Abstract
The keystone zooplankton Daphnia magna has recently been used as a model system for understanding host-microbiota interactions. However, the bacterial species present and functions associated with their genomes are not well understood. In order to understand potential functions of these species, we combined 16S rRNA sequencing and shotgun metagenomics to characterize the whole-organism microbiota of Daphnia magna. We assembled five potentially novel metagenome-assembled genomes (MAGs) of core bacteria in Daphnia magna. Genes involved in host colonization and immune system evasion were detected across the MAGs. Some metabolic pathways were specific to some MAGs, including sulfur oxidation, nitrate reduction, and flagellar assembly. Amino acid exporters were identified in MAGs identified as important for host fitness, and pathways for key vitamin biosynthesis and export were identified across MAGs. In total, our examination of functions in these MAGs shows a diversity of nutrient acquisition and metabolism pathways present that may benefit the host, as well as genomic signatures of host association and immune system evasion.
Similar content being viewed by others
Introduction
Organisms are hosts to complex communities of microorganisms collectively known as the microbiota. Species in the microbiota may provide beneficial functions to the host and other species in the microbiota, including nutrient acquisition and uptake1, production of accessible metabolites2, host immune system priming3, and direct pathogen protection4. However, characterizing beneficial host-microbe and microbe-microbe interactions in biologically relevant systems is difficult due to the number of core species present in the microbiota and the variation in an individual organism’s microbiota over time from dietary changes or differential environmental exposure5.
The zooplankton Daphnia magna provides a useful model for studying functional relationships between microbes and their hosts. Daphnia species are used as a model system in ecology, ecotoxicology, and host-parasite dynamics due to their well-documented life cycle and rapid asexual reproduction6. The ability to raise Daphnia magna clonally allows for the use of genetically identical hosts, reducing the impact of genetic variation on the microbiota. Furthermore, their indiscriminate filter feeding allows for control over food input. Daphnia are colonized with bacteria throughout their entire body cavity and gut7,8. Composition of the Daphnia microbiota appears to be similar in spatially unique populations8,9, suggesting mechanisms of acquisition and cultivation of these microbes by the host. The Daphnia microbiota is relatively simple at the class level, with β-proteobacteria, γ-proteobacteria, and Flavobacteriia consistently identified at high relative abundances9,10. However, relatively little work has extended past coarse-level 16S rRNA analysis for identification and characterization of bacterial taxa present in the Daphnia microbiota7. The genomic content of Daphnia-associated bacteria is largely unknown due to the amplicon sequencing methods used in prior research, and we cannot make statements about the metabolic potential of these bacteria due to the relative scarcity of full genomes for close relative species.
Beyond identifying the composition of the microbiota, it is clear that this composition affects host fitness. The Daphnia microbiota has been implicated in nutrient acquisition and breakdown of toxic compounds11, and survival and growth are affected by the presence of, and perturbations to, the microbiota12,13,14. Host fecundity has been specifically tied to Limnohabitans, an abundant genus in the Daphnia microbiota15. However, the functions underlying the beneficial Limnohabitans-Daphnia association are unknown, and how other species in the Daphnia microbiota contribute to host life history and what functions these species may be providing is entirely unclear.
Here, we use shotgun metagenomics to characterize the bacterial species present in the Daphnia magna microbiota. The metagenome-assembled genomes (MAGs) generated from this data were then used to examine potential metabolic functions of the microbiota in total and for each species assembled. This study is the first to report MAGs of bacteria in the Daphnia magna microbiota and their associated gene content. This increased resolution allows us to formulate testable hypotheses about the metabolic interactions happening between the host and microbes, and among microbes, that impact host fitness. This functional knowledge will provide a new lens for studying this important ecological model system.
Results
Shotgun sequencing, assembly, and binning
Shotgun sequencing resulted in 20.19 Mbp of paired-end Illumina read data, which was then reduced to 9.64 Mbp after quality trimming and host genome filtering. A co-assembly of all samples generated 174,991 contigs (N50 of 1,923 bp; longest assembled contig 226,522 bp). Summaries of assembly statistics for the co-assemblies and the individual sample assemblies can be found in Table 1. Identification of high-quality reads and ≥1000 bp assembled contigs using Kraken, Kaiju, and MetaPhlAn2 indicated 18 genera that were present as more than 1% of the sample (Supplementary Fig. 1). Of the 18 genera, only Limnohabitans was identified by all three tools in reads and contigs. Other abundant genera included Pedobacter, Flavobacterium, Polaromonas and unclassified Burkholderiaceae. 16S rRNA sequencing of the library preparation and DNA extraction kits resulted in <50 reads (Fig. 1a). 16S rRNA community profiles of the Daphnia magna food source, Chlamydomonas reinhardtii, the COMBO culture media for Daphnia magna, and samples of 5 healthy adult Daphnia magna showed differences in composition (Fig. 1a), which were then confirmed using an unweighted principal coordinate analysis on the unweighted UniFrac distances between samples (Supplementary Fig. 2). Chlamydomonas samples showed reduced relative abundance of Proteobacteria as compared to adult Daphnia and COMBO; COMBO showed higher relative abundance of Actinobacteria; and healthy Daphnia magna were primarily colonized by Proteobacteria and Bacteroidetes.
Comparison of bacterial community profiles of Daphnia magna, Chlamydomonas reinhardtii, COMBO media, and negative controls using 16S rRNA sequencing. (a) Phylum-level 16S rRNA profiles of Chlamydomonas reinhardtii (Chlamy, n = 4), COMBO culture media (Culture, n = 4), and the DNA extraction kit and library preparation kit used for sequencing (Kit (−), n = 2). (b) Relative abundance of phyla generated from 16S rRNA community profiles in Chlamydomonas reinhardtii, healthy adult Daphnia magna, and the culture media (all n = 4).
