Abstract
Co-expression of Yin Yang 1 (YY1) is required for the full function of the transcription factor, PLZF, which is essential for the development of natural killer T cell (NKT cell) effector functions. Discordant expression of YY1 and PLZF, therefore, might define NKT cell subsets with distinct effector functions. A subset of NKT cells was identified that expressed low levels of YY1. YY1lo NKT cells were found in all tissues, had a mature phenotype and, distinct from other NKT cells, expressed almost no ThPOK or Tbet. When activated, YY1lo NKT cells produced little IL-4 or IFN-γ. YY1lo NKT cells were found to constitutively transcribe IL-10 mRNA and, accordingly, produced IL-10 upon primary activation. Finally, we find that tumor infiltrating NKT cells are highly enriched for the YY1lo subset. Low YY1 expression, therefore, defines a previously unrecognized NKT cell subset that is committed to producing IL-10.
Similar content being viewed by others
Introduction
Invariant Natural Killer T cells (NKT cells) are a highly conserved population of αβ T cells associated with many types of immune responses1. Mouse NKT cells are defined by the expression of an invariant TCR α-chain (TCRVα14Jα18) that pairs with a limited repertoire of TCRVβ chains including TCRVβ8.2, TCRVβ7, and TCRVβ22. This TCR complex recognizes glycolipid antigens presented in the context of CD1d3. Primary activation of NKT cells, either by the recognition of lipid antigens or inflammatory cytokines leads to rapid and copious cytokine production4. The potent production of this multitude of cytokines plays a major role in the ability of NKT cells to execute such diverse effector functions.
Differential expression of key transcription factors has been used to define NKT cell subpopulations. NKT1 cells, for example, express T-box 21 (Tbet) and are skewed towards the production of Th1 cytokines including IFN-γ, NKT2 cells, which express higher levels of GATA binding protein-3 (GATA3), produce IL-4 and retinoic acid receptor-related orphan nuclear receptor gamma (RORγt) expressing NKT17 cells, produce the cytokine IL-175,6. Although these three subgroups are convenient for comparison of NKT cell responses and development, it has been shown that cytokine production by NKT cells is actually astonishingly diverse. In one particularly extensive study, nearly 20 different cytokines were detected post activation, including IFN-γ, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-13, IL-17, IL-21, GM-CSF, TNF, MIP-1α, RANTES, IL-12, and MCP-17. The variety in the expression of these different cytokines suggests that there might actually be a multitude of distinct NKT cell effector subsets. It is not clear, however, if distinct cytokine expression patterns are defined during development or are a consequence of differentiation following activation. Interestingly, many of the gene loci for these cytokines are constitutively in an “open” configuration in resting NKT cells, resulting in a continuous basal level of mRNA transcription8,9. The one clear commonality between all NKT cell effector subsets is the requirement for the expression of the BTB-ZF transcription factor, PLZF10,11. In the absence of PLZF, NKT cells develop, but fail to acquire nearly all of their unique defining characteristics, including the capacity to rapidly produce cytokines upon primary activation. Importantly, the hallmark constitutive transcription of cytokine genes is dependent upon PLZF12.
Among the many cytokines made by NKT cells, the ability to produce IL-10 is of particular interest. IL-10 is a potent immunomodulator that, for example, impacts the growth of tumors and controls inflammatory bowel and autoimmune diseases13. Although many leukocytes can make IL-10, NKT cell specific IL-10 production has been associated with multiple immune responses. For example, control of homeostasis in the visceral adipose tissue is dependent on IL-10 from NKT cells14, as are some aspects of cardiac remodeling15. Autoimmune diseases ranging from alopecia areata16 to insulin-dependent diabetes mellitus17 are associated with IL-10 producing NKT cells18,19, as is the growth of intestinal polyps that can lead to cancer20.
The origin of IL-10 producing NKT cells (sometimes referred to as NKT10), however, is not clear21,22. Unlike the NKT1, NKT2, and NKT17 subpopulations6, a definitive transcription factor expression pattern that specifically identifies NKT10 cells has not been identified. Furthermore, the study of IL-10 producing NKT cells, to this point, has largely relied upon first activating NKT cells, followed by reactivation of the cells to detect IL-1021,22. Studies such as these have actually shown that all of the defined NKT cell effector subgroups can be induced to express IL-1021. One study suggests that pre-existing “natural NKT10” cells are present in the mouse22. This study, however, reports that these cells can only be detected 16 hours after in vivo activation with the strong agonist ligand, αGalCer. Therefore, the consensus is, that similar to conventional T cells, IL-10 production by NKT cells is a consequence of activation induced differentiation.
In this study, we find that a population of NKT cells, primed to secrete IL-10 following primary activation, is marked by low expression of the transcription factor Yin Yang-1 (YY1). A polycomb group protein, YY1 is thought to modulate chromatin architecture both by mediating the formation of promoter-enhancer loops and recruiting HDAC and HAT proteins to specific genetic loci23,24. Multiple biological processes have been shown to be dependent upon YY1, including oncogenisis25, hematopoeisis26 and heart development27 as well as several facets of T cell biology. During T cell development, YY1 is required for suppression of p53 mediated apoptosis of immature thymocytes28. In mature CD4 T cells, YY1 cooperates with lineage defining transcription factors such as FoxP3 in Tregs29,30 and Gata3 in Th2 cells31 to coordinate each effector lineages specific gene expression program. YY1 has also been found to play an important role during the commitment of CD8 T cells to effector lineages as opposed to the memory lineage32.
In a recent publication, we found that YY1 is required for NKT cell effector functions. YY1 deficient NKT cells failed to produce the burst of cytokines characteristic of primary NKT cell activation, despite expressing wild type levels of PLZF33. Therefore, YY1 co-expression in NKT cells is required for the function of PLZF. The control of YY1 by PLZF may be direct, since the transcription factors were shown to be in a complex. To further explore how PLZF and YY1 might cooperate to regulate effector functions in NKT cells, we examined expression of the two proteins. These studies identified a small population of NKT cells that expressed low levels of YY1 as compared to PLZF. In the thymus, YY1lo NKT cells mostly had a mature, Stage 3, phenotype. Also, consistent with being mature cells, YY1lo NKT cells were found in all examined tissues, including the spleen, liver and lungs. Despite the mature phenotype, YY1lo cells expressed very low levels of both Tbet and ThPOK. Most interestingly, when activated, YY1lo NKT cells produced little IL-4 and IFN-γ, but rather, produced IL-10. Finally, we find that YY1lo NKT cells selectively accumulate in tumors. Thus, our data identify a subset of invariant NKT cells that is dedicated for producing IL-10.
