Abstract
Auxins are one of the most important and studied phytohormones in nature. Auxin signaling and perception take place in the cytosol, where the auxin is sensed. Then, in the nucleus, the auxin response factors (ARF) promote the expression of early-response genes. It is well known that not all plants respond to the same amount and type of auxins and that the response can be very different even among plants of the same species, as we present here. Here we investigate the behavior of ARF in response to various auxins in Agave angustifolia Haw., A. fourcroydes Lem. and A. tequilana Weber var. Azul. By screening the available database of A. tequilana genes, we have identified 32 ARF genes with high sequence identity in the conserved domains, grouped into three main clades. A phylogenetic tree was inferred from alignments of the 32 Agave ARF protein sequences and the evolutionary relationship with other species was analyzed. AteqARF 4, 15, 21, and 29 were selected as a representative diverse sample coming from each of the different subclades that comprise the two main clades of the inferred phylogenetic reconstruction. These ARFs showed differential species-specific expression patterns in the presence of indole-3-acetic acid (IAA) and 2,4-dichlorophenoxyacetic acid (2,4-D). Interestingly, A. angustifolia showed different phenotypes in the presence and absence of auxins. In the absence of auxin, A. angustifolia produces roots, while shoots are developed in the presence of IAA. However, in the presence of 2,4-D, the plant meristem converts into callus. According to our results, it is likely that AteqARF15 participates in this outcome.
Similar content being viewed by others
Introduction
Auxins are a very simple group of plant growth regulators with a very complex set of activities1. Auxins appear to be a set of molecules with limitless powers; they give rise to a vast range of specific developmental outputs2. The canonical auxin signaling allows auxin to respond in different ways depending on concentration and context3,4. The auxin binds to the TIR1/AFB family of auxin receptors, where it works as a cement between the Aux/IAA transcriptional repressor family and the TIR1/AFB receptor5. There are different possible Aux/IAA TIR1/AFB combinations that give rise to the wide range of effects produced by auxins2,3. Auxins are involved in numerous physiological and developmental processes, such as organogenesis, cellular elongation, apical dominance, gravitropism, root formation, flower morphogenesis, embryo development, and others6,7,8. The auxins indole-3-acetic acid (IAA), indole-3-butyric acid (IBA), phenyl acetic acid (PAA) and 4-chloro-indole-3-acetic acid (4-Cl-IAA)9,10,11 are all auxins previously characterized in plant tissues. However, there are also auxin analogues, such as naphthalene-1-acetic acid (NAA) and 2,4-dichlorophenoxyacetic acid (2,4-D)1. At different concentrations, auxins can be used to induce dedifferentiation and redifferentiation of tissues, and also to promote rooting in plants12,13,14,15,16. The auxinic herbicides, such as dicamba (3,6-dichloro-2-pyridinecarboxylis acid), dichloroprop [(±)-2-(2,4-dichlorophenoxy) propionic acid], 2,4-D [(2,4-dichlorophenoxy) acetic acid], and several more are described as synthetic auxins or growth regulators with herbicidal action or herbicides with growth regulator activity11,17,18. It has been reported that the effect of 2,4-D is 300 times more potent than IAA on the growth of plants11. The major characteristic shared by auxins and auxinic herbicides is the fact that the positive charge of the indole nitrogen is separated by 5.5 Å from a negatively charged substituent (the carboxyl group) on a flat lipophilic group/ring system11.
The genes that encode the enzymes and proteins needed for synthesis, transport, and signaling of auxins have been identified mainly in maize, rice and Arabidopsis thaliana19,20,21,22. Some of the early genes transcribed due to auxin response are Auxin/IAA (Aux/IAA), Gretchen Hagen 3 (GH3) and SMALL AUXIN UP RNA (SAUR), which regulate the physiology of the plant by modulating the interaction of the auxin-responsive elements (AuxRes) in some genes23,24,25. The Aux/IAA proteins contain four conserved domains (I-IV). The role of domain I has been reported as a potential active repression domain in Arabidopsis26. Domain II is required for Aux/IAA degradation, which is essential for normal auxin signaling27,28. Domains III and IV work like a domain of protein-protein interaction allowing the formation of homo- and heterodimers among Aux/IAA and repressing the activity of the Auxin Response Factors (ARFs)26,29.
ARFs are transcription factors that bind to the AuxRes, regulating the transcription of downstream genes25,30. The protein structures of the 23 ARFs reported in Arabidopsis are divided into three main domains. One is an N-terminal B3-type DNA binding domain (DBD), which binds to the AuxRes, followed by a domain rich in glutamine-serine-leucine (QSL) or serine-proline-leucine/glycine (SPLG) that functions as an activation or repression domain, respectively. The third domain is the carboxy-terminal dimerization domain, which is homologue to the domains III and IV in the Aux/IAA proteins to form heterodimers31. Of the 23 ARFs, five (ARF5–8 and 19) have activation domains and, therefore, can activate the transcription of auxin response genes, while the rest of the ARFs (18) function as transcriptional repressors31,32,33.