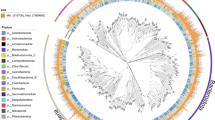
CONCOCT clustering of contigs from the master co-assembly resulted in 19 bins of draft and incomplete genomes. Manual curation, refinement, and merging using Anvi’o resulted in 15 bins, with 7 meeting the ≥50 quality metric. The adult-only co-assembly generated 12 bins, which further refined to 5 bins, with 4 meeting the ≥50 quality metric (Supplementary Fig. 5). Three of the four high-quality bins were also present in the master co-assembly and one unique bin was identified. The juvenile-only co-assembly generated 15 bins, which further refined to 7 bins, with 4 meeting the ≥50 quality metric (Supplementary Fig. 6). Because the master co-assembly generated bins with higher coverage or higher completeness, we used GTDB-Tk on the seven high- and medium-quality bacterial MAGs identified from the master co-assembly only. Two MAGs were placed in the Limnohabitans genus, one in the Polaromonas genus, one unable to be placed below the Burkholderiaceae family, one in the Pedobacter genus, one in the Emticicia genus, and one unable to be placed below the Chitinophagales order (Supplementary Table 1; Supplementary Fig. 4). None of the MAGs mapped to previously identified species in the GTDB-Tk database. Average nucleotide identity of the two Limnohabitans MAGs was measured at 86.6% similarity. This is less than the 95% cutoff generally accepted for strain-levels similarity, suggesting the MAGs are likely separate species16.
To examine potential functions of the Daphnia magna microbiota, we focused on the three most complete MAGs: unknown Burkholderiaceae, 99.28% complete, 11.5X mean coverage; Pedobacter sp., 98.56% complete, 14.87X mean coverage; and Polaromonas sp., 82.73% complete, 15.3X mean coverage. We also examined the two Limnohabitans MAGs: sp1, 78.42% complete, 29.99X coverage; sp2, 60.43% complete, 14.16X coverage (Fig. 2). Though the Limnohabitans MAGs were not over 90% complete, coverage of both exceeded 30x in the juvenile D. magna samples and prior work has indicated the importance of this genus in Daphnia, suggesting unique functions may be present in these genomes. We extracted the nearest matching reference genome for each of the five MAGs above and calculated ANI for each to further investigate if these MAGs were novel or just new strains of already sequenced species (Table 2).
Anvi’o display of the two adult and two juvenile samples with the metagenome-assembled genomes highlighted in color. The innermost dendogram represents similarity among contigs based on sequence composition, GC content (green inner ring), and differential coverage (black bars across the four grey rings). The outermost layer shows the genome bins. Genome bin identification performed by GTDB-Tk.
Functional profiling of the five high- and medium-quality MAGs
Contigs from the five MAGs were analyzed for potential coding sequences (CDS) using Prokka. All identified CDS were then queried against the KEGG database and identified orthologs were grouped into KEGG functional categories (Fig. 3). We focused on pathways associated with respiration, carbohydrate metabolism, amino acid metabolism, other energy metabolism, and transport. These functional categories indicate not only general bacterial metabolism but also potential functions involved in host association. From the five MAGs’ total 7,453 CDS that mapped to KEGG pathways, 707 (9.48%) were associated with carbohydrate metabolism, 642 (8.71%) were associated with amino acid metabolism, and 358 (4.80%) were associated with other energy metabolism (Fig. 3a,b; Supplementary Fig. 3). Although C. reinhardtii has a relatively high concentration of lipids17, only 188 (2.52%) of all CDS were associated with lipid metabolism. To understand how many genes were shared among MAGs, we clustered identified genes with OrthoVenn. The two Limnohabitans MAGs shared the highest number of genes (641), and across all MAGs 325 genes were shared (Fig. 4). The Pedobacter MAG had the most unique genes in its gene set (117), while the Burkholderiaceae had so much overlap with other MAGs that it had relatively few unique genes (21).
Annotated genes in each of the five MAGs associated with specific KEGG metabolic pathways. (a) Number of genes associated with carbohydrate metabolism pathways in the five MAGs. Genes within each MAG bin were annotated with Prokka, assigned KEGG orthology, and mapped to KEGG metabolic pathways. Genes assigned to pathways were then counted. (b) Number of genes associated with KEGG amino acid metabolism pathways in the five MAGs.
Shared and unique genes with KEGG orthologs in each MAG. Unique gene sets are highlighted in gold; shared genes are in red. Intersections between MAGs, or numbers of genes in each MAG’s gene set shared with the other MAGs connected by lines, are noted in the bottom matrix. The total number of genes in each MAG is listed on the left side of the matrix.
After examining broad gene content of the five MAGs, we examined complete metabolic pathways annotated by KEGG. Here, we describe some of the complete pathways annotated, specifically focusing on pathways involved in critical functions for bacteria such as nutrient uptake and biosynthesis, as well as pathways that may be involved in host or environment interaction. A description of each MAG’s complete pathways can be found in Supplementary Materials and in Supplemental Tables 2–5. Here, we separate pathway descriptions into those encoded by all or multiple MAGs, indicating shared pathways that could indicate functional redundancy or common metabolites accessible to multiple species, and pathways uniquely encoded by single MAGs, indicating potential niche differentiation within the microbiota18. Finally, we highlight some of the functions as signals of host association, providing potential explanations for the presence of these genera in the Daphnia magna microbiota.
Shared functions of the Daphnia magna microbiota
Nutrient uptake and major biosynthesis pathways
All five MAGs shared genes involved in some key metabolic pathways (Supplementary Tables 2, 3, & 5), including those for a complete TCA cycle. Transporters for multiple TCA cycle intermediates were present across the MAGs, including a C4-dicarboxylate transport system and transporters for alpha-glucosides, malate, fumarate, 2-oxogultarate, succinate, and aspartate. Genes encoding a cytochrome c oxidase were identified, suggesting all have the capacity for aerobic respiration. All encoded for lipopolysaccharide transport and lipoprotein release. All MAGs except the Pedobacter encoded for a complete glyoxylate cycle, and the non-oxidative phase of the pentose phosphate pathway was present in all MAGs except the Limnohabitans species.