Results
A population of NKT cells in the thymus expresses low levels of YY1
Nearly all of the most potent effector functions of NKT cells, including the exceptionally rapid response to activation, resulting in the production of a massive burst of cytokines, require expression of the transcription factor PLZF11,12. We recently showed, however, that many of the functions of PLZF are dependent upon expression of YY1, a transcription factor itself, that we find is in association with PLZF33. Of particular interest, YY1 deficient NKT cells, we found, express wild type levels of PLZF, but do not produce cytokines following primary activation33.
NKT cell subpopulations with distinct cytokine production profiles express different levels of PLZF6. Since YY1 is required for PLZF function, we reasoned that noncoordinate expression of the two transcription factors might correlate with distinct functionality. Expression levels of the two transcription factors were quantified in NKT cells from the thymuses from 8 week old C57BL/6J mice. By comparison of PLZF to YY1 expression levels, three subsets of NKT cells were identified (Fig. 1a). YY1 expression was highest in NKT cells that also had the highest level of PLZF (Fig. 1a–c). These YY1hiPLZFhi NKT cells accounted for roughly one third of the total NKT cell population (Fig. 1d). The largest subpopulation, making up more than half of the NKT cells (Fig. 1d), expressed somewhat lower levels of YY1 and, also, lower levels of PLZF. Therefore, in the bulk of NKT cells, levels of PLZF and YY1 mimicked each other. However, the smallest of these subpopulations expressed much lower levels of YY1, but only slightly lower levels of PLZF (Fig. 1a–c). The frequency and cell numbers of the three populations, including the small, YY1loPLZFlo population (ranging from 3–5% of total NKT cells) were consistent from mouse to mouse (Fig. 1d). Differential YY1 expression was also observed in other thymic T cell populations (Supp. Figure 1). Therefore, although YY1 is ubiquitously expressed in all NKT cells, the level of expression varied among these newly defined subsets. Furthermore, in one population, the expression of the two factors was found to be discordant.
A population of NKT cells in the thymus expresses low levels of several transcription factors known to regulate NKT cell effector functions. Thymuses were collected from C57BL/6J (WT) mice and analyzed by FACS with the indicated antibodies. (a–d) YY1 and PLZF expression levels were determined in WT NKT cells (MHCII−, CD3+, CD1d tet+, CD24−). (a) NKT cells segregated into three populations on the basis of YY1 and PLZF expression: YY1lo PLZFlo (red), YY1hi PLZFlo (black), and YY1hi PLZFhi (blue). (b) Mean fluorescence intensity (MFI) of YY1 and (C) PLZF expression in the three subsets and (d) the percentage and absolute cell number of NKT cells in each of the three subpopulations. YY1 MFI measurements were normalized to expression in CD4 single positive thymocytes stained in the same experiment. PLZF MFI measurements were normalized to YY1lo NKT cells stained in the same experiment. (e–g) Intracellular staining for (e) Tbet and PLZF or (f) RORγt and PLZF in all thymic NKT cells (left), YY1hi NKT cells (middle), and YY1lo NKT cells (right). (g) Distribution of YY1lo and YY1hi NKT cells in the NKT1, NKT2, and NKT17 subpopulations as defined in (e,f). (h,i) ThPOK expression was compared between YY1lo and YY1hi NKT cells by FACs. Representative histogram in (h) and cumulative data in (i). (j) TCRVβ usage, measured by FACs, was compared between YY1lo, YY1hi PLZFlo, and YY1hi PLZFhi NKT cells. FACS plots show representative results from 3–5 independent experiments. (b,c,f,g,i,j) show compiled data from at least 3 independent experiments. For (i) N = 5, (b–d,j) N = 6, (g) N = 7. Each dot depicts one mouse. The horizontal lines indicate the mean (±s.e.m.). *P < 0.05, **P < 0.01 ***P < 0.001 determined by One-Way Anova (b,c,d,j) or Mann-Whitney U Test (g,i).
NKT cells require Tbet to fully acquire their characteristic effector/memory cell surface phenotype and to express the cytokine IFN-γ34. RORγt expression is necessary for mature NKT cells to produce the inflammatory cytokine IL-1735,36. Mature NKT cells can be segregated into effector subpopulations including NKT1 (Tbethi PLZFlo), NKT2 (Tbetlo PLZFhi) or NKT17 (RORγt+)6. The majority of YY1hi NKT cells (for simplicity, the two YY1hi populations are combined here) were found to be Tbethi PLZFlo similar to NKT1 cells. (Fig. 1e,g) A small percentage were Tbetlo PLZFhi consistent with the NKT2 phenotype (Fig. 1e,g). In contrast, YY1lo NKT cells were mostly Tbetlo PLZFlo (Fig. 1e,g) and, therefore, based on transcription factor expression were distinct from the well-described NKT cell effector subpopulations. ThPOK has also been shown to be essential for NKT cell effector functions37,38. Most notably, ThPOK deficient NKT cells do not produce IL-4 following primary activation. Interestingly, NKT cells in ThPOK KO mice are skewed towards the NKT17 lineage. YY1lo NKT cells expressed substantially lower levels of ThPOK as compared to YY1hi NKT cells (Fig. 1h and Supp Fig. 1). The YY1lo NKT cells, however, did not express RORγt (Fig. 1f,g).
TCR Vβ gene segment usage has been shown to correlate with the effector lineage adopted by developing NKT cells. We determined if YY1lo NKT cells had differential TCR Vβ usage that might be associated with their unique characteristics39. 60% of YY1lo NKT cells expressed TCR Vβ8.2, 15% of YY1lo NKT cells expressed TCR Vβ7, and 5% of YY1lo NKT cells expressed TCR Vβ2 (Fig. 1i). These TCR Vβ usage frequencies were similar to YY1hiPLZFlo NKT cells and YY1hiPLZFhi NKT cells, suggesting that differential TCRVβ usage did not impact the unique transcription factor expression pattern of the YY1lo NKT cells.
YY1lo NKT cells in the thymus are not at an immature or transient population
Based on the expression of several key transcription factors, our data suggested that the YY1lo NKT cells might either be an immature or a functionally distinct population of NKT cells. As NKT cells develop, they upregulate cell surface markers associated with effector/memory T cells including CD44, CD69, and CXCR3 as well as markers typically found on NK cells such as NK1.140,41. Since differential expression of these cell surface markers on NKT cells is associated with maturation, we analyzed expression on the YY1lo subset.