ARFs have important functions in lateral root growth34,35, embryogenesis36,37, leaf expansion38,39 and fruit development40,41, and they have also been studied in response to different auxin concentrations42. In an Arabidopsis transcriptomic study, Paponov et al.42 found that in response to different concentrations of auxins, from very low (0.1 μM) to very high (10 μM), ARFs 4, 16 and 19 were up-regulated in response to auxin. Of these three, only ARF19 was sensitive to a very low auxin concentration. ARF19 is considered to be a transcriptional activator due to its QSL-rich characteristic domain31 and it has been also considered to be a tissue-specific amplifier of the auxin signal42. The study of different ARFs in plant tissue culture has been justified by the use of exogenous auxins in order to develop either organogenesis or embryogenesis in important plants37,43,44,45,46. However, ARFs in non-model plants or from different species from the same genus have been poorly investigated.
Agaves are of great importance for their role as a source of raw material in the fiber industry, such as A. fourcroydes (pentaploid). In the liquor industry, e.g., the production of tequila47, A. tequilana Weber (Var. Azul, diploid) has been used with great success, and additionally is designated as a plant that originated in Mexico (NOM-V-1978). A. angustifolia Haw. (Var. Bacanora, pentaploid) has been used for mezcal production48. Although the genome of Agaves has not been sequenced due to its large size and complexity49,50, there are reports describing transcriptome analysis in A. tequilana49,51, A. deserti51 and A. sisalana52. The aim of this work is to determine how different auxins affect the growth and phenotype of different Agave species (A. angustifolia, A. fourcroydes and A. tequilana) and to elucidate the effects on short- and long-term cultivation of IAA and 2,4-D targeted toward two ARF with domains related to repression, AteqARF4 and 21, and two ARF related to the activation of expression, AteqARF15 and 29.
Results
Identification of ARF in Agave spp. revealed by gene structure and protein analysis
Identification of the ARFs present in Agave was done from an Agave transcriptome database51 by homology to known ARFs. Using as probes the 23 well-characterized members of the A. thaliana ARF family, we found 32 putative Agave ARF transcripts (Table 1). These sequences were aligned to find the conserved domains (Fig. 1). Three characteristic ARF domains were found: the DNA binding domain (DBD; B3), the auxin response domain, and the AUX-IAA domain. According to Zhang et al.53, sequences that do not have the DBD domain are considered to be pseudogenes and cannot be functional. Therefore, we scanned the proteins encoded by the transcripts to annotate the presence of conserved domains and discarded those that did not contain a DBD domain. Of the 32 putative ARF sequences, only 23 have the complete DBD domain, three have an incomplete DBD sequence and six do not have a DBD domain. ARFs with incomplete or missing domains could be pseudogenes or be due to technical problems of the transcriptome assembly.
Graphical representation of the Multiple Sequence Alignment (MSA) of the 32 putative ARF proteins from A. tequilana. Yellow boxes: Conserved domains predicted by Interproscan against Pfam signatures database; Gray to black boxes: blocks of conserved amino acids (darker = more conserved); thin horizontal lines: alignment gaps. The top histogram indicates percentage of identity at each position.
Phylogenetic analysis reveals ARF homology with other plants
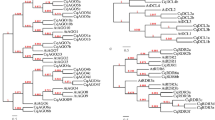
A phylogenetic analysis divided 30 of 32 transcripts into three major clades (Fig. 2). AteqARF18 and AteqARF13 were not used in the analysis due to their truncated structure (Fig. 1). The phylogenetic analysis of the ARF of Agave was performed by comparing these to the ARF from seven others already reported from different plant species, including 15 ARF of Cucumis sativus54, 19 in Vitis vinifera55, 20 in Solanum lycopersicum56, 22 in A. thaliana57, 20 in Oryza sativa58, and 31 in Zea mays59 (Fig. 3). Interestingly, 13 of the Agave ARF genes were grouped with the ARF of V. vinifera (Vv). Specifically, AteqARF12 and AteqARF21 were mainly grouped with VvARF15; AteqARF4 and AteqARF25 were grouped in the same clade with VvARF13; AteqARF24 and AteqARF28 were grouped with VvARF6; AteqARF22 and AteqARF30 were grouped with VvARF1; AteqARF6 was grouped with VvARF9; AteqARF29 was grouped with VvARF3; and AteqARF5, AteqARF16, and AteqARF17 were grouped with VvARF18 (Fig. 3).
Phylogenetic analysis of the ARF of Agave tequilana. Phylogeny was calculated from a MSA of the 30 putative A. tequilana ARFs (two highly truncated ARFs were removed). The evolutionary history was inferred using the Minimum Evolution method81. The optimal tree with the sum of branch length = 11.98949640 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches82. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method80 and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1.219). The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm83 at a search level of 2. The Neighbor-joining algorithm84 was used to generate the initial tree. The analysis involved 30 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 74 positions in the final dataset. Evolutionary analyses were conducted in MEGA785. ARFs selected for gene expression analyses are marked by solid black circles.