Transport and biosynthesis of other essential molecules were shared across MAGs. All MAGs except Pedobacter shared transport systems for phospholipids, phosphate, and branched-chain amino acids. All except for Limnohabitans MAG 2 encoded for the elongation step of fatty acid biosynthesis, but only the Burkholderiaceae, Polaromonas, and Pedobacter MAGs encoded for the initiation step. The Limnohabitans MAGs shared multiple sialic acid TRAP transporter genes (siaM, siaT, siaQ).
All MAGs encoded for the transport of multiple necessary vitamins, including riboflavin (B2, ribX, ribY, ribZ), pantothenate (B5, panS), and cobalamin (B12, btuB). Genes involved in the cobalamin salvage pathway were present in both Limnohabitans MAGs and in the Polaromonas MAGs (cobO, cobP). Some species were able to biosynthesize vitamins: the Pedobacter and Polaromonas MAGs encoded for biotin biosynthesis, and all MAGs except Pedobacter encoded for tetrahydrofolate biosynthesis. All MAGs encoded for a pyridoxine 5-phosphate synthase (PdxJ), indicating likely biosynthesis of pyridoxine (B6). Biosynthesis and transport of amino acids varied across MAGs, with no shared amino acid transport systems present across all species. However, all MAGs except Pedobacter encoded for arginine and ornithine biosynthesis pathways. The Burkholderiaceae, Limnohabitans 1, and Polaromonas MAGs encoded for threonine, cysteine, lysine, proline, phenylalanine, tyrosine, and glutathione biosynthesis.
Host and environment interaction
Superoxide dismutase (sodA, sodB) and catalase-peroxidase (katG) were found in all MAGs, potentially acting as defense mechanisms against host-produced reactive oxygen species. The universal minimal Tat system was encoded by all MAGs (tatA, tatC), allowing all of these species to transport folded proteins across cell membranes. Both Limnohabitans MAGs and the Polaromonas MAG encoded for complete adhesin transport systems and a gene associated with type IV pilus biosynthesis (pilQ). These MAGs also encoded two membrane proteases related to aminoglycoside resistance (HtpX, FtsH)19. All MAGs except Pedobacter encoded for the QseC-QseB quorum sensing regulatory system. Though the Polaromonas MAG was the only genome to encode for a complete flagellar assembly pathway, the Limnohabitans MAGs encoded for multiple genes involved in flagellar assembly (flhA, flhB, flgB – L). A gene for chemotaxis protein CheA was present in these three MAGs.
Some amino acid importers were present across the MAGs. These include arginine (artM), proline (proP), cystine (yecS) in Limnohabitans MAGs, glutamine (glnQ, glnM) and glutathione (gsiC) in Limnohabitans MAGs and in the Polaromonas MAG, and histidine (hisP, hisM, hisQ) in the Burkholderiaceae MAG and Limnohabitans MAG 1. It is unclear whether food (e.g., Chlamydomonas reinhardtii) or other bacteria is the source of these imported amino acids. As some MAGs are able to biosynthesize the amino acids others require, cross-feeding among bacteria here may be occurring.
Environmental stress tolerance mechanisms were shared among genomes. All encoded for the phosphate starvation response regulatory system PhoR-PhoB. The Limnohabitans MAGs and the Polaromonas MAG shared the EnvZ-OmpR osmotic stress response system, allowing the bacterial cells to respond to changes in osmolality. Limnohabitans MAG 2 and the Polaromonas MAG shared CusS-CusR, a copper tolerance regulatory system.
MAG-specific functions of the Daphnia magna microbiota
Nutrient uptake and major energy pathways
Other respiratory pathways were encoded by some MAGs. The Burkholderiaceae encoded a nitrate/nitrite transporter (narK), a respiratory nitrate reductase (narGHI), and the nitrate respiration two-component regulatory system NarX-NarL. The Polaromonas encoded for thiosulfate transport (cysT, cysW, cysP) and potential use via a subunit of the Sox complex (soxA). We found a supplementary bacteriochlorophyll biosynthesis energy pathway in the Limnohabitans MAGs (pufLM, bchY). This pathway has been documented in other Limnohabitans species20.
Differences in carbohydrate metabolism among MAGs was apparent. The Pedobacter MAG was the only high-quality genome to encode for glycolysis and glycogen biosynthesis; the Burkholderiaceae MAG for the Leloir pathway; and Limnohabitans MAG 2 for the Entner-Doudoroff pathway. The Polaromonas MAG encoded for a complete beta-oxidation system, enabling fatty acid metabolism. Different carbohydrate transporters were found in the MAGs. Limnohabitans MAG 1 uniquely encoded for the transport of glucose, mannose, and glycerol (gtsABC, malK), L-arabinose (araPQ), and for a semiSWEET general sugar transporter. The Burkholderiaceae MAG encoded for methyl-galactoside transport (mglABC) and a galactose processing pathway. The Pedobacter MAG uniquely encoded for chitin degradation to fructose-6-phosphate (chiA, chb, nagB), suggesting potential utilization of chitin.
Other pathways for vitamin and amino acid import, biosynthesis, and degradation were uniquely present in some genomes. Limnohabitans MAG 1 encoded for taurine import (tauA, tauB). This MAG also encoded for degradation of histidine to glutamate. Limnohabitans MAG 2 could transport thiamine into the cell via a putative thiamine transport system (KEGG Module M00192). Uniquely, Polaromonas could synthesize cobalamin (cobA, cobQ, cbiB, cobP, cobC), and potentially could synthesize riboflavin via the purine biosynthesis pathway.
Host and environment interaction
Multiple genes and pathways involved in antibiotic resistance and detoxification were found in the bacterial MAGs, as were a multitude of secretion and regulatory systems. Pedobacter encoded the multidrug efflux pump MdlAB/SmdAB and genes for macrolide export (macA, macB). The Polaromonas MAG encoded an AcrAB-TolC/SmeDEF efflux pump. Polaromonas was the only MAG to encode for a type I secretion system (RaxAB-RaxC). The Burkholderiaceae MAG encoded the BaeS-BaeR envelope stress response two-component regulatory system, as well an alpha-hemolysin and cyclolysin secretion system and a type IV secretion system (virB1–5, 10, 11). It also encoded for an osmotically-inducible protein, OsmY, part of an osmoprotectant ABC transporter complex, and a putrescine transporter (potFGHI).