Thymic NKT cells from 8 week old C57BL/6J mice were stained for the surface markers CD44 and NK1.1 and the transcription factors YY1 and PLZF. NKT cells from all three YY1/PLZF subsets as defined in (Fig. 1a) were found to be distributed among all three of the described stages of development (Fig. 2a,b). The YY1hi PLZFhi population was mostly in Stage 1 (CD44−NK1.1−) or Stage 2 (CD44 + NK1.1−), with some cells progressing to Stage 3 (CD44 + NK1.1+). The majority of YY1hi PLZFlo cells, were concentrated in Stage 3, consistent with Tbet+ NKT1 type cells. Interestingly, despite little Tbet expression, the YY1lo NKT cells largely had a Stage 3 phenotype (Fig. 2a,b). The expression of CD44 and NK1.1 strongly suggested that YY1lo NKT cells were a mature population of NKT cells.
YY1lo NKT cells are not an immature or transient population. Thymuses were collected from C57BL/6J (WT) mice and analyzed by FACS with the indicated antibodies. (a,b) NKT cells (MHCII−, CD3+ CD1dtet+ CD24−) were stained for the transcription factors PLZF and YY1, as well as, the cell surface markers CD44 and NK1.1. Expression of CD44 and NK1.1 was compared between the YY1/PLZF subpopulations defined in (Fig. 1a). Representative FACs plots are shown in (a) and cumulative data concerning the distribution of NKT cells in different stages are plotted in (b). The NKT cell stages were defined as: Stage 1 (CD44− NK1.1−), Stage 2 (CD44+ NK1.1−), and Stage 3 (CD44+ NK1.1+). (c,d) Thymus NKT cells isolated from 2 week old, 10 week old, and 16 week old WT mice were stained for YY1 and PLZF. (c) Representative FACs plots depicting YY1 and PLZF expression are shown. (d) Cumulative data comparing the frequency of each YY1/PLZF defined NKT cell subpopulation between mice of different ages. FACS plots show typical results from indicated specimen and graphs show compiled data from 7 (b) or 6 (d) mice, examined in 3 or more independent experiments. The horizontal lines indicate the mean (±s.e.m.). ***P < 0.001 determined by One Way Anova (b,d).
YY1 and PLZF expression was profiled in thymic NKT cells isolated from 2 week, 10 week and 16 week old C57BL/6J mice. An increase in the frequency of YY1lo NKT cells would imply that this was an induced phenotype. The frequency of YY1lo NKT cells, however, was consistent between mice of different ages (Fig. 2c,d). In contrast, the frequency of YY1hiPLZFhi NKT cells and YY1hiPLZFlo NKT cells changed substantially as mice aged (Fig. 2c,d). YY1hiPLZFhi NKT cells were overrepresented in younger mice, whereas YY1hiPLZFlo NKT cells were found at a significantly higher frequency in older mice, consistent with previous results12. The stable representation of YY1lo NKT cells in both young and old mice implied that this subset is not a derivative of the other populations.
YY1lo NKT cells are found in secondary lymphoid and non-lymphoid tissues
The abundance of different NKT cell effector subpopulations differs between tissues. 90% of the NKT cells found in the liver belong to the NKT1 subpopulation, while the majority of NKT cells in the lung are NKT17 cells and the adipose tissue is enriched for IL-10 producing NKT cells42. Given the variability in representation of NKT cell effector subpopulations in different tissues, we reasoned that the percent of YY1lo NKT cells might fluctuate between tissues. NKT cells were isolated from the spleen, visceral adipose tissue, lung, and liver of 8 week old C57BL/6 mice. The expression of YY1 and PLZF in these NKT cells was measured by flow cytometry. All three YY1/PLZF based subpopulations, as defined in (Fig. 1a), were identified in all tissues (Fig. 3a). The majority of NKT cells in each tissue were YY1hiPLZFlo (Fig. 3a,b). Approximately 3%-10% of NKT cells in each tissue belonged to the YY1loPLZFlo subpopulation (Fig. 3a,b), indicating that the relative abundance of YY1lo NKT cells was not highly variable between tissues.
YY1lo NKT cells are found in lymphoid and nonlymphoid tissues and share a common transcription factor expression pattern. Lymphocytes were isolated from the spleen, visceral adipose, lung, and liver of 12 week old, C57BL/6J (WT) mice and analyzed by FACS with the indicated antibodies. Expression of PLZF and YY1 in the NKT cells (CD45.2+, MHCII−, CD3+ CD1dtet+ CD24−) was determined. (a) Representative FACs plots and (b) cumulative data comparing the frequency of the YY1/PLZF subpopulations between tissues (c–e). Intracellular staining for (c) Tbet and PLZF or (d) RORγt and PLZF in total spleen NKT cells (left), YY1hi NKT cells (middle), and YY1lo NKT cells (right). (e) Distribution of spleen YY1lo and YY1hi NKT cells in the NKT1, NKT2, and NKT17 subpopulations as defined in (c,d). (f,g) ThPOK expression was compared between spleen YY1lo and YY1hi NKT cells by FACs. A representative histogram is shown in (f) and cumulative data are summarized in (g). (h,i)Expression of CD44 and NK1.1 was compared between the YY1/PLZF subpopulations. Data in (a,c,d,f,h) are representative of at least 4 independent experiments. Each dot in (b,e,g,i) represents an individual mouse, for (b) N ≥ 5 for each tissue and for (e,g) N = 4, for (i) N = 6. The horizontal lines indicate the mean (±s.e.m.). *P < 0.05 **P < 0.01, ***P < 0.001 determined by one-way ANOVA (b,i) or Mann-Whtiney U Test (e,g).
The frequency of NKT cell effector subpopulations also differs between mouse strains. NKT2 cells, for example, are found at a higher frequency in BALB/cJ mice as compared to C57BL/6J mice, whereas NKT1 cells are more abundant in C57BL/6J mice6. The frequency of YY1lo NKT cells in the thymuses and spleens of BALB/cJ mice and C57BL/6J mice was, however, indistinguishable (Supplemental Fig. 2).
Similar to what was found for thymic NKT cells, few splenic YY1lo NKT cells expressed high levels of Tbet (Fig. 3c) and they also did not express RORγt (Fig. 3d). Therefore, YY1lo spleen NKT cells also appeared to be distinct from NKT1 cells, NKT2 cells, or NKT17 cells (Fig. 3e), which was in contrast to YY1hi NKT cells (Fig. 3c–e). In addition, the YY1lo NKT cells in the spleen expressed low levels of ThPOK (Fig. 3f,g). These data demonstrate that, similar to the thymus, YY1lo NKT cells in the spleen have a distinct transcription factor expression pattern. The majority of the YY1lo NKT cells in the spleen expressed NK1.1 (Fig. 3h,j) suggesting that some YY1lo NKT cells were recent thymic emigrants, while others had been in the periphery for an extended period.