Phylogenetic relationship of the ARF of Agave to other species. Phylogeny was calculated from an MSA of the 30 putative A. tequilana ARFs (marked by solid blue circles) plus 127 ARFs from seven previously reported plant species (15 ARF of Cucumis sativus54, 19 of Vitis vinifera55, 20 of Solanum lycopersicum56, 22 of A. thaliana57, 20 in Oryza sativa58, and 31 in Zea mays59). The evolutionary history was inferred using the Minimum Evolution method81. The optimal tree with the sum of branch length = 52.16578416 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method80 and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1.011). The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm83 at a search level of 2. The Neighbor-joining algorithm84 was used to generate the initial tree. The analysis involved 158 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1827 positions in the final dataset. Evolutionary analyses were conducted in MEGA785.
Auxins display phenotypic differences in Agave spp
In order to know how different auxins affect the growth and phenotype of different Agave species (A. angustifolia, A. fourcroydes and A. tequilana), all plants were exposed to the same conditions during days 3 and 21 (see Materials and Methods). These concentrations and time points were selected based on previous experiments done in our group60,61. All Agave plantlets at the beginning of the experiment (control, day 0), were selected to have the same height, on average 2.5 cm (Fig. 4A). After being in contact with 0.5 μM of IAA and 2,4-D for days 3 and 21, the weight, height and number of leaves were recorded (Fig. 4B). Due to its robust nature, plantlets of A. fourcroydes have more leaves than A. angustifolia or A. tequilana. After 3 days in contact with IAA and 2,4-D, Agave species started to show important differences in comparison with the control (without auxins). For instance, while A. fourcroydes started to develop roots in the absence of auxins, in the presence of either IAA or 2,4-D there was no root development, although these plants were thinner than the control (Fig. 4A). On the other hand, A. angustifolia showed substantial growth variation after 3 days without auxins. It was found that while some A. angustifolia plantlets did not grow, others grew taller, reaching more than 5 cm in height (Fig. 4B). In the presence of 2,4-D, A. angustifolia and A. fourcroydes plantlets grew better and had a greener phenotype than in the presence of IAA. The opposite result happened in A. tequilana, where the plants grew better in the presence of IAA than in 2,4-D. The most interesting results were observed at 21 days in the presence and absence of the auxins. For instance, in the absence of auxins, A. angustifolia developed roots, in the presence of IAA the plantlets developed shoots, and in the presence of 2,4-D, the plantlets developed callus (Fig. 4A). It is very likely that the formation of callus contributed to this condition, having more weight (0.586 g) than in the presence of IAA (0.479 g) or the control (0.351 g) (Fig. 4B). These phenotype characteristics did not appear in A. fourcroydes or A. tequilana, where roots only developed in the absence of auxins. Although the roots in the control of A. fourcroydes emerged at day 3, by day 21 A. tequilana presented the longest roots. It is worth noting that A. tequilana in the presence of 2,4-D developed plants with wide leaves.
Effect of IAA and 2,4-D on different Agave species. (A) Plantlets of A. angustifolia, A. fourcroydes and A. tequilana in the presence and absence (control) of auxins at 0, 3 and 21 days. The plantlets were treated with 0.5 μM of IAA and 2,4-D. (B) Phenotypic variation of weight, height and number of leaves in A. angustifolia, A. fourcroydes and A. tequilana in the presence and absence (control) of auxins at 0, 3 and 21 days.
Gene expression of ARF revealed species-specific sensibility to auxins
To determine the possible effects over time of IAA and 2,4-D on gene expression, we analyzed the expression pattern of two ARF with domains related to repression, AteqARF4 and 21, and two ARF related to activation of expression, AteqARF15 and 29, after 3 and 21 days in the absence and presence of auxins (Fig. 5). These ARFs were selected as a representative diverse sample coming from each of the different subclades that comprise the two main clades inferred from the phylogenetic reconstruction (Fig. 2).
Expression profiles of ARF in Agave angustifolia, A. fourcroydes and A. tequilana. (A) Expression of AteqARF4, 15, 21 and 29 as shown by RT-PCR analysis of total RNA samples isolated from plantlets treated with 0.5 μM of IAA and 2,4-D at days 0, 3 and 21. UBQ11 was used as a reference gene. (B) Densitometric analysis of the gene expression shown in A. Relative expression of AteqARF4, 15, 21 and 29 were normalized to the constitutive gene UBQ11.
Unexpectedly, A. fourcroydes did not present important expression changes in the absence or presence of IAA or 2,4-D. Interestingly, in the case of A. angustifolia and A. tequilana, the presence of IAA or 2,4-D provoked differential expression for different ARFs. For instance, AteqARF21 had very low expression in A. angustifolia without auxins at day 21, while in A. tequilana low expression was detected in the presence of both IAA and 2,4-D at day 3. AteqARF4 showed low expression only in the absence of auxins in A. tequilana at day 21. In the case of AteqARF29, the transcript levels were very low in 2,4-D at day 21 in A. angustifolia and in the presence of both IAA and 2,4-D in A. tequilana. On the other hand, AteqARF15 only showed low expression in the presence of 2,4-D in A. angustifolia at day 21 (Fig. 5). Therefore, ARF15 is affected by 2,4-D but only in A. angustifolia. This opens a new avenue to study auxin regulation in a species-specific manner that will help to develop new strategies to understand plant development.