Genomic signs of host association in the Daphnia magna microbiota
Because the Daphnia magna microbiota has been shown to differ significantly from the surrounding aquatic environment at the genus level9,21, we examined the MAGs for any potential indicators of host association. Multiple instances of host immune system evasion or tolerance were noted in the MAGs. Both Limnohabitans MAGs encoded for the stealth protein CpsY, implicated in host immune system evasion22. Genes involved in dTDP-L-rhamnose biosynthesis were present in all MAGs, indicating the potential for use of this alternative cell wall polysaccharide. L-rhamnose has been linked to bacterial viability and virulence23, and similarly structured polysaccharides have been shown to modulate host immune systems24. A gene involved in quorum quenching (signaling between other microbial species or between microbes and the host) was identified in Limnohabitans MAG 1 and the Pedobacter MAG (ytnP). Other virulence factors and regulators were identified in both Limnohabitans and the Pedobacter MAG (cvfB, bvgS/bvgA).
A potential benefit these bacterial species may provide is amino acid and vitamin biosynthesis and export for host use, as eukaryotes must acquire essential molecules from their diet or from heterotrophic microorganisms. Six amino acids have been demonstrated as essential for Daphnia magna’s close relative, Daphnia pulex: arginine, histidine, leucine, phenylalanine, isoleucine, and tryptophan25, and are likely essential for Daphnia magna. All of the these are biosynthesized by at least one of the MAGs, and some may export them as well. The Limnohabitans MAGs and the Burkholderiaceae MAG encode for an arginine exporter (argO), Limnohabitans sp. 1 encodes for a threonine exporter (rhtA), and the Burkholderiaceae MAG a threonine-serine exchanger (steT). A beneficial vitamin for Daphnia magna is cobalamin, which is not produced by Chlamydomonas reinhardtii26. However, the Polaromonas MAG encodes for cobalamin biosynthesis. Other vitamins are biosynthesized by at least one of the MAGs, including tetrahydrofolate, biotin, and pyridoxine. Supplementation of Daphnia magna growth media with biotin and cobalamin has been shown to increase host fitness27, though it is unclear if these vitamins are essential for Daphnia survival.
Other host-microbe interactions were also present in the MAGs. The Limnohabitans MAG 2 encoded for heme binding proteins and a heme transport system (ccmB, ccmC), implicated in use of host-synthesized heme28. The Pedobacter MAG encoded for an N-acetylneuraminate lyase (nanA) and all five MAGs contain genes involved in transport of sialic acids. Sialic acids are found in complex host tissue29, indicating potential cleavage of sialic acids from host cells for import and use by the bacteria. Pathways involved in host invasion and colonization were also present in the MAGs. The Polaromonas MAG encoded a suite of type IV pilus and fimbriae associated genes, including type IV pilus biogenesis factors (pilY1, pilQ), fimbrial proteins (pilE), the type IV fimbriae synthesis two-component regulatory system PilS-PilR, and a complete set of flagellar assembly genes. This MAG, along with the two Limnohabitans MAGs, also encoded for twitching motility (pilT). Both Limnohabitans MAGs encoded for swarming motility (swrC, rssA), and all three for other genes involved in chemotaxis (pctA, cheA, cheY, tar, cheB, mcp4, tsr, cheW). Though genes for adhesin production were not identified, genes involved in adhesin transport were identified in both Limnohabitans MAGs and in the Burkholderiaceae MAG (bmaC, ehaG, ata).
Discussion
To date, studies examining the Daphnia magna microbiota have only sequenced the 16S rRNA marker gene to understand broad-level interactions and functions of the microbiota. Here, we were able to assemble the first metagenome-assembled genomes from the Daphnia magna microbiota to elucidate genome-specific functions associated with highly abundant members of the bacterial community. Our 16S rRNA gene level data shows that the Daphnia magna bacterial community is structured differently than the surrounding culture environment and from the microbiome of their food, which agrees with results from earlier studies9,30. Our shotgun sequencing revealed five MAGs that were distinct from each other and distinct from their closest sequenced relatives based on average nucleotide identity. While generalizations of the importance of these species to the host may be specific to the single host genotype used in our study, Limnohabitans spp., Pedobacter spp. and other betaproteobacteria have been well-documented as abundant constituents of the Daphnia magna microbiota in laboratory cultures of different host genotypes sourced from different geographical locations and in field samples, suggesting that these are common across Daphnia magna genotypes31. We found that these MAGs were high- or medium-quality, meaning they contained some or most of the single-copy genes found in all bacteria. With metagenomic short-read shotgun sequencing it is unlikely that the MAGs assembled here contained the full gene sets associated with these species; however, bins of relatively high-quality suggest that we found many important genes within each assembled genome. Most studies of the Daphnia magna microbiota using marker-based sequencing have identified more OTUs or ASVs and higher diversity than the 12 bins we identified, likely due to the higher sequencing depth necessary for shotgun sequencing to identify rare taxa32. However, the five MAGs assembled from our shotgun sequencing are mostly consistent with genera found in 16S rRNA sequencing from other studies and from our own sequencing.
The Burkholderiales order has been demonstrated as the most abundant in the Daphnia gut and whole organism microbiota9,13, and four of the five MAGs assembled here were identified to families or genera within this order. Two Limnohabitans MAGs were assembled and found at high coverage in juveniles; this genus has been reported to be highly abundant in the Daphnia microbiota and has been implicated in increasing host fecundity12,15. Here, we show that there are two distinct Limnohabitans MAGs that encode for different metabolic pathways. Only one other study has definitively identified more than one OTU in this genus13. We also identify two other MAGs in the Burkholderiales order, including a Polaromonas species and one unclassified Burkholderiaceae. Surprisingly, we also identify a Pedobacter MAG, which has previously only been reported as a rare taxon in the Daphnia microbiota33.