YY1lo NKT cells produce little IL-4 and IFN-γ following activation
Low expression of Tbet, PLZF, and ThPOK suggested that YY1lo NKT cells might have distinct effector functions as compared to other NKT cells. To test this, NKT cells were activated in vivo by intravenous (IV) injection of mice with 10 µg of the NKT cell specific agonist ligand, α-galactosylceramide (α-GalCer). 90 minutes after injection, splenocytes were harvested and stained for the transcription factors PLZF and YY1, as well as the cytokines IL-4 and IFN-γ. The percentage of YY1hi NKT cells that produced IL-4 and IFN-γ (both the PLZFhi and PLZFlo subsets) was consistently 2–3 fold higher than the YY1lo NKT cells (Fig. 4a,b). Furthermore, the amount of IL-4 produced by the responding YY1lo cells, as measured by mean fluorescence intensity (MFI), was less than half that of the YY1hi cells (Fig. 4c). The MFI of IFN-γ response was not statistically different (Fig. 4d). The difference in ability to produce IL-4 and IFN-γ following primary activation was even more exaggerated following activation with PMA plus ionomycin in vitro. These experiments resulted in almost no IL-4 and IFN-γ production from YY1lo NKT cells as compared to YY1hi NKT cells (Fig. 4e,f). YY1lo NKT cells also failed to produce IL-17a following in vivo activation with α-GalCer (Supp Fig. 3a).
YY1lo NKT cells produce less IL-4 and IFN-γ as compared to YY1hi NKT cells. (a–d) C57BL/6J (WT) mice were injected I.V. with either α-GalCer (10ug) suspended in saline or saline only (control). 90 minutes after injection, splenic NKT cells were harvested and stained for IL-4 and IFN-γ. (a) Representative FACs plot for IL-4 and IFN-γ expression in each WT activated NKT cell (MHCII−, CD3+ CD1dtet+ CD24−) subset: YY1lo PLZFlo, YY1hi PLZFlo, and YY1hi PLZFhi. (b) Cumulative data from 6 mice, depicting the % of IL-4+ and IFN-γ + double positive cells. The IL-4 MFI of IL-4+ cells (d) and IFN-γ MFI of IFN-γ+ cells (d) from each NKT cell subset is plotted. The IL-4 MFI and IFN-γ MFI of each experimental sample was normalized to the IL-4 MFI and IFN-γ MFI of control, unactivated NKT cells stained in the same experiment. (e–f) NKT cells were isolated from the spleens of WT mice and activated in vitro for 5 hours by culture with PMA (50 ng/mL) and Ionomycin (500n/mL). Activated NKT cells were stained for IL-4, IFN-γ, and YY1. (e) Representative histograms and (f) and aggregate data compare the production of IFN-γ (left) and IL-4 (right) between YY1lo and YY1hi NKT cells following in vitro activation. Data are representative of 4 (a–d) or 3 (e–f) independent experiments. Each dot represents one mouse, N = 6 for (b–d), N = 3 for (f). The horizontal lines indicate the mean (±s.e.m.). *P < 0.05 **P < 0.01, ***P < 0.001 determined by one-way ANOVA (b–d).
YY1lo NKT cells include dedicated IL-10 producing cells
NKT cells express many cytokines in addition to IL-4 and IFN-γ, including IL-10. To determine the capacity of YY1lo NKT cells to produce IL-10, mice were injected IV with α-GalCer. 90 minutes after injection, mice were scarified and total spleen NKT cells were purified and a cell surface capture assay was used to detect IL-10 producing NKT cells. A substantial percentage of YY1lo NKT cells were found to produce IL-10, while none was detected from YY1hi NKT cells (Fig. 5a,b). Purified spleen NKT cells activated in vitro with PMA/ionomycin for 3 hours showed similar results (Fig. 5c,d). Interestingly, thymic NKT cells isolated and activated in vitro with PMA/ionomycin were also found to secrete IL-10 (Fig. 5e,f). Therefore, low expression of YY1 marks a population of NKT cells primed to secrete IL-10 following primary activation. Importantly, this subset exists in the thymus, suggesting it is not an induced phenotype caused by antigen activation.
YY1lo NKT cells produce IL-10 following primary activation. (a–c) C57BL/6J (WT) mice were injected I.V. with α-GalCer (10ug) suspended in saline. 90 minutes after injection, spleens were harvested and NKT cells (MHCII−, CD3+ CD1dtet+ CD24−) were enriched for by automacs. A cell surface capture assay was used to determine IL-10 production by YY1lo PLZFlo, YY1hi PLZFlo, and YY1hi PLZFhi NKT cells. (a) Representative histogram depicting IL-10 production by each subset. (b) Cumulative data from 5 mice, depicting the % of IL-10+ cells. (c–f) NKT cells were isolated from the spleens (c,d) or thymuses (e,f) of WT mice and activated in vitro for 3 hours by culture with PMA (50 ng/mL) and Ionomycin (500n/mL). NKT cell IL-10 production was measured by a cell surface capture assay. Representative histograms (c,e) and aggregate data (d, f), compare the production of IL-10 between YY1lo PLZFlo, YY1hi PLZFlo, and YY1hi PLZFhi NKT cells following in vitro activation. (g–h) C57Bl/6 (WT) mice were IV injected with 10 ug of α-GalCer or vehicle. Mice were sacrificed 72 hours or four weeks after injection. Spleen NKT cells (MHCII−, CD3+ CD1dtet+ CD24−) YY1 and PLZF expression were measured by FACs. (g) Representative FACS plots and (h) cumulative data. (i) C57Bl/6 (WT) mice were IV injected with 10 ug of α-GalCer or vehicle. 4 weeks later, NKT cells were enriched from spleens, activated in vitro with PMA/Ion, and IL-10 production was measured using a cell surface capture assay. Graph shows YY1 MFI and whether or not the cells produced IL-10. Data in histograms are representative of at least 3 independent experiments representing 3 or more biological replicates. For (b) N = 5, (d) N = 3, (f,h,i) N = 4. The horizontal lines indicate the mean (±s.e.m.). *P < 0.05determined by One-Way Anova (b,d,f,h,i).
Primary activation induces the differentiation of NKT cells such that upon secondary activation a large percentage produce a wide range of cytokines, including IL-1021. We tested the possibility that this increase in the frequency of IL-10 producing NKT cells following secondary activation was the consequence of an expansion of YY1lo NKT cells following primary activation. We injected mice with 10 ug of α-GalCer and analyzed YY1 expression levels in NKT cells either 72 hours and or 4 weeks later. The frequency of YY1lo NKT cells remained similar to vehicle injected mice at both time points (Fig. 5g,h). Treatment with α-GalCer did, however, result in a substantial increase in the frequency of YY1hiPLZFhi NKT cells and a corresponding decrease in the frequency of YY1hiPLZFlo NKT cells in the spleen at both time points measured (Fig. 5g,h).
