Abstract
The transcription factor Pax6 is crucial for the development of the central nervous system, eye, olfactory system and pancreas, and is implicated in human disease. While a single Pax6 gene exists in human and chicken, Pax6 occurs as a gene family in other vertebrates, with two members in elephant shark, Xenopus tropicalis and Anolis lizard and three members in teleost fish such as stickleback and medaka. However, the complement of Pax6 genes in jawless vertebrates (cyclostomes), the sister group of jawed vertebrates (gnathostomes), is unknown. Using a combination of BAC sequencing and genome analysis, we discovered three Pax6 genes in lampreys. Unlike the paired-less Pax6 present in some gnathostomes, all three lamprey Pax6 have a highly conserved full-length paired domain. All three Pax6 genes are expressed in the eye and brain, with variable expression in other tissues. Notably, lamprey Pax6α transcripts are found in the pancreas, a vertebrate-specific organ, indicating the involvement of Pax6 in development of the pancreas in the vertebrate ancestor. Multi-species sequence comparisons revealed only a single conserved non-coding element, in the lamprey Pax6β locus, with similarity to the PAX6 neuroretina enhancer. Using a transgenic zebrafish enhancer assay we demonstrate functional conservation of this element over 500 million years of vertebrate evolution.
Similar content being viewed by others
Introduction
Pax6 is an evolutionarily conserved, pleiotropic transcription factor with key roles during embryonic and postnatal development as well as in adult tissue maintenance. In both vertebrates and invertebrates Pax6 acts as a master regulator controlling multiple genetic networks that drive differentiation and cell type specification. In vertebrates Pax6 is essential for proper development of the central nervous system (CNS), the eye, and the olfactory system1,2,3,4. It is also crucial for the development of the pancreas and subsequent insulin production from its endocrine secretory cells5,6,7,8. A correct dosage of Pax6 is essential for proper eye development. Haploinsufficiency leads to the congenital eye malformation aniridia in humans and the small eye mutation in mice, whereas loss of both alleles causes a complete lack of eye development and results in congenital lethality9. Pax6 is expressed in all tissues of the embryonic eye. Spatio-temporally restricted ablation of the gene in conditional mouse mutants has revealed tissue-specific requirements in lens and retina development10,11,12. Interestingly, while human, mouse and chicken possess a single Pax6 gene (Pax6.1), other gnathostomes such as elephant shark (Callorhinchus milii), Xenopus tropicalis and Anolis lizard contain two Pax6 genes13, known as Pax6.1 and Pax6.2 (the latter is also referred to as Pax1014). The two genes in the latter taxa have been attributed to two rounds of whole-genome duplication (WGD), commonly referred to as 2 R, that occurred at the base of vertebrates15,16, followed by secondary loss of two paralogs13. One more gene was subsequently lost in the lineage leading to mammals and birds. A third Pax6 paralog is present in many teleost fish such as fugu (Takifugu rubripes), medaka (Oryzias latipes) and stickleback (Gasterosteus aculeatus), where, as a result of a further teleost-specific WGD and subsequent separate gene losses, a variable complement of Pax6 paralogs is present in contemporary lineages13,14. However, the number of Pax6 genes in cyclostomes, the jawless vertebrates, is currently unknown.
Inter-species sequence comparisons of the Pax6 genomic region have aided in the identification of a large number of enhancer elements, revealing a highly complex cis-regulatory landscape surrounding the gene. The importance of these cis-regulatory elements (CREs) is exemplified by a subset of aniridia patients in whom, even though the PAX6 sequence itself remains intact, regulation of the gene is disrupted due to nearby chromosomal breaks17. A large number of these CREs can be traced back to the common ancestor of gnathostomes, as shown by their presence in the elephant shark Callorhinchus milii13,18. In this study, we traced back further in evolution and investigated the presence of Pax6 genes and their regulatory landscapes in cyclostomes.
Cyclostomes are the sister group of the gnathostomes. They split from the jawed vertebrates very early during vertebrate evolution, estimated to be around 500 million years ago. Cyclostomes are a monophyletic group19 whose only extant members are the lampreys and hagfishes. As these species represent the earliest branching lineage of the vertebrates, they form a key resource for understanding the molecular events that occurred during the early evolution of vertebrates. Evaluation of the gene content in lampreys and hagfishes might therefore shed light on the pattern of gene and genome duplications in early vertebrates. However, the interpretation of the findings might be complicated due to recent reports suggesting a third round of WGD in lampreys20, although evidence from another study instead points towards a scenario of segmental duplications of several, but not all, genomic loci21,22.