Analysis of annotated genes and pathways across the five MAGs showed overlap and differences in metabolism. Key pathways such as the TCA cycle were shared across all MAGs, and multiple ABC transporters of key vitamins and amino acids were identified. Many genes encoding for the use of different carbohydrates were encoded across the MAGs. Microalgae are generally rich in carbohydrates34 and serve as a major food source for Daphnia magna35, potentially allowing these microbes easy access to myriad nutrient sources. The two Limnohabitans MAGs shared a high proportion of annotated genes, suggesting some functional similarity between them. Indeed, multiple metabolic pathways were shared between these two MAGs, including the glyoxylate cycle, bacteriochlorophyll biosynthesis, and biosynthesis of some vitamins and amino acids. The Limnohabitans MAGs also shared many genomic features with the Polaromonas MAG and the Burkholderiaceae MAG, including multiple TCA cycle intermediate importers and several transport systems. There is also potential flexibility among the taxa in encoded respiration, notably in the Polaromonas MAG’s ability to use thiosulfate and the Burkholderiaceae’s ability to utilize nitrate under hypoxic conditions. Along with the variety of two-component regulatory systems found within each MAG, the wide range of potential respiratory pathways may allow functions to sustain through different bacterial species even when stressful environments cause fluctuations in the abundance of species within the microbiota.
The difference between taxa identified in the Daphnia magna microbiota and its culture environment suggest that there are some key interactions between the host and its associated microbes in order to establish and maintain these microbial populations. Furthermore, there may be some interactions between bacterial species in the microbiota that could impact the host. We found many genes in the Limnohabitans MAGs and the Polaromonas MAG involved in flagellar assembly, type IV pilus biogenesis and production, and biofilm formation, all of which have been implicated in host colonization and successful adhesion to host-associated surfaces36,37,38. All MAGs encode for l-rhamnose production, which has been implicated in adhesion to other cells23. Genes in secretion systems implicated in host cell adhesion, particularly Type I, II, and IV were also encoded39. We found many genes involved in host immune system evasion or modification, which may allow these bacteria to persist within the host species40. Notably, superoxide dismutase and catalase were encoded by multiple MAGs, suggesting the bacteria could defend against radical oxygen species produced by the host as a defense mechanism41. Also present across MAGs are genes involved in the detoxification of antibiotics and toxins, including multidrug efflux transporters and pumps (MdlAB/SmdAB, AcrAB-TolC/SmeDEF), macrolide export (macA, macB), and stress tolerance to antibiotics (BaeS-BaeR).
Biosynthesis and provision of amino acids by bacteria to their host is a well-documented set of interactions that is known to confer fitness benefits to the host42,43,44. Here, we find that the Limnohabitans MAGs and the Burkholderiaceae MAG encode for the export of arginine and threonine, essential amino acids for the host Daphnia25. Similarly, biosynthesis or metabolism of vitamins and minerals by bacteria and provisioning to the host has been well-documented in the microbiota of other organisms45,46,47. Many genes involved in vitamin B biosynthesis were found across all MAGs. Media supplemented with cobalamin (B12) is used to successfully culture Daphnia magna48, and we find the Polaromonas MAG encoded for a complete cobalamin biosynthesis pathway, suggesting potential vitamin provisioning to the host from this species. We also find a potential microbe-microbe interaction, where the Pedobacter MAG encodes for cleavage of sialic acids from host tissue, where it can then be transported and utilized by other species as a carbohydrate source via a sialic acid TRAP transporter. The breakdown of sialic acid to metabolites that are accessible to the host by microbiome-associated species has been shown to increase host fitness49. If Limnohabitans are able to use sialic acid as a nutrient source, this may be the basis for a microbe-host-microbe interaction, where Limnohabitans provides essential amino acids to the host using energy generated from metabolism of molecules provisioned from the host.
In total, our data shows that there is much versatility in metabolism among the MAGs, but some overlap in function. As Daphnia magna are indiscriminate filter feeders, they may feed on a wide variety of particulate matter with variable nutrient profiles. The versatility in metabolism encoded by these MAGs indicates that they are able to utilize this unpredictable range of nutrients both in the digestive tract and on the carapace. The specific functions of certain MAGs, particularly in amino acid and vitamin biosynthesis and export, seem critical in providing nutritional benefits to the host zooplankton.
Conclusions
Daphnia magna is an important model system for multiple facets of ecology, and has recently become an organism of interest for understanding fundamental questions about the microbiota. Our metagenomic sequencing and subsequent analysis characterizes the Daphnia magna microbiota to the species level and finds some genomic features that allow core bacterial species to acquire and biosynthesize nutrients, and to potentially interact with their host via amino acid and vitamin export. By examining this relatively simple microbiota via MAGs, we can begin to investigate metabolic interactions between the host and its associated microbes. Future work to further elucidate functions of these MAGs will involve long-read metagenome sequencing to complete genome assemblies and pure, single-isolate sequencing to understand strain variation within the microbiota. Furthermore, transcriptomics and metabolomics could be used to understand which of these encoded genes are functioning under different environmental and host conditions, and will direct future hypotheses on host-microbiota interactions. For example, how much of the differences in Daphnia life history and population dynamics across food environments50 can be attributed to differences in microbiota composition? These results will help to inform future work studying the effects of the microbiota on host health and population dynamics across ecological contexts. Moreover, as more populations of Daphnia and their microbiota are sequenced, it will become possible to examine the coevolutionary relationships between hosts and microbiota, and this functional information will be essential for making sense of those relationships.
Methods
Sample collection and extraction
Daphnia magna clone 8A, isolated from a pond at Kaimes Farm, Leitholm, Scottish Borders51, was used in this sequencing effort. Two samples of 100 21-day-old adult Daphnia magna and two samples of 6-day-old juvenile Daphnia were collected from laboratory cultures of between 20–40 individuals maintained in 400 mL jars in defined COMBO medium48. Laboratory cultures were fed a standardized volume calculated with the Biotek Epoch Microplane Spectrophotometer of green algae Chlamydomonas reinhardtii (CPCC 243) to provide 0.25 mg C/ml/day. Laboratory cultures were maintained in a 19 °C controlled environment under a 16 h:8 h light:dark cycle. Samples were immediately ground after collection using in sterile 1.5 mL microcentrifuge tubes. To separate bacterial cells from host cells, a modified protocol from Benson et al. was used52. Samples were suspended in 2 mL PBE buffer and layered on a cushion of 50% sucrose, then centrifuged at 4,000 g for 10 minutes. DNA was extracted from the pellets at the base of the sucrose fractions following the Qiagen DNEasy Blood & Tissue Kit spin-column protocol of total DNA from animal tissues (Qiagen, Hilden, Germany).