Discussion
The family of the ARF genes is large and its members have functional redundancy, which creates additional challenges when trying to understand ARF function. Finet et al.62 reported a complete phylogenetic analysis covering 21 species, including A. thaliana, Carica papaya, Glycine max, Medicago truncatula, O. sativa, V. vinifera, Z. mays and others in order to find the similitude between and among the ARF of different species. In that analysis, it was found that all ARFs were grouped in three clades. We also found that the 23 ARFs in Agave can be grouped in these three major clades (Fig. 2). With a phylogenetic analysis, we found that AteqARF15 was in the same clade as ARF27 of Z. mays, ARF7 of Solanum lycopersicum, ARF7 and 19 of A. thaliana, ARF10 of V. vinifera and ARF3 of C. sativus (Fig. 3). This similitude seems to be the result of duplication events among the genes and a domain rearrangement due to alternative splicing63,64, which has modified the number of ARF genes in different species53,62. An expression analysis in silico of ZmARF27 showed that this gene in maize is mainly expressed in the seed, tassel and ear59, tissues affected by auxin concentration65,66,67. For instance, Chen et al.65 found that there is a positive correlation between IAA concentration and ear height. On the other hand, Erkoyuncu et al.67 reported that high concentrations of 2,4-D increase callus formation in maize embryos. We found that 2,4-D was associated with callus formation in A. angustifolia after 21 days, the day at which AteqARF15 was downregulated (Fig. 5). A similar conclusion was made in a study related to callus formation in Arabidopsis and rice, in which it was found that AtIAA14-ARF7/19 in Arabidopsis is not required for callus initiation but in rice a homolog gene is strictly required68.
The number of ARF genes in Agave (Table 1) is consistent with the number of ARF found in other monocotyledonous species62. Studies in Arabidopsis have revealed that ARFs show gene redundancy69. Therefore, we selected four representative ARF, two repressor-related and two activator-related, to investigate whether different species exposed to auxin could interfere with ARF gene expression (Fig. 5). Here, we show that plants from the same genus but different species not only can be susceptible to a different degree in the presence or absence of auxins but also depending on the auxin (IAA or 2,4-D). In all cases, the effect on the expression of the ARFs would be different (Fig. 5).
In vitro propagation of Agave has solved many of the problems that conventional Agave farming has faced, such as a long maturation period and lack of high-quality materials to work with and genetic improvement programs70,71,72. In vitro culture represents an excellent alternative for producing a massive number of plants with superior market qualities in a short period of time. However, plant growth regulators have been a challenge to work with in order to determine the best doses for plant growth and propagation, and auxin concentrations have been particularly challenging. The use of concentrations of 2,4-D between 0.05 and 0.5 μM have been related to phenotypic variation in commercial cultivars such as strawberry73, soybean74 and cotton75. In the case of Agave, the concentration of 0.5 μM of 2,4-D provoked the formation of callus in A. angustifolia while in A. tequilana and A. fourcroydes it promoted robustness of the leaves, a finding that indicates different auxin regulation functions in different species. In A. thaliana, it has been reported that ARF19 increases its expression as a function of the concentration – the greater the concentration, the higher the expression – and the time in contact with external IAA42.
Given that only A. angustifolia showed callus formation in response to the same auxin stimulus experienced by A. tequilana and A. fourcroydes, it is clear that a different mechanism of callus initiation must exist between species. It is important to answer how the mechanism for auxin response differs in one species versus another species and how ARFs can be regulated in order to achieve high-quality plants resistant to developing challenges.
Materials and Methods
Plant material and culture conditions
Three different in vitro-propagated Agave species (Agave angustifolia Haw., A. fourcroydes Lem., and A. tequilana Weber var. Azul) were used to perform all the analyses. All Agave plantlets were cultured in multiplication media supplemented with 2.22 μM BA and 0.1 μM 2,4-D76 under photoperiod (12/12 hr) conditions. After six weeks, plantlets 1.5–2 cm in height were selected in order to have a homogenous group of plants. Once plants of the same size from each species were selected, these were kept for eight weeks in Magenta culture boxes filled with 50 mL of Murashige and Skoog media with reduced nitrogen, solidified with 1.75 g/L of Gelrite and without growth regulators in order to avoid contact with exogenous growth regulators before the treatments. After a two-month period without auxins, each Agave species was cultured with either of two different types of auxins (IAA and 2,4-D) at 0.5 μM and without auxins (Control). In each condition for all Agave species the weight, height and number of leaves were evaluated at days 0, 3 and 21. These concentrations and time points were selected based on previous experiments done in our group60,61.
