Fig. S1). Owing to its higher specific activity, leucine is a more sensitive tracer than lysine for assessing bacterial growth particularly at lower rates measured in the sheath-water. Consequently, we base our assessments of bacterial metabolic activity and growth on the results of experiments with the leucine tracer.
The amino acid uptake rate of a bacterial cell indicates its rate of protein synthesis and overall cellular metabolic activity. Therefore leucine clearance rates (the volume of water cleared of dissolved, bioavailable leucine by a cell in an hour) allows direct comparison of cell-specific metabolic activity of bacterioplankton living in the surface (Fig. 5a, Supplementary Table S7) and sheath- waters (Fig. 5b). The leucine clearance rates demonstrate that the metabolic activity of the sheath-water bacterioplankton cell is relatively high and, on average, 7.4% of the metabolic activity of the surface water bacterioplankton cell. However, because the sheath-water bacterioplankton standing stock is only 7.5% of the bacterioplankton standing stock in the surface waters (Fig. 3c, Supplementary Table S1), the standing stock-specific uptake of leucine in the sheath-water (Fig. 5d) is merely 0.074% of the uptake in the surface waters (Fig. 5c, Supplementary Table S7), underlining the challenge of measuring bacterioplankton growth in the deep-ocean waters.
Bacterioplankton growth in the surface and sheath- waters of Tropic Seamount. Comparison of cellular clearance rates (a,b), bacterioplankton uptake rates of leucine (Leu) (c,d) and bacterioplankton linear doubling time (LDT) (e,f) between the surface mixed layer (25 m) above Tropic Seamount (Stations 2, 10–12) and in periphery (Stations 1 and 13) of the seamount (a,c,e) and in the sheath-water (b,d,f).
According to the measurements of leucine uptake (42 ± 30 pmol l−1 h−1, mean ± SD, Fig. 5c), the surface waters above Tropic Seamount are about three times more productive than the surface waters of the oligotrophic Atlantic gyres (13 ± 3 pmol l−1 h−1) and of comparable productivity to the tropical surface waters (40 ± 14 pmol l−1 h−1)42,43 unaffected by upwelling. This independently supports our earlier conclusion that Tropic Seamount has virtually no effect on bacterioplankton in the overlying waters. The leucine uptake rates were converted into bacterial production and the linear doubling time according to the Eqs. 1–3 using corresponding concentrations (Fig. 3c, Supplementary Table S1) and leucine uptake rates of bacterioplankton (Fig. 5c,d, Supplementary Table 7).
The calculated mean doubling time (Fig. 5e, LDT = 6.4 ± 3.6 days, EDT = 4.4 ± 2.5 days) of bacterioplankton in the surface waters above Tropic Seamount is comparable to bacterioplankton doubling in the open ocean (11 ± 15 d)44, in the temperate North Atlantic (7.1–12.5 d)45 or in the oligotrophic ocean (10–20 d)46. Compared to the surface waters bacterioplankton in the sheath-water of Tropic Seamount double ninety times slower, once every 543 ± 348 days (Fig. 5f). The mean LDT = 1.5 ± 1.0 years or EDT = 1.0 ± 0.66 years is within the broad range of 0.1–30 years47,48 estimates for deep-ocean bacterioplankton. Comparatively high cellular metabolic activity of the sheath-water bacterioplankton (5a, b) in conjunction with their slow growth (Fig. 5f) is in agreement with the proteomics results of effective intracellular recycling of main proteins rather than active bacterial reproduction (Fig. 4c).
Based on the determined LDT of 1.5 years and concentration difference (1.5× and 1.8× in the bottom two layers, Fig. 3c; on average ∆Nsp = 1.6×), it would take bacterioplankton in the seamount periphery 0.7 = 1.5 × ln(1.6) years and 1.2 = 1.5 × 1.6/2 years to grow to the concentration of bacterioplankton in the sheath-water using the exponential (Eq. 6) or linear (Eq. 5) model, respectively. The exponential model gives an estimate for the fastest, unrestricted bacterioplankton growth, whilst the more realistic linear model accounts for such restrictions28 and therefore should better approximate bacterioplankton growth in the seamount sheath-water.