In all analysed gnathostome species Pax6 exhibits a highly tissue-specific expression pattern. Control of Pax6 gene expression in lamprey is of interest from an eye evolution perspective as the eyes of adult lampreys are already similar to jawed-vertebrate eyes, possessing a lens, iris and three-layered retina. On the other hand, the hagfish eye appears more basic and lacks a proper lens and cornea, and has a simpler retina. This is outwardly similar to the simpler eyes found in the larval stage of lampreys, before they dramatically metamorphose into the adult form23. As Pax6 is a master control gene for eye development, with its strict spatio-temporal expression pattern controlled by several highly conserved CREs, we were curious to investigate the evolutionary origin of eye-specific enhancers. Pax6 also plays a key role in the development and maintenance of the endocrine pancreas5,7,8, a vertebrate-specific organ which exists in a simple form in cyclostomes where it is usually referred to as the islet organ24. Whereas in hagfish the islet organ is represented by scattered follicles, lampreys have a discrete islet organ associated with the gut. We were therefore interested to investigate whether Pax6 expression is associated with pancreas development in lampreys.
We had previously generated a whole-genome sequence for the Japanese lamprey (Lethenteron japonicum aka Lethenteron camtschaticum)20. Using a combination of whole-genome sequence analysis and sequencing of BAC clones we identified three Pax6 paralogs in the Japanese lamprey genome and designated them as Pax6α, Pax6β and Pax6γ. Analysis of the recently published germline genome assembly of the sea lamprey (Petromyzon marinus) showed that the three Pax6 genes are also present in this lamprey22. A search for ancient conserved noncoding elements (aCNEs) identified a single element in the Japanese lamprey Pax6β locus. In contrast, no CNEs are detected in sequence comparisons between vertebrate and Amphioxus Pax6 loci. Using a transgenic zebrafish reporter assay, we show that the conserved lamprey element is capable of driving specific expression in the neuroretina, revealing a remarkable functional conservation dating back to the origin of vertebrates.
Results
Pax6 genes in the Japanese lamprey genome
In order to identify Pax6 genes in Japanese lamprey we searched its germline genome assembly20 using known Pax6 protein sequences as TBLASTN queries. These searches identified three distinct Pax6 gene fragments, present on scaffold 194 (1,091,192 bp), scaffold 23 (4,607,062 bp), and a third one distributed across multiple smaller scaffolds (scaffolds 7303, 22485, 72958, 1356, 12381 and 20282) (Fig. 1a). To obtain contiguous sequence for each of these loci, we used the identified scaffolds to design probes for screening of Japanese lamprey BAC libraries. We identified several BACs, of which three were sequenced completely. One of these (LJT240I23) covers the first Pax6 locus (scaffold 194) whereas the remaining two (LJT73L19 and LJT210A8) are overlapping BACs covering the second Pax6 locus (scaffold 23). We were unable to identify a BAC clone for the third Pax6 locus. To obtain contiguous sequence for the third Pax6 locus we used a combination of RT-PCR and RACE to identify and orient scaffolds belonging to the same Pax6 gene and used genomic PCR to fill the intervening gaps. Synteny and sequence analysis confirmed that these three genes are distinct Pax6 genes showing some level of conserved synteny with human and elephant shark Pax6 gene loci (Fig. 1b). Since phylogenetic analysis was unable to assign clear orthology of the three lamprey genes to specific Pax6 family members in gnathostomes (see Phylogenetic analysis section), we named the three Japanese lamprey Pax6 genes as Pax6α, Pax6β and Pax6γ. Lamprey Pax6β had been sequenced and characterized in a previous study25,26.
The lamprey genome contains three Pax6 genes. (a) The three Pax6 loci in the Japanese lamprey, L. japonicum. Lamprey Pax6 genes are named Pax6α, Pax6β and Pax6γ. The BAC clones sequenced are shown below. LjPax6γ resides on a short (66 kb) scaffold containing no other genes. (b) Gene synteny comparison of the three Japanese lamprey (Jlamprey) and sea lamprey Pax6 loci with Pax6 loci from selected gnathostomes.
Pax6 is a transcription factor containing two distinct DNA-binding domains: the 128 amino acid long paired domain located at the N-terminal and the more centrally located homeo-domain. These are connected via a linker region while the C-terminus harbours a PST-rich transactivation domain3. All three lamprey Pax6 protein sequences showed very high sequence similarity with each other as well as with the human PAX6 over the paired- and homeo-domains (Fig. 2a), with lower similarity in the linker region and higher similarity again in the PST-domain. Alignment of the lamprey Pax6 protein sequences revealed 79.7% identity between Pax6α and Pax6β, 58.3% identity between Pax6α and Pax6γ and 59.4% between Pax6β and Pax6γ.