Bioinformatic and phylogenetic analysis
Putative ARF transcripts in the reported transcriptome of A. tequilana at NCBI’s database51 were identified by means of a homology search using the BLASTN algorithm and the 23 A. thaliana ARFs as queries. The theoretical coding sequences and proteins for the transcripts were predicted with FGENESH+77, using the closest A. thaliana ARF protein sequence as a homologous reference for each putative transcript. A multiple sequence alignment (MSA) of the putative 30 A. tequilana ARF proteins, plus the previously reported ARFs from seven other plant species [15 ARF of Cucumis sativus54, 19 of Vitis vinifera55, 20 of Solanum lycopersicum56, 22 of A. thaliana57, 20 in Oryza sativa58, and 31 in Zea mays59], was then obtained using the MAFFT algorithm78 implemented on the software Geneiuos® R7 (http://www.geneious.com) with option E-INS-I79. A phylogeny of the A. tequilana and A. thaliana ARF proteins was inferred by the Minimum Evolution method using the JTT matrix-based model80 with a gamma distribution (shape parameter α = 1.011), and 1,000 bootstrap repetitions as a test of confidence level. The best amino acid substitution model was found using the best-fit test implemented in MEGA7. There were 1,827 amino acid positions on the dataset. Ambiguous data were removed for each sequence pair. For structural analysis and determination of conserved domains, the aligned sequences were tested against the databases of Pfam (http://pfam.xfam.org/), Gene3D (http://gene3d.biochem.ucl.ac.uk/Gene3D/), Interpro (http://www.ebi.ac.uk/interpro/), SMART (http://smart.embl-heidelberg.de/), PANTHER (http://www.pantherdb.org/), and Superfamily (http://supfam.org/SUPERFAMILY/hmm.html), using the InterProScan plugin for Geneious® R7.
Highly truncated A. tequilana ARF proteins missing entire domains due to incomplete transcripts were removed from the original multiple alignment as well as all non-A. tequilana ARF proteins. A second phylogeny was inferred from the resulting dataset as above with the following changes: gamma distribution α = 1.219 and complete deletion of ambiguous or missing data (gaps). There was a total of 30 amino acid sequences with 74 positions in the final dataset.
Primer design
Primers for the different ARF genes of the three selected Agave species were designed based on the identified sequences in the A. tequilana transcriptome51. Primer sequences for RT-PCR are listed in Table S1.
RT-PCR analysis
Total RNA was extracted from 0.3 g of leaf tissue from each plant (A. angustifolia, A. fourcroydes, and A. tequilana) and every condition (control, IAA, and 2,4-D) at day 0, 3, and 21 using the BRL Trizol reagent (Invitrogen) and re-purified with the Qiagen RNeasy Mini Kit, following the manufacturer’s instructions. RT reactions were performed in a 20 µl volume containing 5 µg of total RNA and 200 units of the M-MLV Reverse Transcriptase (Invitrogen), following the manufacturer’s conditions. Platinum Taq polymerase (Invitrogen) was used for the PCR reaction, and conditions were as follow: 5 min at 95 °C, followed by 35 cycles of 95 °C for 40 s, 62 °C for 40 s, and 72 °C for 2 min, and a final cycle of 72 °C for 10 min. Ubiquitin (UBQ11) was used as a constitutive gene control. The primer sequences of the ARF genes are listed in Table S1. The PCR products were electrophoresed in a 1.5% agarose gel and the identity was confirmed through sequencing (Clemson University Genomic Institute). Each RT-PCR was conducted twice with three biological replicates.
The densitometric analyses for band intensities were performed using ImageQuant™ software. Intensities of ARFs in each Agave species were normalized versus the UBQ11. The fold induction of each condition was calculated relative to the T0 control sample.
Statistical analysis
All data were analyzed using the Origin 8 program. The analysis of variance (ANOVA) was used to calculate the statistical significance and means ± SE. Significant differences were determined using the Tukey’s test at P < 0.05.
References
Bennett, T. & Leyser, O. In Auxin and Its Role in Plant Development (eds. Eva Zažímalová, Jan Petrášek, & Eva Benková) Ch. 1, 3–19 (Springer Vienna, 2014).
Napier, R. M. In Auxin and Its Role in Plant Development (eds. Eva Zažímalová, Jan Petrášek, & Eva Benková) Ch. 6, 101–116 (Springer Vienna, 2014).
Calderón Villalobos, L. I. A. et al. A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485, https://doi.org/10.1038/nchembio.926 (2012).
Kieffer, M., Neve, J. & Kepinski, S. Defining auxin response contexts in plant development. Current Opinion in Plant Biology 13, 12–20, https://doi.org/10.1016/j.pbi.2009.10.006 (2010).
Tan, X. et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645, http://www.nature.com/nature/journal/v446/n7136/suppinfo/nature05731_S1.html (2007).