Library preparation and sequencing
Shotgun sequencing libraries from the two adult samples and two technical replicates of one juvenile sample were prepared and multiplexed using the Illumina Nextera XT kit and protocol. Input DNA was quantified using the Qubit dsDNA system. Libraries were checked using the Agilent Technology 2100 Bioanalyzer. Libraries were manually normalized due to a final library yield under 10 nM. Paired-end sequencing was performed on an Illumina MiSeq using a MiSeq Reagent Kit V2.
Quality filtering and metagenome assembly
Reads were demultiplexed using the built-in Illumina MiSeq Reporter. Quality of demultiplexed reads was checked using FastQC v0.11.5. Reads were trimmed using Trimmomatic v0.3653 with the commands: ILLUMINACLIP:NexteraPE-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:25 MINLEN:36 to remove adapter sequences and to remove segments of reads where quality fell below 25. Trimmed reads were mapped to the Daphnia magna draft genome54 using BWA55, Samtools56, and BEDtools57, and reads with greater than 80% identity over 50% of the read were filtered. Remaining paired unmapped reads were assembled de novo using metaSPAdes in SPAdes v3.1158. A co-assembly of all four samples, a co-assembly of the adult samples, a co-assembly of the juvenile samples, and individual assemblies for each sample were created.
Taxonomy-independent sequence binning and identification of individual MAGs
To resolve genomes of identified organisms and to discover genomes of organisms not present in read-based taxonomy identification programs or entirely new organisms, contigs from the master co-assembly with reads mapped from each sample were binned using CONCOCT within Anvi’o59, accepting contigs over 2500 bp in length. Contigs greater than 20 kb in length were split into 20 kb fragments prior to running CONCOCT. Bins from the master co-assembly were assessed using Anvi’o, using Parks et al.’s quality score (genome completeness - 5x estimated redundancy or contamination) of ≥50 as a cutoff for further refinement59. Bins meeting this quality cutoff were manually curated within Anvi’o, where contigs within a bin that deviated dramatically from the mean GC content or mean coverage of the bin were removed from the bin. Bins that increased completeness when merged, did not increase redundancy above 10%, and were similar in GC content were merged. Bins that did not have high (>90% completion) or medium quality (>50% completion) after merging and refining were not analyzed further. After merging and refining, bins that still met the quality score cutoff were assigned taxonomy using GTDB-Tk60,61,62. GTDB-Tk uses average nucleotide identity and genome topology to find the closest genomic relative in its database. The same process was repeated for the juvenile co-assembly and the adult co-assembly to confirm species presence and attempt to resolve species identity at different host life stages. Similarity between MAGs was calculated using the average nucleotide identity tool in Pyani63.
Read-based taxonomic classification
Kaiju v1.564, Kraken v1.065, and MetaPhlAn2 v2.666 were used to assign taxonomy to reads. We used all three identifiers due to due to Kaiju’s high rate of false identification67 and MetaPhlAn2’s use of specific marker genes from reference organisms rather than entire genomes. All programs were used with their built-in databases. Kraken results were confirmed via cross-comparison of abundant species with both Kaiju and MetaPhlAn2. All three taxonomy profilers were also used to assign taxonomy to contigs assembled from each sample. Visualization of each sample’s community composition was performed in R.
Functional profiling
Contigs from each MAG and from the adult, juvenile, and master co-assemblies were annotated using Prokka v1.1268. Genes annotated in Prokka were assigned KOs (KEGG Orthologs) using the GhostKOALA tool on the Kyoto Encyclopedia of Genes and Genomes69. KOs identified from GhostKOALA were mapped to standard KEGG categories and metabolic pathways using the KEGG Pathway Mapper & KEGG Module tools to examine pathway completeness and identify pathways of interest70. Genes identified using KEGG and GhostKOALA in each MAG were compared using OrthoVenn71. Overlapping and unique orthologs were compared using custom R scripts and with the ‘UpSetR’ package72.
16S rRNA gene sequencing and identification of contaminant taxa
The V4 hypervariable region of the 16S rRNA gene was sequenced on the Illumina MiSeq using a MiSeq Reagent Kit V2 and the same Qiagen DNEasy Blood & Tissue Kit and reagents as in the shotgun sequencing sample processing. Four samples of five adult Daphnia magna were sequenced to compare community composition found in 16S sequencing to that found in shotgun sequencing, along with four samples of the COMBO media Daphnia magna cultures are raised in, four samples of Chlamydomonas reinhardtii, and two samples of the DNA sequencing kit and library preparation kit as negative controls. Paired-end reads were analyzed in R using the ‘dada2’ package to trim primer sequences, identify amplicon sequence variants, and assign taxonomy73. Taxonomy was assigned using the RefSeq + RDP taxonomic training data set formatted for dada274. Further analysis of community composition and visualization were carried out using the ‘phyloseq’ package in R75.
Data availability
The datasets supporting the conclusions of this article are available at the NCBI BioProject Portal under IDs PRJNA543317 (shotgun sequences) and PRJNA543842 (16S rRNA sequences). Raw metagenomic sequencing reads from each sample are deposited under accession numbers SAMN11660785 and SAMN11660786. 16S rRNA sequencing data can be found under accession numbers SAMN11784745 - SAMN11784837. All scripts for data analysis and visualization are available on GitHub (https://github.com/reillyowencooper/daphnia_magna_metagenome).
References
Hacquard, S. et al. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe 17, 603–616 (2015).
Wahlström, A., Sayin, S. I., Marschall, H. & Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 24(1), 41–50 (2016).