Next, we isolated NKT cells from the spleens of C57Bl/6J mice 1 month after IV injection of α-GalCer. These cells were reactivated in vitro with PMA and ionomycin for 3 hours. Data show that in addition to YY1lo cells producing IL-10, prior activation with α-GalCer also enables some YY1hi cells to produce IL-10 (Fig. 5i). Therefore, while YY1lo NKT cells are committed IL-10 producing NKT cells, activation induced differentiation of YY1hi NKT cells into IL-10 producing cells also occurs.
Having demonstrated YY1lo NKT cells to be dedicated IL-10 producers, we next determined the expression of transcription factors and cell surface markers associated with other IL-10 secreting NKT cell populations, namely arNKT cells and NKT10 cells, in YY1lo NKT cells. In accordance with previous studies, we found that no NKT cells, including YY1lo NKT cells, expressed the Treg lineage defining transcription factor FoxP3 (Supplemental Fig. 3b). Like arNKT cells and NKT10 cells, YY1lo NKT cells isolated from both the thymus and spleen were found to express high levels of PD-1 (Fig. 6a,b). Unlike NKT10 cells, YY1lo NKT cells were found to express low levels of NRP1, CD49d and ICOS (Supplemental Fig. 3c). Differential expression of these three surface markers potentially can be attributed to differences in activation state between YY1lo NKT cells, which were freshly isolated from naive mice and NKT10 cells, which were activated in vitro for four hours with PMA/Ionomycin.
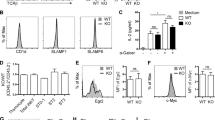
YY1 deficient NKT cells do not produce IL-10. (a,b) NKT cells from the thymus and spleen of C57BL/6J (WT) mice were stained for PD-1. Representative histogram in (a) and cumulative data from 4 mice in (b). (c–e) C57Bl/6 (WT) and PLZF-Cre YY1 flx.flx (YY1 cKO) mice were IV injected with 10 ug of α-GalCer. 90 minutes after injection, mice were scarified, NKT cells were enriched from spleens, and an IL-10 cell surface capture assay was used to measure NKT cell IL-10 production. (c) Representative FACs plot and (d) cumulative data. (e) NKT cells were FACs sorted from WT and YY1 cKO mice, RNA was isolated from sorted cells, and reverse transcribed into cDNA. qPCR measured β2 M and IL-10 expression in the sorted NKT cell populations. Data are from 4 or more independent experiments representing 4 biological replicates. The horizontal lines indicate the mean (±s.e.m.). *P < 0.05, ***P < 0.001, determined Mann Whitney U-Test (b,d,e).
YY1 deficient NKT cells lose the capacity to rapidly produce IL-10
Since low YY1 expression marked an NKT cell population primed to rapidly produce IL-10, we tested the possibility that YY1 deficient NKT cells would have an enhanced capacity to produce IL-10. YY1 deficient NKT cells isolated from PLZF-cre YY1 flox/flox mice (YY1 cKO mice), however, produced little or no IL-10 following in vitro activation with PMA/ionomycin (Fig. 6c,d). To determine if YY1 deficient NKT cells failed to rapidly produce IL-10 as a consequence of losing their preformed IL-10 mRNA, mRNA was isolated from sorted YY1 cKO NKT cells, and qPCR experiments were performed. While preformed IL-10 message was detected in wildtype NKT cells, substantially less was detected in YY1 deficient NKT cells (Fig. 6e). YY1, therefore, like for other cytokines33, is required for rapid production of IL-10 in NKT cells, likely by opening the gene locus.
YY1lo NKT cell are enriched in tumors
Recently, pre-cancerous colon polyps were shown to be enriched for IL-10 producing NKT cells43. We reasoned that IL-10 producing, YY1lo NKT cells might be enriched in tumors. To test this possibility, 5 ×105 CT26 colon cancer cells44 were implanted intradermally into BALB/cJ mice. When tumors reached an area between 8 by 8 millimeters and 10 by 10 millimeters (approximately 12–16 days after injection), mice were sacrificed, and hematopoietic cells were isolated from tumors. In contrast to all other studied tissues, nearly 40% of the NKT cells isolated from CT26 tumors were found to be YY1lo (Fig. 7A,B). We next intradermally injected C57BL/6J mice with either 5 ×105 B16F10 melanoma cells45 or 5 ×105 D4M3A melanoma cells46,47. For both tumors, we once again found that a high percentage (more than 35%) of the tumor infiltrating NKT cells were YY1lo (Fig. 7A,B). Therefore, enrichment for YY1lo NKT cells occurs in different tumor types and in different mouse genetic backgrounds. Efforts to detect IL-10 expression by the tumor infiltrating NKT cells were not, however, successful. It is possible that YY1lo NKT cells infiltrate tumors quickly after implantation and only secrete IL-10 for a short period of time48. We attempted, therefore, to isolate leukocytes from tumors at earlier time points (3–5 days post tumor implantation) but were unable to collect sufficient cells for analysis.
Tumors are enriched for YY1lo NKT cells. 5×105 tumor cells were injected intradermally into mice. When tumors reached an area between 8×8 millimeters and 10×10 millimeters mice were sacrificed, tumors were harvested, and leukocytes were isolated from tumors. CT26 colon cancer cells were injected into BALB/CJ mice, while B16F10 melanoma cells and D4M3A melanoma cells were injected into C57BL/6J mice. (a,b). YY1 and PLZF expression in NKT cells (CD45.2+, CD19−, MHCII−, CD3+, CD1dtet+) isolated from tumors was determined by flow cytometry. A representative FACs plot for each tumor type is shown in (a) and cumulative data are shown in (b). Data are from 2 or more independent experiments representing 7 (b). The horizontal lines indicate the mean (±s.e.m.). *P < 0.05, **P < 0.01, determined by Mann One-Way Anova (b).
Discussion
Functionally distinct NKT cell effector subpopulations, primed to secrete unique combinations of cytokines and defined by particular transcription factor expression patterns, potently modulate disparate aspects of immunity49,50,51,52. In this present study, we uncovered a small population of NKT cells that expressed low levels of the transcription factor YY1. YY1lo NKT cells, irrespective of their tissue or mouse strain of origin, shared a distinct transcription factor expression pattern (PLZFlo, Tbetlo, RORγT−, and ThPOKlo) as compared to NKT1, NKT2, or NKT17 cells. When activated, YY1lo NKT cells produced little of the typical cytokines, IL-4 and IFN-γ, but instead secreted IL-10. Based upon the unique transcription factor expression pattern and cytokine production capacity of YY1lo NKT cells, we propose that we have identified a novel NKT cell effector subpopulation.