Protein sequences and expression patterns of the Japanese lamprey Pax6 genes. (a) Comparison of the amino acid sequences of LjPax6α, LjPax6β and LjPax6γ with human (Hs) PAX6. All three LjPax6 genes encode a highly conserved paired domain (solid black line), in contrast to known Pax6.2 (also referred to as Pax1014) genes which lack the sequences coding for this domain, as well as a highly conserved homeodomain (dotted grey line) and C-terminal transactivation domain. No evidence was found for the presence of the alternative exon 5a in the lamprey genes. The positions of the exon boundaries (black arrow head, phase 0 intron; open arrow head, phase 1 intron) are conserved between the human and lamprey genes. (b) qRT-PCR analysis using a panel of adult lamprey tissues showing the tissue-specific expression pattern of the LjPax6 genes. All three genes are highly expressed in the eye and brain, with lower and variable expression in other tissues.
Gene structure of the Japanese lamprey Pax6 genes
All three lamprey Pax6 genes contain both the paired- and homeo-domains. This contrasts with gnathostome species that carry multiple Pax6 genes in their genome, in which the Pax6.2 gene lacks the N-terminal paired domain13. The intron-exon structure of the three genes is also fully conserved between the lamprey genes and the canonical human PAX6 gene (Fig. 2a; Fig. S1). Both lamprey Pax6α and Pax6γ have only a few amino acids in their first coding exon before the start of the paired domain. This is equivalent to mammalian Pax6 where the first coding exon (exon 4) encodes just three/four amino acids with the paired domain being encoded by exons 5, 6 and part of 7 (Fig. 2a).
Absence of exon 5a in Japanese lamprey Pax6 genes
Two major isoforms of the full-length Pax6 exist in tetrapods, teleost fishes and elephant shark. These isoforms, called Pax6 and Pax6(5a), differ by a stretch of 12 to 14 amino acids which are present in the latter isoform as an insertion in the PAI subdomain within the paired domain and change its binding site recognition characteristics27,28. A study searching EST and other databases for the Pax6(5a) isoform had previously found this isoform only in gnathostome species29. To check for the potential presence of this alternative exon in the Japanese lamprey Pax6 genes we closely inspected the genomic sequence between exons 5 and 6 in the three Pax6 genes. However, we were unable to find any sequence homologous to exon 5a in any of the three Pax6 genes (Fig. 2a). RT-PCR products from the three genes also lack the bases coding for this exon. It is therefore most likely that exon 5a is an innovation that is specific to the gnathostomes.
Pax6 genes in the sea lamprey genome
Following the recent completion of the germline genome assembly of the sea lamprey22, we also searched its genome sequence for Pax6 genes by TBLASTN using Japanese lamprey and other representative Pax6 protein sequences as query. These two lamprey species are estimated to have diverged about 10 to 40 million years ago30. We identified three Pax6 genes in the sea lamprey that are homologous to the Pax6 genes of Japanese lamprey (Fig. 1b). They are present on scaffold_2: 14371676-14388243 (Pax6α), scaffold_14: 14097046-14127107 (Pax6β) and scaffold_55: 4629581-4632980 (Pax6γ). Some exons of Pax6γ are found on a short scaffold (scaffold_699, 41.7 kb). Since these genes are either incomplete or contain frame shifts and other errors, we could not predict reliable full-length protein sequences for them.
Synteny relationships
Next we compared the synteny of genes at the three Japanese lamprey Pax6 loci with those from sea lamprey and representative gnathostomes (Fig. 1b). Lamprey Pax6α is flanked by Eif3m and Caluα at the 5′ end and Elp4 at the 3′ end, similar to the gnathostome Pax6.1 (e.g. human PAX6) synteny region. The lamprey Pax6β locus contains Caluβ and Fbxo47 genes. Linkage with Calu is seen for Pax6.2 in a number of gnathostome species such as lizard, Xenopus, zebrafish and elephant shark, but only in the latter is the Fbxo47 gene found between Pax6.2 and Calu. None of the immediate flanking genes in the sea lamprey Pax6γ locus (Fig. 1b) are conserved in any of the gnathostome Pax6 loci.