Dinesh, D. C., Villalobos, L. I. A. C. & Abel, S. Structural biology of nuclear auxin action. Trends in Plant Science 21, 302–316, https://doi.org/10.1016/j.tplants.2015.10.019 (2016).
Kasahara, H. Current aspects of auxin biosynthesis in plants. Bioscience, Biotechnology, and Biochemistry, 1–9, https://doi.org/10.1080/09168451.2015.1086259 (2015).
Aloni, R., Aloni, E., Langhans, M. & Ullrich, C. I. Role of auxin in regulating Arabidopsis flower development. Planta 223, 315–328, https://doi.org/10.1007/s00425-005-0088-9 (2006).
Bhatla, S. C. In Plant Physiology, Development and Metabolism Ch. 15, 569–601 (Springer Singapore, 2018).
Cook, S. D. An historical review of phenylacetic acid. Plant and Cell Physiology 60, 243–254 (2019).
Sterling, T. M. & Hal, J. Mechanism of action of natural auxins and the auxinic herbicides. Rev. Toxicol. 1, 111–142 (1997).
Casimiro, I. et al. Auxin transport promotes Arabidopsis lateral root initiation. The Plant Cell 13, 843–852, https://doi.org/10.1105/tpc.13.4.843 (2001).
Du, Y. & Scheres, B. Lateral root formation and the multiple roles of auxin. Journal of Experimental Botany 69, 155–167 (2017).
Gutierrez, L. et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. The Plant Cell 24, 2515–2527, https://doi.org/10.1105/tpc.112.099119 (2012).
Kumari, S. & Panigrahi, K. C. S. Light and auxin signaling cross-talk programme root development in plants. Journal of Biosciences 44, 26, https://doi.org/10.1007/s12038-018-9838-2 (2019).
Pacurar, D. I., Perrone, I. & Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiologia Plantarum 151, 83–96, https://doi.org/10.1111/ppl.12171 (2014).
Fedtke, C. Biochemistry and physiology of herbicide action. (Springer Science & Business Media, 2012).
Fletcher, W. W. & Kirkwood, R. C. Herbicides and plant growth regulators. (Granada Publishing Ltd., 1982).
Strader, L. C. & Zhao, Y. Auxin perception and downstream events. Current Opinion in Plant Biology 33, 8–14, https://doi.org/10.1016/j.pbi.2016.04.004 (2016).
Leyser, O. Auxin signaling. Plant Physiology 176, 465–479, https://doi.org/10.1104/pp.17.00765 (2018).
Casanova-Sáez, R. & Voß, U. Auxin metabolism controls developmental decisions in land plants. Trends in Plant Science 24, 741–754, https://doi.org/10.1016/j.tplants.2019.05.006 (2019).
Li, Z., Li, P. & Zhang, J. Expression analysis of PIN-formed auxin efflux transporter genes in maize. Plant Signaling & Behavior 14, 1632689, https://doi.org/10.1080/15592324.2019.1632689 (2019).
Abel, S. & Theologis, A. Early genes and auxin action. Plant Physiology 111, 9–17, https://doi.org/10.1104/pp.111.1.9 (1996).
Chen, X. et al. Three cis-regulatory motifs, AuxRE, MYCRS1 and MYCRS2, are required for modulating the auxin- and mycorrhiza-responsive expression of a tomato GH3 gene. Plant and Cell Physiology 58, 770–778, https://doi.org/10.1093/pcp/pcx013 (2017).
Roosjen, M., Paque, S. & Weijers, D. Auxin Response Factors — output control in auxin biology. J. Exp. Biol. 69, 179–188, https://doi.org/10.1101/129122 (2018).
Tiwari, S. B., Hagen, G. & Guilfoyle, T. J. Aux/IAA proteins contain a potent transcriptional repression domain. The Plant Cell 16, 533–543, https://doi.org/10.1105/tpc.017384 (2004).
Ramos, J. A., Zenser, N., Leyser, O. & Callis, J. Rapid degradation of Auxin/Indoleacetic Acid proteins requires conserved amino acids of domain II and is proteasome dependent. The Plant Cell 13, 2349–2360, https://doi.org/10.1105/tpc.010244 (2001).
Worley, C. K. et al. Degradation of Aux/IAA proteins is essential for normal auxin signalling. The Plant Journal 21, 553–562, https://doi.org/10.1046/j.1365-313x.2000.00703.x (2000).
Liscum, E. & Reed, J. W. In Auxin Molecular Biology (eds. Catherine Perrot-Rechenmann & Gretchen Hagen) 387–400 (Springer Netherlands, 2002).
Guilfoyle, T., Hagen, G., Ulmasov, T. & Murfett, J. How does auxin turn on genes? Plant Physiology 118, 341–347, https://doi.org/10.1104/pp.118.2.341 (1998).
Guilfoyle, T. J. & Hagen, G. Auxin response factors. Current Opinion in Plant Biology 10, 453–460, https://doi.org/10.1016/j.pbi.2007.08.014 (2007).
Tiwari, S. B., Hagen, G. & Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell 15, 533–543, https://doi.org/10.1105/tpc.008417 (2003).