The signals that drive NKT cells to adopt different effector lineages remain incompletely described. One potential mechanism is differential TCRVβ chain usage, as certain NKT cell effector lineages display greater usage of specific TCRVβ chains39. However, no substantial TCRVβ usage differences were observed between YY1lo and YY1hi NKT cells. Alternatively, some NKT cell effector lineages, such as the T follicular helper NKT (NKTfh) cell, appear to be induced when mature NKT cells are activated under very specific conditions53. The YY1lo NKT cell phenotype does not seem to be inducible in mature NKT cells, as the frequency of YY1lo NKT cells was not affected by age, tissue of origin, or in vivo activation. Rather, mature YY1lo NKT cells are likely derived from immature, stage 1 YY1lo NKT cells found in the thymus. Why some stage 1 NKT cells express low levels of YY1 remains to be determined. It should be noted that our data do not rule out the possibility that YY1lo NKT cells in the thymus are actually mature cells that have returned. Should this be the case, it may impact the interpretation that YY1lo NKT cells are a distinct lineage.
Consequential differences in YY1 expression levels between helper T cells and Tregs have previously been reported30. Knockdown of YY1 in Th2 cells, which usually express high levels of YY1, dramatically reduced their expression of several Th2 cytokines including IL-4, IL-5, and IL-1331. Conversely, over expression of YY1 in Tregs, which usually express relatively low levels of YY1, led to their functional impairment and limited their production of IL-1030. Experimental evidence from other systems including embryos54 and hematopoietic stems cells26 provide additional evidence that YY1 dosage is critical for the normal development and function of cells. Our findings, which demonstrate that functionally distinct NKT cell effector populations express different levels of YY1, are therefore consistent with previous studies.
YY1lo NKT cells appear to be a “natural” source of IL-10, as the cells constitutively expressed IL-10 mRNA and rapidly secreted the cytokine following primary activation. Steady state production of IL-10 by YY1lo NKT cells may modulate the general tone of the immune system, serving such functions as maintaining adipose tissue homeostasis. This would be analogous to the constitutive production of IL-4 by NKT cells, which drives the development of “innate-like” CD8 T cell populations55,56. Furthermore, the rapid production of IL-10 by YY1lo NKT cells after primary activation may serve a critical function in limiting immune-mediated tissue injury.
YY1lo NKT cells were found at a high frequency in tumors, irrespective of the tumor type or recipient mouse strain. Given the capacity of YY1lo NKT cells found in other tissues to produce IL-10, we hypothesized that YY1lo NKT cells would secrete IL-10 in tumors, but at least at the time point tested, which was ~12 days after tumor implantation, the cells did not express IL-10. It is possible that YY1lo NKT cells infiltrate tumors soon after implantation, and secrete IL-10 for only a brief period. This would be similar to what is found recently found during the immune response to flu, where NKT cell produced IL-4 is made from 3–7 days post infection, but not after day 948. An alternative explanation is that YY1lo NKT cells found in tumors are functionally distinct and unrelated to YY1lo NKT cells found in other tissues.
In accordance with previous studies, we found that a higher frequency of NKT cells secreted IL-10 after secondary activation21. Activation, however, does not cause the expansion of the YY1lo NKT cell population, but rather results in an IL-10 producing NKT cell population that expressed high levels of YY1. These induced IL-10 producing NKT cells, therefore, appear to be distinct from the YY1lo NKT cells reported here.
In summary, we found that the ubiquitously expressed transcription factor, YY1, is differentially expressed in NKT cells. Low YY1 expression marks a functionally distinct NKT cell effector lineage that is a natural source of IL-10. It remains to be determined what factors drive the development of this lineage, but we have ruled out differential TCRVβ usage. Furthermore, activation does not lead to the specific expansion of YY1lo NKT cells, but rather induces YY1hi NKT cells to produce IL-10. Our data also suggest that YY1lo NKT cells might impact the tumor microenvironment, but future studies will have to explore this more thoroughly.
Methods
Animals
C57Bl/6J mice and BALB/cJ mice were obtained from Jackson Labs. YY1 flox/flox mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and, also from a Dr. Michael Verzi (Rutgers University). PLZF-Cre mice were developed in our lab as reported14. IL-10 GFP mice were obtained from Jackson Laboratories. All mouse strains were bred and maintained in the animal facility of the Child Health Institute of New Jersey. All experimental protocols and procedures were approved by the Institutional Animal Care and Use Committee of the Child Health Institute of New Jersey. Animal care and experimental procedures were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of Rutgers University and the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Tumor cell lines
The CT26 and B16F10 cell lines were obtained from Dr. Andrew Zloza (Rush University). The D4M3A cells were provided by Dr. David Mullins (Dartmouth University). Cells were cultured as monolayers at 37 degrees Celsius and 5% CO2 in RPMI (CT26 and D4M3A) or DMEM (B16F10) supplemented with 10% heat-inactivated bovine serum and 0.5% penicillin G-streptomycin sulfate (Corning). 0.25 trypsin EDTA (Corning) was used to detach adherent cells.
Tumor implantation into mice
5×105 CT26 colon cancer cells were injected intradermally into BALB/c mice. 5×105 B16F10 melanoma cancer cells or D4M3A melanoma cancer cells were injected intradermally into C57Bl/6 mice. Tumor growth was monitored daily. Mice were sacrificed and tumors were harvested when they reached an area between 8×8 millimeters and 10×10 millimeters.
Preparation of single cell suspensions from lymphoid and non-lymphoid tissues
Thymus, spleen, liver, and lung were harvested and dissociated between glass slides and filtered through a 40-μm mesh. Spleen and thymus samples were treated with Red Blood Cell Lysing Buffer (Sigma) and counted on a hemocytometer. Liver homogenate was suspended in Percoll (Sigma) to a final concentration of 30% Percoll. Liver homogenates were then centrifuged for 20 min at 500 r.c.f. in a Sorvall RTH-750 rotor. Lymphocytes were then collected from the buffy layer, washed once with FACS buffer, treated with Red Blood Cell Lysing Buffer (Sigma) and counted on a hemocytometer. Lung homogenate was underlayed with lympocyte seperation medium (MP Biomedicals) and then centrifuged for 20 min at 500 r.c.f. in a Sorvall RTH-750 rotor. Lymphocytes were then collected from the buffy layer, washed once with FACS buffer, and counted on a hemocytometer. Visceral adipose tissue was harvested from mice and minced. Minced adipose tissue was suspended in RPMI medium containing collagenase Type IV (Worthington) at a concentration of 1 mg/ML and rotated at 37 degrees Celsius for 45 minutes. After rotation, cells were washed with RPMI. Tumors were collected from mice when they reached an area between 8×8 millimeters and 10×10 millimeters. Tumors were suspended in RPMI medium containing collagenase Type IV (Worthington) at a concentration of 1 mg/ML. Tumors were dissociated using gentleMACs (Miltenyi Biotec) program m_impTumor_01_01. Dissociated tumors were incubated with rotation for 45 minutes at 37 degrees Celsius and washed with FACs buffer and filtered through 40-μm filters.