Sansone, C. L. et al. Microbiota-Dependent Priming of Antiviral Intestinal Immunity in Drosophila. Cell Host Microbe 18, 571–581 (2015).
Kaltenpoth, M., Göttler, W., Herzner, G. & Strohm, E. Symbiotic Bacteria Protect Wasp Larvae from Fungal Infestation. Curr Biol. 15, 475–479 (2005).
Spor, A., Koren, O. & Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290 (2011).
Ebert, D. A Genome for the Environment. Science 331, 539–540 (2011).
Qi, W., Nong, G., Preston, J. F., Ben-Ami, F. & Ebert, D. Comparative metagenomics of Daphnia symbionts. BMC Genom. 10, 21 (2009).
Eckert, E. M. & Pernthaler, J. Bacterial epibionts of Daphnia: a potential route for the transfer of dissolved organic carbon in freshwater food webs. ISME J. 8, 1808–1819 (2014).
Freese, H. M. & Schink, B. Composition and Stability of the Microbial Community inside the Digestive Tract of the Aquatic Crustacean Daphnia magna. Microb Ecol. 62, 882–894 (2011).
Sison-Mangus, M. P., Mushegian, A. A. & Ebert, D. Water fleas require microbiota for survival, growth and reproduction. ISME J. 9, 59–67 (2015).
Macke, E., Callens, M., Meester, L. D. & Decaestecker, E. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat Commun. 8, 13 (2017).
Peerakietkhajorn, S., Tsukado, K., Kato, Y., Matsuura, T. & Watanabe, H. Symbiotic bacteria contribute to increasing the population size of a freshwater crustacean, Daphnia magna. Environ. Microbiol. Rep. 7, 364–372 (2015).
Motiei, A. et al. Disparate effects of antibiotic-induced microbiome change and enhanced fitness in Daphnia magna. Preprint at, https://doi.org/10.1101/586669 (2019).
Callens, M., Hajime, W., Yasuhiko, K., Miura, J. & Decaestecker, E. Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia. Microbiome 6, 56 (2018).
Peerakietkhajorn, S., Kato, Y., Kasalicky, V., Matsuura, T. & Watanabe, J. Betaproteobacteria Limnohabitans strains increase fecundity in the crustacean Daphnia magna: symbiotic relationship between major bacterioplankton and zooplankton in freshwater ecosystem. Environ Microbiol. 18, 2366–2374 (2016).
Richter, M. & Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106, 19126–19131 (2009).
Légeret, B. et al. Lipidomic and transcriptomic analyses of Chlamydomonas reinhardtii under heat stress unveil a direct route for the conversion of membrane lipids into storage lipids. Plant Cell Environ. 39, 834–847 (2016).
Hernandez-Agreda, A., Gates, R. D. & Ainsworth, T. D. Defining the Core Microbiome in Corals’ Microbial Soup. Trends Microbiol. 25, 125–140 (2017).
Hinz, A., Lee, S., Jacoby, K. & Manoil, C. Membrane Proteases and Aminoglycoside Antibiotic Resistance. J Bacteriol. 193, 4790–4797 (2011).
Kasilicky, V. et al. Aerobic Anoxygenic Photosynthesis Is Commonly Present within the Genus. Limnohabitans. Environ Microbiol. 84, 13 (2018).
Mariadassou, M., Pichon, S. & Ebert, D. Microbial ecosystems are dominated by specialist taxa. Ecol Lett. 18, 974–982 (2015).
Sperisen, P., Schmid, C. D., Bucher, P. & Zilian, O. Stealth Proteins: In Silico Identification of a Novel Protein Family Rendering Bacterial Pathogens Invisible to Host Immune Defense. PLOS Comput Biol. 1, 0492–0499 (2005).
Mistou, M. Y., Sutcliffe, I. C. & van Sorge, N. M. Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol Rev. 40, 464–479 (2016).
Fanning, S. et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA 109, 2108–2113 (2012).
Fink, P., Pfilitsch, C. & Marin, K. Dietary Essential Amino Acids Affect the Reproduction of the Keystone Herbivore Daphnia pulex. PLoS ONE 6, 8 (2011).
Helliwell, K. E., Wheeler, G. L., Leptos, K. C., Goldstein, R. E. & Smith, A. G. Insights into the Evolution of Vitamin B12 Auxotrophy from Sequenced Algal Genomes. Mol Biol Evol. 28, 2921–2933 (2011).
Elendt, B. P. & Bias, W. R. Trace nutrient deficiency in Daphnia magna cultured in standard medium for toxicity testing. Effects of the optimization of culture conditions on life history parameters of D. magna. Water Res. 24, 1157–1167 (1990).
Tong, Y. & Guo, M. Bacterial heme-transport proteins and their heme-coordination modes. Arch Biochem Biophys. 481, 1–15 (2009).
Severi, E., Hood, D. W. & Thomas, G. H. Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822 (2007).
Callens, M. et al. Food availability affects the strength of mutualistic host-microbiota interactions in Daphnia magna. ISME J. 10, 911–920 (2016).
Sullam, K. E., Pichon, S., Schaer, T. M. M. & Ebert, D. The Combined Effect of Temperature and Host Clonal Line on the Microbiota of a Planktonic Crustacean. Microb Ecol. 76, 506–517 (2018).
Ranjan, R., Rani, A., Metwally, A., McGee, H. S. & Perkins, D. L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophysis. Res Commun. 469, 967–977 (2016).
Poehlein, A., Daniel, R. & Simeonova, D. D. Genome sequence of Pedobacter glucosidilyticus DD6b, isolated from zooplankton Daphnia magna. Stand Genomic Sci. 10, 100 (2015).
Work, V. H. et al. Increased Lipid Accumulation in the Chlamydomonas reinhardtii sta7-10 Starchless Isoamylase Mutant and Increase Carbohydrate Synthesis in Complemented Strains. Eukaryot Cell 9, 1251–1261 (2010).
Taipale, S. J. et al. Terrestrial carbohydrates support freshwater zooplankton during phytoplankton deficiency. Sci Rep 6, 30897, https://doi.org/10.1038/srep30897 (2016).