Bacterioplankton concentration in the seamount periphery is, however, still 1.5× higher than in the deep-ocean waters unaffected by seamounts, ∆Npd = 1.5× (Fig. 3a). Therefore, it would take the deep-ocean bacterioplankton between ERT = 1.3 = 1.5 × ln(1.6 × 1.5) years and LRT = 1.8 = 1.5 × (1.6 × 1.5)/2 years to grow to reach the concentration of bacterioplankton in the sheath-water. To enable bacterioplankton to grow from the deep-ocean to the seamount sheath-water concentrations Tropic Seamount should retain the deep-water in its sheath for 1.3–1.8 years. This provides the original, experimentally-derived estimate of the years-long interaction between a representative seamount and the deep-ocean water flow.
Even the maximal 1.8-year retention is a short time compared with the 1,000 year global thermohaline circulation1. However, considering that the North Atlantic deep-water components of northern origin spread throughout the western North Atlantic within 25–30 years49, 1.8-year seamount sheath-water retention time is significant. Higher concentrations of bacterioplankton cells in the sheath-water (Fig. 3b,c) could explain what controls growth of deep-ocean bacterioplankton. Because there is no extra source of dissolved organic molecules in the sheath-water of Tropic Seamount, bacterioplankton consume bio-available, dissolved molecules. The only difference between the sheath- and surrounding deep- water is the complex sheath-water dynamics that includes intensive turbulent mixing of the former7,8 compared to laminar-flowing deep-water in seamount-free areas. Therefore, it would be turbulent water mixing that ultimately controls bacterioplankton growth in the deep ocean: growth of the deep-ocean bacterioplankton is limited by the bioavailability of organic molecules in cell vicinity and turbulent mixing alleviates that limitation. Furthermore, the longer the sheath-water is retained by a seamount, the more organic molecules in the retained water are consumed by resident bacterioplankton. The consequences of such interactions and their impact on the seamount surface is worth exploring in follow-up studies.
Data availability
The Ion Torrent-generated libraries of bacterial 16S rRNA gene sequences were archived at the European Nucleotice Archive (ENA) (https://www.ebi.ac.uk/ena) of The European Bioinformatics Institute (EMBL-EBI) with the International Nucleotide Sequence Database Collaboration (INSDC) accession number PRJEB35653. Flow Cytometry datasets are archived at the FlowRepository database (https://flowrepository.org), ID: FR-FCM-Z2D9. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.ebi.ac.uk/pride) via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD016702.
References
Rahmstorf, S. Ocean circulation and climate during the past 120,000 years. Nature 419, 207–214 (2002).
Hillier, J. K. & Watts, A. B. Global distribution of seamounts from ship-track bathymetry data. Geophys. Res. Lett. 34, https://doi.org/10.1029/2007GL029874 (2007).
Yesson, C., Clark, M. R., Taylor, M. L. & Rogers, A. D. The global distribution of seamounts based on 30 arc seconds bathymetry data. Deep. Sea Res. Part. I: Oceanographic Res. Pap. 58, 442–453, https://doi.org/10.1016/j.dsr.2011.02.004 (2011).
Wessel, P., Sandwell, D. T. & Kim, S.-S. The global seamount census. Oceanography 23, 24–33 (2010).
White, M., Bashmachnikov, I., Arístegui, J. & Martins, A. In Seamounts: ecology, fisheries & conservation. Blackwell Fish and Aquatic Resources Series (eds Tony J Pitcher et al.) 65–84 (Blackwell Publishing (2007).