Flow cytometry, antibodies, automacs enrichment, and cell sorting
Single cell suspensions were blocked in FACS buffer with 2% normal mouse serum, 0.1% anti-Fcγ antibody, and 0.1ug/ml streptavidin for 15 minutes at 4 C followed by staining with primary antibodies for 20 minutes at 4 C. Intracellular staining for transcription factors and cytokines was accomplished using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). All intracellular staining was performed at room temperature. IL-10 was detected using the Mouse IL-10 Secretion Assay - Detection Kit (Miltenyi Biotech). The following antibodies were used in this study: anti-CD3 (500A2), Anti-CD4 (RM4-5), anti-CD8α (53–6.7), anti–IL-17A (Ebio17B7), anti-CD19 (1D3), anti-CD24 (M1/69), anti-CD44 (IM7), CD45.2 (104), anti-CD69 (H1.2F3), anti-TCRVβ 8.2 (MR5-2), anti-TCRVβ7 (TR310), anti-TCRVβ2 (B20.6), anti-IFN-γ(XMG1.2), anti-IL4 (11B11), anti-MHCII (212.A1), PD-1 (J43), ICOS (15F9), CD49d (R1-2), NRP1 (3DS304M), anti-NK1.1 (PK136), anti-PLZF (Mags.21F7), anti-RORγT (B2D), anti-T-bet (eBio4B10), anti-YY1 (sc-7341), anti-ThPOK (2POC), FoxP3 (FJK-16s). The PBS57-loaded CD1d-tetramer was provided by the National Institutes of Health tetramer core facility. Dead cells were excluded when possible, by DAPI staining and doublet events were eliminated by comparing FSC-W to FSC-H and SSC-W to SSC-H. Events were acquired on a LSRII cytometer (BD Biosciences, San Jose, CA), and the data were analyzed with the FlowJo software (TreeStar, Ashland, OR).
NKT cell enrichment was performed using an AutoMACS (Miltenyi Biotec). Cells were stained with the PE-labeled CD1d tetramer, incubated with anti-PE microbeads (Miltenyi Biotec), and separated on the AutoMacs (Miltenyi Biotec). Samples were at least 80% pure following enrichment.
Cell sorting was performed at the Flow Cytometry/Cell Sorting & Confocal Microscopy Core Facility at Rutgers EOSHI or at the Cancer Institute of NJ.
In vivo and In vitro activations
Mice were injected intravenously via the retro orbital route with 10 ug of the NKT cell specific agonist α-galactosylceramide (α-GalCer). 90 minutes after injection, splenocytes were harvested.
Following enrichment by Automacs (Miltenyi Biotech) NKT cells were activated in vitro by incubation with phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (500 ng/ml) for 3 hours or 5 hours when detecting IL-10 or IL-4/IFN-γ respectively. The Mouse IL-10 Secretion Assay - Detection Kit (Miltenyi Biotech) was then utilized to assess the production of IL-10 by NKT cells.
RNA isolation and quantitative PCR analysis
Sorted NKT cells were lysed in Trizol solution at a concentration of 1×106 cells per mL. RNA was isolated from the trizol solution using the Direct-zol RNA MicroPrep Kit(Zymo Research). Reverse transcription was performed using the GoScript Reverse Transcription System (Promega). The resultant cDNA was utilized to perform TaqMan (Life Technologies) based qPCR. Taqman Universal PCR Master Mix No AmpErase UNG (Life Technologies) was used. The following TaqMan probes were used: B2M (Mm00437762_m1) and IL-10 (Mm01288386) Samples were run on a QuantStudio6 Flex Real Time PCR System(Life Technology). Relative expression of IL-10 was calculated by using the ΔΔCT method, using B2M as the internal housekeeping control gene.
General experiment design and statistical analysis
Data from at least three samples in three or more independent tests were collected as detailed in the figure legends. Statistical analysis was performed using Microsoft Excel or GraphPad Prism. All data was subjected to analysis with Mann Whitney U-Test or One-Way Anova.
References
Kronenberg, M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol 23, 877–900 (2005).
Matsuda, J. L. et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med 192, 741–754 (2000).
Brossay, L. et al. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med 188, 1521–1528 (1998).
Kohlgruber, A. C., Donado, C. A., LaMarche, N. M., Brenner, M. B. & Brennan, P. J. Activation strategies for invariant natural killer T cells. Immunogenetics 68, 649–663 (2016).
Carr, T. et al. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. J Exp Med 212, 793–807 (2015).
Lee, Y. J., Holzapfel, K. L., Zhu, J., Jameson, S. C. & Hogquist, K. A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 14, 1146–1154 (2013).
Coquet, J. M. et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci USA 105, 11287–11292 (2008).
Govindarajan, S. et al. Stabilization of cytokine mRNAs in iNKT cells requires the serine-threonine kinase IRE1alpha. Nat Commun 9, 5340 (2018).
Stetson, D. B. et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med 198, 1069–1076 (2003).
Kovalovsky, D. et al. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol 184, 6746-6755.
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol 9, 1055–1064 (2008).
Iyer, S. S. & Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 32, 23–63 (2012).
Lynch, L. et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol 16, 85–95 (2015).
Wang, H. X. et al. CD1d-dependent natural killer T cells attenuate angiotensin II-induced cardiac remodelling via IL-10 signalling in mice. Cardiovasc Res 115, 83–93 (2019).
Ghraieb, A. et al. iNKT cells ameliorate human autoimmunity: Lessons from alopecia areata. J Autoimmun 91, 61–72 (2018).
Hammond, K. J. et al. alpha/beta-T cell receptor (TCR)+CD4-CD8- (NKT) thymocytes prevent insulin-dependent diabetes mellitus in nonobese diabetic (NOD)/Lt mice by the influence of interleukin (IL)-4 and/or IL-10. J Exp Med 187, 1047–1056 (1998).
Tard, C., Rouxel, O. & Lehuen, A. Regulatory role of natural killer T cells in diabetes. Biomed J 38, 484–495 (2015).
Wilson, M. T. & Van Kaer, L. Natural killer T cells as targets for therapeutic intervention in autoimmune diseases. Current pharmaceutical design 9, 201–220 (2003).
Wang, Y. et al. Unique invariant natural killer T cells promote intestinal polyps by suppressing TH1 immunity and promoting regulatory T cells. Mucosal Immunol 11, 131–143 (2018).