Latasa, C. et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mo Microbiol. 58, 1322–1339 (2005).
Rossez, Y., Wolfson, E. B., Holmes, A., Gally, D. L. & Holden, N. J. Bacterial Flagella: Twist and Stick, or Dodge across the Kingdoms. PLoS Pathog. 11, 1004483, https://doi.org/10.1371/journal.ppat.1004483 (2015).
McLoughlin, K., Schluter, J., Rakoff-Nahoum, S., Smith, A. L. & Foster, K. R. Host Selection of Microbiota via Differential Adhesion. Cell Host Microbe 19, 550–559 (2016).
Kline, K. A., Fälker, S., Dahlberg, S., Normark, S. & Henriques-Normark, B. Bacterial Adhesins in Host-Microbe Interactions. Cell Host Microbe 5, 580–592 (2009).
Foster, T. J. Immune evasion by Staphylococci. Nat Rev Microbiol. 3, 948–958 (2005).
Reveillaud, J., Anderson, R., Reves-Sohn, S., Cavanaugh, C. & Huber, J. A. Metagenomic investigation of vestimentiferan tubeworm endosymbionts from Mid-Cayman Rise reveals new insights into metabolism and diversity. Microbiome 6, 19 (2018).
Hansen, A. K. & Moran, N. A. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA 108, 2849–2854 (2011).
Pérez-Brocal, V. et al. A Small Microbial Genome: The End of a Long Symbiotic Relationship? Science 314, 312–313 (2006).
Douglas, A. E. The microbial dimension in insect nutritional ecology. Funct Ecol. 23, 38–47 (2011).
Degnan, P. H., Taga, M. E. & Goodman, A. L. Vitamin B12 as a Modulator of Gut Microbial Ecology. Cell Metab. 20, 769–778 (2014).
Smith, T. A., Driscoll, T., Gillespie, J. J. & Raghavan, R. A. Coxiella-Like Endosymbiont Is a Potential Vitmain Source for the Lone Star Tick. Genome Biol Evol. 7, 831–838 (2015).
McKenney, E. A., Koelle, K., Dunn, R. R. & Yoder, A. D. The ecosystem services of animal microbiomes. Mol Ecol. 27, 2164–2172 (2018).
Kilham, S. S., Kreeger, D. A., Lynn, S. G., Goulden, C. E. & Herrera, L. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377, 147–159 (1998).
Castanys-Muñoz, E., Martin, M. J. & Vazquez, E. Building a Beneficial Microbiome from Birth. Adv Nutr. 7, 323–330 (2016).
Nelson, W. A., McCauley, E. & Wrona, F. J. Multiple dynamics in a single predator-prey system: experimental effects of food quality. Proc R Soc Lond B. 268, 1223–1230 (2001).
Auld, S. K. J. R., Wilson, P. J. & Little, T. J. Rapid change in parasite infection traits over the course of an epidemic in a wild host-parasite population. Oikos 123, 232–238 (2014).
Benson, A. K. et al. Microbial successions are associated with changes in chemical profiles of a model refrigerated fresh pork sausage during an 80-day shelf-life study. Appl Environ Microbiol. 80, 5178–5194 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Daphnia Genomics. Genome Informatics Lab, Indiana University Biology Department, http://wfleabase.org. Accessed 02 March 2018.
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at, https://arxiv.org/abs/1303.3997 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Eren, A. M. et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, 1319, https://doi.org/10.7717/peerj.1319 (2015).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2, 1533–1542 (2017).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 36, 996–1004 (2018).
Chaumeil, P. A., Hugenholtz, P. & Parks, D. H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Available at, https://github.com/Ecogenomics/GTDBTk (2019).
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, G. J. & Toth, I. K. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8, 12–24 (2016).
Menzel, P., Ng, K. L. & Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 7, 11257, https://doi.org/10.1038/ncomms11257 (2016).
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9, 811–814 (2012).
Piro, V. C., Matschkowski, M. & Renard, B. Y. MetaMeta: integrating metagenome analysis tools to improve taxonomic profiling. Microbiome 5, 101 (2017).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol. 428, 726–731 (2016).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Wang, Y., Coleman-Derr, D., Chen, G. & Gu, Y. Q. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43, W78–W84 (2015).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583 (2016).
Alishum, A. DADA2 formatted 16S rRNA gene sequences for both bacteria & archaea. Version 1 Refseq+RDP. Available at, https://doi.org/10.5281/zenodo.2541239 (2019).
McMurdie, P. J. & Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 8, 61217, https://doi.org/10.1371/journal.pone.0061217 (2013).
Acknowledgements
We are grateful for Dr. Andrew K. Benson, Mallory Van Haute, and Qinnan Yang’s assistance with sequencing preparation and machine use, and to Brady Bathke for assistance with bioinformatic analyses. This work was completed utilizing the Holland Computing Center of the University of Nebraska, which receives support from the Nebraska Research Initiative. This work was supported by funds from the University of Nebraska and a Maude Hammond Fling Faculty Research Fellowship to CEC.
Author information
Authors and Affiliations
Contributions
R.O.C. and C.E.C. designed the study. R.O.C. collected and analyzed the metagenomic data. R.O.C. wrote the first draft of the manuscript, and R.O.C. and C.E.C. revised the manuscript and approved its final form.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cooper, R.O., Cressler, C.E. Characterization of key bacterial species in the Daphnia magna microbiota using shotgun metagenomics. Sci Rep 10, 652 (2020). https://doi.org/10.1038/s41598-019-57367-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-57367-x
This article is cited by
-
Dynamics of Gut Bacteria Across Different Zooplankton Genera in the Baltic Sea
Microbial Ecology (2024)
-
Natural populations of the putative ancient asexual Darwinula stevensoni (Crustacea, Ostracoda) differ in their microbiomes
Hydrobiologia (2023)
-
Microplastic exposure across trophic levels: effects on the host–microbiota of freshwater organisms
Environmental Microbiome (2022)
-
Disentangling Diet- and Medium-Associated Microbes in Shaping Daphnia Gut Microbiome
Microbial Ecology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.