Boehlert, G. W. Current-topography interactions at mid-ocean seamounts and the impact on pelagic ecosystems. GeoJournal 16, 45–52, https://doi.org/10.1007/BF02626371 (1988).
van Haren, H., Groenewegen, R., Laan, M. & Koster, B. High sampling rate thermistor string observations at the slope of Great Meteor Seamount. Ocean Sci. 1, 17–28, https://doi.org/10.5194/os-1-17-2005 (2005).
van Haren, H. & Gostiaux, L. A deep-ocean Kelvin-Helmholtz billow train. Geophys. Res. Lett. 37, https://doi.org/10.1029/2009GL041890 (2010).
Beckmann, A. & Mohn, C. The upper ocean circulation at Great Meteor Seamount. Ocean Dyn. 52, 194–204, https://doi.org/10.1007/s10236-002-0018-3 (2002).
Mendonça, A. et al. Is there a seamount effect on microbial community structure and biomass? The case study of seine and sedlo seamounts (northeast atlantic). PLoS One 7, e29526, https://doi.org/10.1371/journal.pone.0029526 (2012).
Boehlert, G. W. & Genin, A. In Seamounts, islands and atolls Vol. 43 Geophysical Monograph Series (eds Barbara H. Keating, Patricia Fryer, Rodey Batiza, & George W. Boehlert) 319-334 (American Geophysical Union (1987).
Lusty, P. A., Hein, J. R. & Josso, P. Formation and occurrence of ferromanganese crusts: Earth’s storehouse for critical metals. Elements: An. Int. Mag. Mineralogy, Geochemistry, Petrology 14, 313–318 (2018).
Genin, A. & Dower, J. F. In Seamounts: ecology, fisheries & conservation. Blackwell Fish and Aquatic Resources Series (eds Tony J Pitcher et al.) 85–100 (Blackwell Publishing (2007).
Lemos, A. T., Ghisolfi, R. D. R. & Mazzini, P. L. F. Annual phytoplankton blooming using satellite-derived chlorophyll-a data around the Vitória-Trindade Chain, Southeastern Brazil. Deep. Sea Res. Part. I: Oceanographic Res. Pap. 136, 62–71, https://doi.org/10.1016/j.dsr.2018.04.005 (2018).
Dower, J., Freeland, H. & Juniper, K. A strong biological response to oceanic flow past Cobb Seamount. Deep. Sea Res. Part. A. Oceanographic Res. Pap. 39, 1139–1145, https://doi.org/10.1016/0198-0149(92)90061-W (1992).
Mourino, B. et al. Variability and seasonality of physical and biological fields at the Great Meteor Tablemount (subtropical NE Atlantic). Oceanol. Acta 24, 167–185, https://doi.org/10.1016/S0399-1784(00)01138-5 (2001).
Vilas, J. et al. Seamounts and organic matter—Is there an effect? The case of Sedlo and Seine Seamounts: Part 1. Distributions of dissolved and particulate organic matter. Deep. Sea Res. Part. II: Topical Stud. Oceanography 56, 2618–2630, https://doi.org/10.1016/j.dsr2.2008.12.023 (2009).
Comeau, L. A., Vézina, A. F., Bourgeois, M. & Juniper, S. K. Relationship between phytoplankton production and the physical structure of the water column near Cobb Seamount, northeast Pacific. Deep. Sea Res. Part. I: Oceanographic Res. Pap. 42, 993–1005, https://doi.org/10.1016/0967-0637(95)00050-G (1995).
Genin, A. & Boehlert, G. W. Dynamics of temperature and chlorophyll structures above a seamount: an oceanic experiment. J. Mar. Res. 43, 907–924, https://doi.org/10.1357/002224085788453868 (1985).
Mohn, C. & White, M. Seamounts in a restless ocean: Response of passive tracers to sub-tidal flow variability. Geophys. Res. Lett. 37, https://doi.org/10.1029/2010GL043871 (2010).
Zubkov, M. V., Tarran, G. A. & Burkill, P. H. Bacterioplankton of low and high DNA content in the suboxic waters of the Arabian Sea and the Gulf of Oman: abundance and amino acid uptake. Aquat. Microb. Ecol. 43, 23–32 (2006).