Expression pattern of lamprey Pax6 genes
Pax6 genes exhibit a strictly defined expression pattern9,17,31, with major sites of expression seen in specific areas of the developing eye and central nervous system (CNS). To obtain some initial insight into the expression patterns of the lamprey Pax6 genes and to assess if the three genes would be distinguished by differences in their levels and tissue-specificity of expression we performed qRT-PCR analysis in a number of tissues. Consistent with other vertebrates, strong expression of all three lamprey genes was found in the eye, and at much lower levels also in brain tissue (Fig. 2b). In addition, expression of lamprey Pax6α and Pax6β was observed in the ovary, whereas Pax6β and Pax6γ expression was seen in a number of other tissues: kidney, intestine and ovary for Pax6β; and skin for Pax6γ (Fig. 2b). Previous in situ hybridization studies had shown that Japanese lamprey Pax6β is expressed in the eye, the nasohypophysial plate, the oral ectoderm and the fore- and hindbrain of embryos25,26.
We had previously generated RNA-seq data from the pancreas of juvenile (8 to 11 cm long) brook lamprey (Lampetra planeri) (GenBank accession number PRJNA369595)32. The pancreas in lampreys exists as a collection of islet-like cells known as the islet organ. We demonstrated expression of Pdx, insulin, glucagon, Slc2a2, Foxa2, Hnf1a, Neurod1 and Prkacb in these cells, key factors involved in pancreas development and insulin secretion in gnathostomes. A TBLASTN search of the assembled transcripts using our newly identified Japanese lamprey Pax6 protein sequences revealed two transcripts for Pax6α with TPM values of 0.08 and 0.22, respectively (see Supplementary Information) but not for Pax6β and Pax6γ. This indicates that Pax6α is specifically expressed in the lamprey pancreas and suggests that Pax6α was co-opted into the pancreas developmental network early during vertebrate evolution.
Phylogenetic analysis
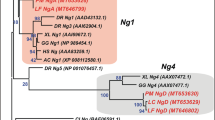
To gain better insight into the relationship between the three lamprey Pax6 genes and the members of the Pax6, Pax4 and Pax3/7 gene families from other chordates, we performed phylogenetic analysis using the Maximum Likelihood method. The ML tree showed that the three lamprey genes cluster with Pax6 genes from other chordates distinct from the Pax4 clade indicating that they are indeed Pax6 genes (Fig. 3). However, the three lamprey Pax6 genes clustered outside of the gnathostome Pax6 clade (Fig. 3), a pattern previously observed for other lamprey gene families such as KCNA, Hox, Runx, and p5320,33,34,35, due to the exceptionally high GC-content in their coding regions that is peculiar to lampreys. The exclusive clustering of the lamprey genes outside the gnathostome clade rendered the analysis uninformative in terms of orthology assignment. We therefore named the three lamprey genes as Pax6α, Pax6β and Pax6γ (Fig. 1a).
Maximum Likelihood tree of vertebrate Pax6 genes. Phylogenetic analysis of the lamprey Pax6 genes with Pax6, Pax4, Pax3 and Pax7 genes from several other chordate species. The Pax3/7 clade was specified as the outgroup. The phylogram is shown on the left and its cladogram on the right. The tree highlights clustering of the three lamprey genes with Pax6 genes from other chordates thereby indicating that they are indeed lamprey Pax6 genes.
Conserved non-coding elements (CNEs) in the lamprey Pax6 loci
Finally, we performed sequence alignments for each of the lamprey Pax6 genomic scaffolds with Pax6 loci from other species using SLAGAN36 to identify putative CREs. An mVISTA visualisation of the alignment of the lamprey Pax6β locus with human and elephant shark Pax6 loci is shown in Fig. 4a. Only one distinct region of sequence similarity outside the exons was identified, located inside the lamprey Pax6β gene. On closer inspection, this element appeared to correspond to the neuroretina enhancer (NRE), a well-known Pax6 enhancer which is located in intron 4 of the human PAX6 gene31,37. The putative Lj_NRE shows 76% identity to the human NRE over an 88 bp core sequence, with 77% to coelacanth and 70% to the elephant shark Pax6.1 NRE core (Fig. 4b). No significant sequence similarity outside the exonic regions was found for the other lamprey Pax6 loci, nor could we detect any CNEs in alignments with the amphioxus Pax6 locus (Fig. 4a). To examine the potential function of this lamprey Pax6β CNE, an 878 bp fragment covering the 88 bp core region plus flanking sequences (Fig. S2) was PCR amplified from the Pax6β locus and inserted into a fluorescence reporter construct for the production of stable transgenic zebrafish. From among many primary transgenic embryos, four independent transgenic lines were established. All expressing lines showed strong and specific GFP fluorescence in the retinae of transgenic fish at 24, 36, 48 and 72 hpf (Fig. 4c), with additional, variable ectopic expression seen in some individual lines due to site of integration of the transgene (Table S1). Fluorescence signal became primarily located to the inner nuclear layer (INL) in 72 hpf embryos (Figs. 4c; and S3). The highly specific retinal expression of the Lj_NRE element is very similar to the expression driven by NRE elements from mouse or elephant shark in zebrafish transgenics13, thus supporting a very ancient role for the NRE element as a retinal enhancer in the ancestral Pax6 locus.