Cameron, G. & Godfrey, D.I. Differential surface phenotype and context-dependent reactivity of functionally diverse NKT cells. Immunol Cell Biol (2018).
Sag, D., Krause, P., Hedrick, C. C., Kronenberg, M. & Wingender, G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest 124, 3725–3740 (2014).
Shi, Y., Seto, E., Chang, L. S. & Shenk, T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377–388 (1991).
Weintraub, A. S. et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588 e1528 (2017).
Castellano, G. et al. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle 8, 1367–1372 (2009).
Lu, Z. et al. Polycomb Group Protein YY1 Is an Essential Regulator of Hematopoietic Stem Cell Quiescence. Cell Rep 22, 1545–1559 (2018).
Morikawa, Y., Leach, J. & Martin, J. F. Yin-Yang 1, a new player in early heart development. Circ Res 112, 876–877 (2013).
Chen, L., Foreman, D. P., Sant’Angelo, D. B. & Krangel, M. S. Yin Yang 1 Promotes Thymocyte Survival by Downregulating p53. J Immunol 196, 2572–2582 (2016).
Kwon, H. K., Chen, H. M., Mathis, D. & Benoist, C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat Immunol 18, 1238–1248 (2017).
Hwang, S. S. et al. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat Commun 7, 10789 (2016).
Hwang, S. S. et al. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc Natl Acad Sci USA 110, 276–281 (2013).
Yu, B. et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat Immunol 18, 573–582 (2017).
Darcy, P. W., Jin, K., Osorio, L., Denzin, L. K. & Sant’Angelo, D. B. Coexpression of YY1 Is Required to Elaborate the Effector Functions Controlled by PLZF in NKT Cells. J Immunol 203, 627–638 (2019).
Townsend, M. J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20, 477–494 (2004).
Michel, M. L. et al. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci USA 105, 19845–19850 (2008).
Venken, K. et al. RORgammat inhibition selectively targets IL-17 producing iNKT and gammadelta-T cells enriched in Spondyloarthritis patients. Nat Commun 10, 9 (2019).
Enders, A. et al. ZBTB7B (Th-POK) regulates the development of IL-17-producing CD1d-restricted mouse NKT cells. J Immunol 189, 5240–5249 (2012).
Engel, I., Zhao, M., Kappes, D., Taniuchi, I. & Kronenberg, M. The transcription factor Th-POK negatively regulates Th17 differentiation in Valpha14i NKT cells. Blood 120, 4524–4532 (2012).
Tuttle, K. D. et al. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat Commun 9, 2650 (2018).
Drennan, M. B. et al. Cutting edge: the chemokine receptor CXCR3 retains invariant NK T cells in the thymus. J Immunol 183, 2213–2216 (2009).
Dao, T. et al. Development of CD1d-restricted NKT cells in the mouse thymus. Eur J Immunol 34, 3542–3552 (2004).
Lee, Y. J. et al. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity 43, 566–578 (2015).
Wang, Y. et al. Promotion or Suppression of Murine Intestinal Polyp Development by iNKT Cell Directed Immunotherapy. Front Immunol 10, 352 (2019).
Wang, M. et al. Active immunotherapy of cancer with a nonreplicating recombinant fowlpox virus encoding a model tumor-associated antigen. J Immunol 154, 4685–4692 (1995).
Fidler, I. J. Biological behavior of malignant melanoma cells correlated to their survival in vivo. Cancer Res 35, 218–224 (1975).
Jenkins, M. H. et al. Multiple murine BRaf(V600E) melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res 27, 495–501 (2014).
Bommareddy, P. K., Zloza, A., Rabkin, S. D. & Kaufman, H. L. Oncolytic virus immunotherapy induces immunogenic cell death and overcomes STING deficiency in melanoma. Oncoimmunology 8, 1591875 (2019).
Gaya, M. et al. Initiation of Antiviral B Cell Immunity Relies on Innate Signals from Spatially Positioned NKT Cells. Cell 172, 517–533 e520 (2018).
Stolk, D., van der Vliet, H. J., de Gruijl, T. D., van Kooyk, Y. & Exley, M. A. Positive & Negative Roles of Innate Effector Cells in Controlling Cancer Progression. Front Immunol 9, 1990 (2018).
Marrero, I. et al. Complex Network of NKT Cell Subsets Controls Immune Homeostasis in Liver and Gut. Front Immunol 9, 2082 (2018).
LaMarche, N. M., Kohlgruber, A. C. & Brenner, M. B. Innate T Cells Govern Adipose Tissue Biology. J Immunol 201, 1827–1834 (2018).
Wang, Y. & Cardell, S. L. The Yin and Yang of Invariant Natural Killer T Cells in Tumor Immunity-Suppression of Tumor Immunity in the Intestine. Front Immunol 8, 1945 (2017).
Chang, P. P. et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol 13, 35–43 (2011).
Affar el, B. et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 26, 3565–3581 (2006).
Verykokakis, M., Boos, M. D., Bendelac, A. & Kee, B. L. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity 33, 203–215 (2010).
Weinreich, M. A., Odumade, O. A., Jameson, S. C. & Hogquist, K. A. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol 11, 709–716 (2010).
Acknowledgements
These studies were supported by NIH NIAID R01 AI083988 and AI059739 (to D.B.S.), the Robert Wood Johnson Foundation (grant #67038) to the Child Health Institute of New Jersey, NCI Comprehensive Core Grant (P30CA072720), the Aresty Undergraduate Research Fellowships (to P.W.D.) and the New Jersey Health Foundation Excellence in Research Award (to D.B.S.). We’d like to thank Joshua Vieth, Arnold Rabson, Agata Krzyzanowska and Stephanie Ji from the CHINJ for critical analysis of the data and/or help with experimental procedures, Arthur Roberts, from the Rutgers Cancer Institute of NJ Flow Cytometry Core Facility for cell sorting (shared resource P30CA072720-5921) and Louis Osorio for help with mouse work. CD1d tetramers were provided by the National Institutes of Health tetramer core facility.
Author information
Authors and Affiliations
Contributions
P.W.D., L.K.D. and D.B.S. conceived the overall project and designed the experimental plan. P.W.D. planned and carried out experiments. Data was discussed and interpreted by P.W.D., L.K.D. and D.B.S. Manuscript was written by P.W.D. and D.B.S. and critically reviewed by L.K.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Darcy, P.W., Denzin, L.K. & Sant’Angelo, D.B. YY1lo NKT cells are dedicated IL-10 producers. Sci Rep 10, 3897 (2020). https://doi.org/10.1038/s41598-020-60229-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60229-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.