Kamennaya, N. A., Kennaway, G., Fuchs, B. M. & Zubkov, M. V. “Pomacytosis”—Semi-extracellular phagocytosis of cyanobacteria by the smallest marine algae. PLoS Biol. 16, e2003502, https://doi.org/10.1371/journal.pbio.2003502 (2018).
Hill, P. G., Warwick, P. E. & Zubkov, M. V. Low microbial respiration of leucine at ambient oceanic concentration in the mixed layer of the central Atlantic Ocean. Limnology Oceanography 58, 1597–1604 (2013).
Yeo, I. et al. Assessment of the mineral resource potential of Atlantic Ferromanganese Crusts based on their growth history, microstructure, and texture. Minerals 8, 327, https://doi.org/10.3390/min8080327 (2018).
Marie, D., Partensky, F., Jacquet, S. & Vaulot, D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR. Green. I. Appl. Env. Microb. 63, 186–193 (1997).
Zubkov, M. V. & Burkill, P. H. Syringe pumped high speed flow cytometry of oceanic phytoplankton. Cytometry Part. A 69A, 1010–1019, https://doi.org/10.1002/cyto.a.20332 (2006).
Zubkov, M. V., Sleigh, M. A., Burkill, P. H. & Leakey, R. J. G. Picoplankton community structure on the Atlantic Meridional Transect: a comparison between seasons. Prog. Oceanogr. 45, 369–386, https://doi.org/10.1016/S0079-6611(00)00008-2 (2000).
Ingraham, J. L., Maaløe, O. & Neidhardt, F. C. in Growth of the Bacterial Cell Ch. 5, 227–265 (Sinauer Associates Inc., 1983).
Pomroy, A. Direct counting of bacteria preserved with Lugol iodine solution. Appl. Env. Microb. 47, 1191–1192 (1984).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, https://doi.org/10.1093/nar/gks808 (2013).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Env. Microb. 75, 7537–7541 (2009).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596 (2012).
Zubkov, M. V., Tarran, G. A. & Fuchs, B. M. Depth related amino acid uptake by Prochlorococcus cyanobacteria in the Southern Atlantic tropical gyre. Fems Microbiol. Ecol. 50, 153–161, https://doi.org/10.1016/j.femsec.2004.06.009 (2004).
Wright, R. R. & Hobbie, J. E. Use of glucose and acetate by bacteria and algae in aquatic ecosystems. Ecology 47, 447–464, https://doi.org/10.2307/1932984 (1966).
Simon, M. & Azam, F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51, 201–213 (1989).
Ruff, S. E. et al. Indications for algae-degrading benthic microbial communities in deep-sea sediments along the Antarctic Polar Front. Deep. Sea Res. Part. II: Topical Stud. Oceanography 108, 6–16, https://doi.org/10.1016/j.dsr2.2014.05.011 (2014).
Learman, D. R. et al. Biogeochemical and microbial variation across 5500 km of Antarctic surface sediment implicates organic matter as a driver of benthic community structure. Front. Microbiol. 7, 284, https://doi.org/10.3389/fmicb.2016.00284 (2016).
Balmonte, J. P., Teske, A. & Arnosti, C. Structure and function of high Arctic pelagic, particle‐associated and benthic bacterial communities. Env. Microbiol. 20, 2941–2954, https://doi.org/10.1111/1462-2920.14304 (2018).
DeLong, E. F. et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496–503, https://doi.org/10.1126/science.1120250 (2006).
Martín-Cuadrado, A.-B. et al. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2, e914, https://doi.org/10.1371/journal.pone.0000914 (2007).
Saito, M. A. et al. Progress and challenges in ocean metaproteomics and proposed best practices for data sharing. J. Proteome Res. 18, 1461–1476, https://doi.org/10.1021/acs.jproteome.8b00761 (2019).