An ancient vertebrate conserved non-coding element is present in the lamprey Pax6β locus. (a) VISTA plot of the SLAGAN alignment of the LjPax6β locus, against the two Pax6 loci from elephant shark and human, as well as the amphioxus locus. Note that there is no PAX6 gene in the virtual ‘human_6.2’ locus. Sequence similarity outside of exons was seen for only a single element, homologous to the PAX6 neuroretina enhancer (NRE). (b) Conservation at the core sequence of the NRE element. (c) Transgenic zebrafish assay of the LjPax6β putative NRE element. The Lj_NRE element was cloned in front of a minimal promoter-eGFP cassette and used to generate stable transgenic fish. Embryos at 24, 36, 48 and 72 hours post fertilisation (hpf) show highly specific and consistent GFP signal in the neuroretina of the developing eye. Ectopic expression in the heart due to site of integration effect was seen in one of the transgenic lines. Embryos were imaged from the lateral side (24 hpf and inset 72 hpf showing a close-up of the eye) or ventral side (36 hpf, 48 hpf and 72 hpf) with the fluorescent confocal signal overlaid on a brightfield view. L, lens; NR, neuroretina; h, heart.
Discussion
In a previous detailed analysis of Pax6 gene loci in gnathostomes13, we found that unlike mammals, genomes of several vertebrate species possess multiple Pax6 genes that are likely to have originated in the 2 R duplications. In the present study we have extended this analysis to the Japanese lamprey, representing the sister group of gnathostomes. We find that the lamprey genome contains three Pax6 genes encoded by three separate genomic loci. The presence of more than two genes in its genome is consistent with a post-2R divergence of lampreys from gnathostomes. However, it has been proposed that an independent third WGD may have occurred in the lamprey lineage20. More recent studies have suggested that the two lineages shared only one WGD followed by a set of segmental duplications in the lamprey lineage21,22. This scenario is supported by analysis of synteny, which suggests similarity between the lamprey Pax6α and gnathostome Pax6.1 loci, with the Rcn1 homolog Caluα and Elp4 flanking lamprey Pax6α/Pax6.1. The presence of Fbxo47 between Caluβ and lamprey Pax6β is reminiscent of the gene content of the elephant shark Pax6.2 locus.
All three lamprey Pax6 genes identified in this study contain the paired box. The Pax6.2 gene found in elephant shark and some other species lacks the paired domain-coding exons13. If lamprey Pax6β is indeed the ortholog of Pax6.2, as suggested by synteny, loss of the paired domain most likely occurred in the gnathostome lineage after the split between the cyclostome and gnathostome lineages, but before the divergence of the cartilaginous fish and bony vertebrate lineages.
In gnathostomes, Pax6 is crucial for proper development and subsequent functioning of the eye, the central nervous system (CNS), the olfactory system and the pancreas1,2,3,4, and this is reflected in its tightly controlled expression in those tissues3,17,31. We found strong expression of all three genes in the eye, confirming the ancient role of Pax6 in ocular development and function. Similarly its well-known importance in the brain is underscored by strong expression of the LjPax6β and LjPax6γ genes in the lamprey brain, with a lower level also seen in the LjPax6α expression pattern. Examination of RNA-seq data from the pancreas of a juvenile brook lamprey (Lampetra planeri) showed expression of Pax6α, but not Pax6β or Pax6γ, indicating a specific function for Pax6α in the lamprey pancreas. In jawed vertebrates, Pax6 is crucial for proper development of the pancreas as well as the functional maintenance of the hormone-producing islet cells. In evolution, the pancreas is a novel endocrine organ that has come into existence with the emergence of the vertebrate lineage. Whereas in tetrapods the pancreas is a distinct organ that combines cells carrying out exocrine and endocrine roles, in cyclostomes the pancreas exists as a conglomeration of diffuse islet nodules associated with the gut area, similar to sharks and most bony fish24,32,38. We have previously shown that a number of key genes known to be crucial for pancreas development in gnathostomes, such as the transcription factors Pdx1, Hnf1a, NeuroD1, as well as insulin and glucagon, are expressed in the lamprey pancreas32. Our observation of Pax6α expression in the lamprey pancreas indicates that Pax6 was also likely recruited early on into the gene network enabling the formation and development of this organ in the common ancestor of vertebrates.