Ulmasov, T., Hagen, G. & Guilfoyle, T. J. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences 96, 5844–5849, https://doi.org/10.1073/pnas.96.10.5844 (1999).
Marin, E. et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. The Plant Cell 22, 1104–1117, https://doi.org/10.1105/tpc.109.072553 (2010).
Pandey, S. K. et al. LBD18 uses a dual mode of a positive feedback loop to regulate ARF expression and transcriptional activity in Arabidopsis. The Plant Journal 95, 233–251, https://doi.org/10.1111/tpj.13945 (2018).
Weijers, D. et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Developmental Cell 10, 265–270, https://doi.org/10.1016/j.devcel.2005.12.001 (2006).
Wójcikowska, B. & Gaj, M. D. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Reports 36, 843–858, https://doi.org/10.1007/s00299-017-2114-3 (2017).
Wilmoth, J. C. et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. The Plant Journal 43, 118–130, https://doi.org/10.1111/j.1365-313X.2005.02432.x (2005).
Wu, L., Tian, Z. & Zhang, J. Functional dissection of Auxin Response Factors in regulating tomato leaf shape development. Frontiers in plant science 9, 957 (2018).
Kumar, R., Tyagi, A. K. & Sharma, A. K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Molecular Genetics and Genomics 285, 245–260, https://doi.org/10.1007/s00438-011-0602-7 (2011).
Liu, S. et al. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Scientific Reports 8, 2971, https://doi.org/10.1038/s41598-018-21315-y (2018).
Paponov, I. A. et al. Comprehensive transcriptome analysis of Auxin responses in Arabidopsis. Molecular Plant 1, 321–337, https://doi.org/10.1093/mp/ssm021 (2008).
Cheng, Z. J. et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiology 161, 240–251, https://doi.org/10.1104/pp.112.203166 (2013).
Yang, J. H., Han, S. J., Yoon, E. K. & Lee, W. S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Research 34, 1892–1899, https://doi.org/10.1093/nar/gkl118 (2012).
Xu, X. et al. High miR156 expression is required for auxin-induced adventitious root formation via MxSPL26 independent of PINs and ARFs in Malus xiaojinensis. Frontiers in Plant Science 8, 1059, https://doi.org/10.3389/fpls.2017.01059 (2017).
Su, Y. H., Liu, Y. B., Zhou, C., Li, X. M. & Zhang, X. S. The microRNA167 controls somatic embryogenesis in Arabidopsis through regulating its target genes ARF6 and ARF8. Plant Cell, Tissue and Organ Culture 124, 405–417, https://doi.org/10.1007/s11240-015-0903-3 (2016).
Torres-Morán, M. I. et al. Assessment of genetic fidelity among Agave tequilana plants propagated asexually via rhizomes versus in vitro culture. Plant Cell, Tissue and Organ Culture 103, 403–409, https://doi.org/10.1007/s11240-010-9777-6 (2010).
Bautista, J. A. & Ramírez Juarez, J. Agricultura y pluriactividad de los pequeños productores de agave en la región del mezcal, Oaxaca, México. Agricultura técnica en México 34, 443–451 (2008).
Simpson, J. et al. Genomic resources and transcriptome mining in Agave tequilana. GCB Bioenergy 3, 25–36 (2011).
Robert, M. L. et al. Wild and agronomically important Agave species (Asparagaceae) show proportional increases in chromosome number, genome size, and genetic markers with increasing ploidy. Bot. J. Linn. Soc. 158, 215–222 (2008).
Gross, S. et al. De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics 14, 563 (2013).
Wang, P. et al. Transcriptome of sisal leaf pretreated with Phytophthora nicotianae Breda. Chinese J. Tropical Crops 35, 576–582 (2014).
Zhang, T. et al. In Biomedical Engineering and Biotechnology (iCBEB), International Conference on. 86–89 (IEEE) (2012).
Liu, S. & Hu, L. Genome-wide analysis of the auxin response factor gene family in cucumber. Genet. Mol. Res 12, 4317–4331 (2013).
Wan, S. et al. Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Reports 33, 1365–1375, https://doi.org/10.1007/s00299-014-1622-7 (2014).
Wu, J. et al. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Reports 30, 2059, https://doi.org/10.1007/s00299-011-1113-z (2011).
Guilfoyle, T. J. & Hagen, G. Auxin Response Factors. Journal of Plant Growth Regulation 20, 281–291, https://doi.org/10.1007/s003440010026 (2001).
Wang, D. et al. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24, https://doi.org/10.1016/j.gene.2007.01.006 (2007).
Liu, Y. et al. Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regulation 63, 225–234, https://doi.org/10.1007/s10725-010-9519-0 (2011).
Duarte-Aké, F. Estudio epigenético de dos especies de Agave spp. en condiciones in vitro y de invernadero M.Sc. thesis, Centro de Investigación Científica de Yucatán (2013).