Hill, P. G., Mary, I., Purdie, D. A. & Zubkov, M. V. Similarity in microbial amino acid uptake in surface waters of the North and South Atlantic (sub-)tropical gyres. Prog. Oceanogr. 91, 437–446, https://doi.org/10.1016/j.pocean.2011.05.006 (2011).
Zubkov, M. V., Tarran, G. A., Mary, I. & Fuchs, B. M. Differential microbial uptake of dissolved amino acids and amino sugars in surface waters of the Atlantic Ocean. J. Plankton Res. 30, 211–220, https://doi.org/10.1093/plankt/fbm091 (2007).
Kirchman, D. L. Growth rates of microbes in the oceans. Annu. Rev. Mar. Sci. 8, 285–309, https://doi.org/10.1146/annurev-marine-122414-033938 (2016).
Zubkov, M. V., Sleigh, M. A. & Burkill, P. H. Heterotrophic bacterial turnover along the 20°W meridian between 59°N and 37°N in July 1996. Deep. Sea Res. Part. II: Topical Stud. Oceanography 48, 987–1001, https://doi.org/10.1016/S0967-0645(00)00105-3 (2001).
Ducklow, H. W., Steinberg, D. K. & Buesseler, K. O. Upper ocean carbon export and the biological pump. Oceanography 14, 50–58 (2001).
Nagata, T. et al. Emerging concepts on microbial processes in the bathypelagic ocean–ecology, biogeochemistry, and genomics. Deep. Sea Res. Part. II: Topical Stud. Oceanography 57, 1519–1536, https://doi.org/10.1016/j.dsr2.2010.02.019 (2010).
Reinthaler, T. et al. Prokaryotic respiration and production in the meso- and bathypelagic realm of the eastern and western North Atlantic basin. Limnol. Oceanogr. 51, 1262–1273, https://doi.org/10.4319/lo.2006.51.3.1262 (2006).
Smethie, W. M. Jr., Fine, R. A., Putzka, A. & Jones, E. P. Tracing the flow of North Atlantic Deep Water using chlorofluorocarbons. J. Geophys. Res. 105, 14297–14323, https://doi.org/10.1029/1999JC900274 (2000).
Acknowledgements
We thank the crew and officers of the RRS James Cook during the JC142 cruise as part of the MarineE-Tech project (NE/M011186/1) of the UK Natural Environment Research Council (NERC). We are obliged to Jeffrey Benson for technical support with the deep-water incubator, the crew of the ROV Isis for water sample collection and Tim Le Bas for the physical map of Tropic Seamount. We also thank Anke Meyerdierks and Greta Reintjes for help with the IonTorrent sequencing as well as Jürgen Bartel and Thomas Sura for help with processing the proteomic samples. This study was supported by the NERC Research Grant NE/M014363/1, the German Science Foundation (DFG, FOR2406), the Hanse Institute for Advanced Studies (Hanse Wissenschaftskolleg,Delmenhorst, Germany) and the Max Planck Society.
Author information
Authors and Affiliations
Contributions
M.V.Z., N.K., D.B., B.J.M., B.M.F. and R.I.A. conceived the experimental study; M.V.Z., A.E. and V.M. designed and built the deep-water in situ incubator; G.G., B.J.M. and M.V.Z. conducted experiments at sea; N.K., G.G., M.V.Z. and B.M.F. performed molecular analyses; N.K., G.G., M.V.Z., A.O. and D.B. performed proteomic analyses; M.V.Z., N.K., G.G. and B.M.F. analysed the data; M.V.Z., B.J.M., D.B. and R.I.A. secured funding; all authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Giljan, G., Kamennaya, N.A., Otto, A. et al. Bacterioplankton reveal years-long retention of Atlantic deep-ocean water by the Tropic Seamount. Sci Rep 10, 4715 (2020). https://doi.org/10.1038/s41598-020-61417-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61417-0
This article is cited by
-
Selfish bacteria are active throughout the water column of the ocean
ISME Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.