In addition to the brain and eye, well-known vertebrate Pax6 expression sites, expression of the lamprey LjPax6β and LjPax6γ genes was detected at low level in a number of other tissues. It remains to be investigated what role the genes play in these tissues. Interestingly, LjPax6β shows expression in the kidney. Kidney expression was also observed for elephant shark Pax6.213. In mammals, Pax6 expression is not observed in the kidney, but it is the main site of expression of the nearby Wilms tumour 1 (Wt1) gene39. We find no evidence for the presence of a Wt1 homolog on our Japanese lamprey Pax6 contigs (scaffolds 194 (1.1 Mb) and 23 (4.6 Mb)). A Wt1 ortholog is present in the sea lamprey, but is located more than 7 Mb away from Pax6β beyond several intervening genes. Our longest contig, around the Pax6α gene, contains closely spaced homologs of Eif3m and Calu (a reticulocalbin (Rcn) family gene). In gnathostomes, including the elephant shark Pax6.1 locus, where no Pax6 kidney expression is seen, Wt1 is situated between Eif3m and Rcn1 (Fig. 1b), suggesting that it either got translocated to this position after the cyclostome divergence, or was lost independently in the lamprey. It is tempting to speculate that non-coding elements enabling kidney expression were present in the wider locus of the ancestral Pax6 gene, which became functionally separated from the Pax6 promoters early in the evolution of the Pax6.1 loci (including the human PAX6 locus), but were captured by a newly inserted Wt1 gene.
The Pax6 genomic region has long been a paradigm locus for understanding the principles of long range gene regulation and the evolution of cis-regulatory landscapes. We have previously studied the conservation of non-coding elements in a wide range of vertebrate Pax6 loci, representing evolutionarily diverged lineages of increasing age13,17,18. Sequence comparisons of mammalian Pax6 with the Pax6 loci of the elephant shark revealed the presence of a large complement of CNEs between the species18. Many of these ancient gnathostome CNEs (agCNEs) have been shown to act as cis-regulatory elements, indicating that the Pax6 cis-regulatory landscape was laid down early in vertebrate evolution. In contrast, comparisons with the tunicate (Ciona intestinalis) or amphioxus Pax6 loci did not yield any recognisable CNEs13. We had therefore anticipated that CNE analysis of the lamprey Pax6 loci could reveal new insights into the hypothesis that the rapid emergence of the multitude of regulatory elements was triggered by the 2 R events40. VISTA analysis in our study uncovered only a single CNE peak mapping to the core sequence of the well characterised neuroretina enhancer (NRE) located in intron 4 of mammalian Pax631,37,41. We show that conservation of this CNE extends to its function, as reporter expression driven by the lamprey Pax6β NRE element in transgenic zebrafish is very similar to the retinal expression driven by the orthologous elements from mouse or elephant shark13. This conservation of sequence and function of the NRE suggests that the element was already present and functional in the ancestral vertebrate locus. Apart from the NRE element, we did not detect any other CNEs in the three lamprey Pax6 loci. Thus, compared to the large number of CNEs in elephant shark18, there appears to be a striking dearth of vertebrate CNEs around the lamprey Pax6 genes (Fig. S4). It is possible that putative ancestral elements have been lost or diverged beyond recognition in the lamprey genome, but a more likely scenario suggests that the large numbers of CNEs were invented in the gnathostome lineage after the split between cyclostome and gnathostome lineages40. Undoubtedly, cis-regulatory elements also exist around the lamprey genes and comparisons between different lamprey species or with the hagfish genome may reveal such cyclostome-specific CNEs, even though the Japanese and sea lamprey are too closely related to be helpful in this respect. Similar to the lack of CNEs between gnathostome Pax6 loci and those from Ciona or Branchiostoma (amphioxus) we detected no CNEs in sequence alignments between lamprey and amphioxus (Fig. 4a). In keeping with this, only a handful of CNEs were found when comparing 50 key developmental loci between cephalochordates and vertebrates42, even though many conserved sequences were identified when comparing the genomes of two cephalochordates, Asymmetron lucayanum and Branchiostoma floridae.