De la Torre-Espinosa, Z. Efecto de la concentración y tipo de auxina en el grado de metilación del ADN en Agave cultivado in vitro. M.Sc. thesis, Centro de Investigación Científica de Yucatán (2013).
Finet, C., Berne-Dedieu, A., Scutt, C. P. & Marlétaz, F. Evolution of the ARF gene family in land plants: old domains, new tricks. Molecular Biology and Evolution 30, 45–56, https://doi.org/10.1093/molbev/mss220 (2013).
Kopelman, N. M., Lancet, D. & Yanai, I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat. Genet. 37, 588–589, http://www.nature.com/ng/journal/v37/n6/suppinfo/ng1575_S1.html (2005).
Su, Z., Wang, J., Yu, J., Huang, X. & Gu, X. Evolution of alternative splicing after gene duplication. Genome Research 16, 182–189, https://doi.org/10.1101/gr.4197006 (2006).
Chen, Y. et al. Proteomic analysis reveals that auxin homeostasis influences the eighth internode length heterosis in maize (Zea mays). Scientific Reports 8, 7159, https://doi.org/10.1038/s41598-018-23874-6 (2018).
Li, Z. et al. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 16, 86–99, https://doi.org/10.1111/pbi.12751 (2018).
Erkoyuncu, M. T., Yorgancılar, M. & Atalay, E. Effect of some auxin types callus induction from mature embryos of maize genotypes. Journal of Molecular Biology and Biotechnology 1, 20–23 (2017).
Guo, F. et al. Callus initiation from root explants employs different strategies in rice and Arabidopsis. Plant and Cell Physiology 59, 1782–1789, https://doi.org/10.1093/pcp/pcy095 (2018).
Okushima, Y. et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell 17, 444–463, https://doi.org/10.1105/tpc.104.028316 (2005).
Robert, M. L., Herrera, J. L., Contreras, F. & Scorer, K. N. In vitro propagation of Agave fourcroydes Lem. (Henequen). Plant Cell, Tissue and Organ Culture 8, 37–48 (1987).
Robert, M. L., Herrera, J. L., Chan, J. L. & Contreras, F. In Biotechnology in Agriculture and Forestry (ed. J. P. Y. Bajaj) 306–329 (Springer- Verlag., 1992).
Robert, M. L., Herrera-Herrera, J. L., Herrera-Alamillo, M. A., Quijano, A. & Balám, U. Manual for the in vitro culture of Agaves. United Nations Industrial Development Organization Technical paper (2004).
Nehra, N. S., Kartha, K. K., Stushnoff, C. & Giles, K. L. The influence of plant growth regulator concentrations and callus age on somaclonal variation in callus culture regenerants of strawberry. Plant Cell, Tissue and Organ Culture 29, 257–268 (1992).
Gesteira, A. S., Otoni, W. C., Barros, E. G. & Moreira, M. A. RAPD-based detection of genomic instability in soybean plants derived from somatic embryogenesis. Plant Breeding 121, 269–271 (2002).
Jin, S. et al. Detection of somaclonal variation of cotton (Gossypium hirsutum) using cytogenetics, flow cytometry and molecular markers. Plant Cell Reports 27, 1303–1316 (2008).
De-la-Pena, C. et al. KNOX1 is expressed and epigenetically regulated during in vitro conditions in Agave spp. BMC Plant Biology 12, 203 (2012).
Solovyev, V. In Handbook of statistical genetics (eds. D. Balding, M. Bishop, & C. Cannings) Ch. 4, 97–159 (Wiley-Interscience, 2007).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797, https://doi.org/10.1093/nar/gkh340 (2004).
Katoh, K., Misawa, K., Kuma, K. I. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30, 3059–3066, https://doi.org/10.1093/nar/gkf436 (2002).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282, https://doi.org/10.1093/bioinformatics/8.3.275 (1992).
Rzhetsky, A. & Nei, M. A simple method for estimating and testing minimum-evolution trees. Molecular Biology and Evolution 9, 945–967 (1992).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Nei, M. & Kumar, S. Molecular evolution and phylogenetics. (Oxford university press, 2000).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425 (1987).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution 33, 1870–1874 (2016).
Acknowledgements
This study was supported by CONSEJO NACIONAL DE CIENCIA Y TECNOLOGÍA (CONACYT), scholarships 284323 to V.C.G., Grants CB2016-285898 to C.D.P. Grant from Cátedras Marcos Moshinsky 2017 to C.D.P.
Author information
Authors and Affiliations
Contributions
V.C.G. raised the in vitro plants for the experiments and performed the experiments. J.R.P. performed bioinformatic analysis. V.C.G., J.R.P. and C.D.P. contributed to the design and interpretation of the research and to the writing of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cancino-García, V.J., Ramírez-Prado, J.H. & De-la-Peña, C. Auxin perception in Agave is dependent on the species’ Auxin Response Factors. Sci Rep 10, 3860 (2020). https://doi.org/10.1038/s41598-020-60865-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60865-y
This article is cited by
-
Auxins, the hidden player in chloroplast development
Plant Cell Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.