While functional genomics of the Mediterranean amphioxus (Branchiostoma lanceolatum) revealed many regions of open chromatin, hinting at the existence of a significant number of putative cis-regulatory elements in its genome, this number of potential cis-elements was nevertheless much lower than that typically found in gnathostomes, in particular when comparing regions around genes with highly restricted expression patterns43. Future detailed functional analysis of the genomic loci of pleiotropic developmental regulatory genes such as Pax6 in lamprey and hagfish may provide insight into the extent of their cis-regulatory complexity. In gnathostomes Pax6 is surrounded by a large array of enhancer elements, many of which drive similar or overlapping expression patterns18. Functional redundancy between cis-elements was demonstrated for two Pax6 lens enhancers by investigation of the consequences of their separate and combined deletions in the mouse10,44,45. Similarly, in addition to the NRE, multiple cis-elements with overlapping enhancer activity in the developing retina have been found in the mammalian Pax6 locus17,46. It remains to be investigated how their deletions in various combinations may affect retinal development, and to what extent these may recapitulate the dramatic impact on eye development seen with the conditional ablation of Pax6 itself from retinal tissues at various stages of development12. Conditional Pax6 deletion using a Cre transgene driven by the mouse NRE enhancer (αCre) led to exclusive amacrine cell formation in the central retina and premature activation of photoreceptor differentiation in the peripheral retina11,47. Ablation at an earlier stage in development using a different Cre driver caused general failure of retinal progenitor cells to proliferate properly, while retinal cells failed to follow a correct differentiation pathway upon post-natal removal of Pax612,48. Formation of correct post-natal cellular circuitry in the retina was shown to be dependent on antagonistic activity on the NRE between Pax6 itself and a complex of LIM domain containing proteins binding to the conserved core of the element49. Overexpression of Pax6 also disrupts aspects of retinal development in a stage-dependent manner48, underlining the critical importance of spatio-temporally precise control of Pax6 dosage. It is conceivable that in lampreys, where a relatively smaller number of cis-regulatory elements is presumed to exist around the individual Pax6 genes (as well as other developmental control genes), precision and robustness in the regulatory control circuitry is dependent on the presence of multiple separate Pax6 genes.
In summary, our work reveals that the lamprey genome contains three Pax6 genes, all possessing a paired domain but lacking an alternatively spliced exon 5a. One of the lamprey Pax6 genes (Pax6α) is specifically expressed in the pancreas, an organ characteristic of vertebrates. We identified only a single ancient CNE, in the lamprey Pax6β locus, and demonstrate its functional conservation as a neuroretina enhancer in a zebrafish reporter assay. This element therefore represents the oldest recognisable vertebrate Pax6 cis-regulatory element, displaying functional conservation over 500 million years of evolution.
Materials and Methods
Identification of Pax6 genes in the Japanese lamprey genome
The genome of the Japanese lamprey, having a relatively small genome size of 1.6 Gb, was recently sequenced by our group20. We searched for Pax6 genes in this assembly (GenBank accession APJL00000000, LetJap1.0) using an available Pax6 protein sequence from Japanese lamprey (GenBank accession BAB62531.125), as well as those from human and elephant shark. Our TBLASTN searches picked up several Pax gene-containing scaffolds of which scaffold 23 (4.6 Mb) and scaffold 194 (1.09 Mb) were positive for Pax6. In addition, we identified fragments of another Pax6 gene distributed across multiple scaffolds. We used the identified Pax6-containing scaffolds to design probes for screening Japanese lamprey BAC libraries. For the third Pax6 gene, we were unable to identify any BAC clones and therefore we performed RT-PCR and RACE (Rapid Amplification of cDNA Ends) using cDNA from eye to generate full-length coding sequence and to orient scaffolds belonging to the same gene, and closed gaps using genomic PCR to obtain the complete sequence of this locus.
Identification and sequencing of BACs
Three different Japanese lamprey BAC libraries (IMCB_Testis1: EcoRI, 92,160 clones, average insert size 100 kb; IMCB_Testis2: HindIII, 165,888 clones, average insert size 115 kb; and IMCB_Blood1: HindIII, 119,808 clones, average insert size 115 kb) were used to identify Pax6-containing BAC clones. The BAC libraries were screened using probes designed from the identified scaffolds and standard radioactive probing methods. Selected positive BACs were sequenced completely using the standard method of shotgun Sanger sequencing and gap filling by PCR or primer walking. Sequencing was done using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) on an ABI 3730xl capillary sequencer (Applied Biosystems, USA). Chromatograms were processed and assembled using Phred-Phrap50 and Consed51. Sequences for the three Japanese lamprey Pax6 loci generated in this study have been submitted to GenBank with accession numbers MH778922-MH778924.
Phylogenetic analysis
Phylogenetic analysis was carried out using Pax6 genes from Japanese lamprey along with orthologues from representative tetrapods (human, mouse, chicken, Anole lizard, Xenopus), coelacanth, teleosts (fugu, medaka, stickleback, zebrafish), spotted gar, cartilaginous fishes (elephant shark and whale shark), inshore hagfish and the cephalochordate (amphioxus). In addition, we included Pax4 and Pax3/7 sequences from representative chordate species. Multiple alignments were generated using MAFFT version 7 web server (https://mafft.cbrc.jp/alignment/server/) using the L-INS-i strategy. Alignments were inspected manually using BioEdit sequence alignment editor52. A Maximum Likelihood (ML) tree was generated for this alignment using TREE-PUZZLE version 5.3.rc1653. We used ‘exact (slow)’ parameter estimation using the ‘quartet sampling plus NJ tree’ option, 10,000 puzzling steps, eight gamma rate categories and a JTT + G + F substitution model as deduced by ModelGenerator version 0.85 for the ML analysis. Amino acid frequencies and the gamma distribution parameter alpha were set to be determined from the dataset. The Pax3/7 clade was specified as the outgroup.
Real-time qRT-PCR
Total RNA of adult Japanese lamprey (from a routine catch from commercial fishermen on the Ishikari River near Ebetsu in Hokkaido, Japan) was extracted from nine tissues (brain, eye, gills, heart, intestine, kidney, ovary, skin and testis) using the Trizol reagent (Life Technologies, Carlsbad, California) according to the manufacturer’s protocol. One µg of total RNA was reverse transcribed into 5′RACE-ready single strand cDNA using the SMART RACE cDNA Amplification kit (Clontech, Palo Alto, CA) and used as template for qRT-PCR using the SYBR Select Master Mix (Life Technologies). qRT-PCR primer sequences are listed in Table S2. The qRT-PCR was performed using the ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA) and SYBR Select Master Mix (Life Technologies) with the following cycling conditions: 50 °C for 2 minutes, 95 °C for 3 minutes, followed by 40 cycles of 95 °C for 3 seconds and 65 °C for 30 seconds. Quantification of gene expression levels was performed using the comparative CT method54. We used three technical replicates per tissue for each of the three Pax6 genes and determined their average expression level (±Standard Error). Expression levels of the Pax6 genes were normalized using the β-actin gene as the reference. The relative expression level of each Pax6 gene between different tissues was estimated using the tissue with the lowest level of expression among the tissues analysed as reference tissue.
Identification and analysis of CNEs
Pax6 loci from Japanese lamprey, amphioxus, elephant shark and human were used for CNE prediction. The amphioxus (Branchiostoma floridae) sequence was extracted from the JGI Genome Portal (https://genome.jgi.doe.gov/), using assembly version Brafl1 with Pax6 present on scaffold_23. Repetitive sequences were masked using the CENSOR web server55. Multiple alignments of the repeat-masked sequences were generated using the global alignment program SLAGAN36 with the Japanese lamprey sequence as the reference. CNEs were predicted using a cut-off of ≥65% identity across >50 bp windows and visualized using VISTA56. In addition, sequences of known CNEs between elephant shark and human were searched against the Japanese lamprey Pax6 loci using BLASTN.
Cloning of the lamprey CNE for the zebrafish enhancer assay
An 878 bp fragment containing the CNE from the Japanese lamprey Pax6β locus plus flanking sequence was cloned by PCR amplification using Phusion high fidelity polymerase (NEB). attB4 and attB1r sequences (underlined in the primers below) were attached to the PCR primers for use with the Gateway recombination cloning system (Invitrogen). The amplified fragment was inserted into the Gateway pP4P1r entry vector using BP clonase and the sequence was verified using M13 forward and reverse primers. Primer sequences used for amplification of the lamprey CNE are:
Lamp6β_NRE_FP-B4:
5′-AACGGGGACAACTTTGTATAGAAAAGTTGGGAGATCGTGATGGAGGTGT-3′ and Lamp6β_NRE_RP-B1r:
5′-AACGGGGACTGCTTTTTTGTACAAACTTGACCCCACGTGTACCGTCTAA-3′ Next, the lamprey CNE-containing pP4P1r entry construct was mixed with a pDONR221 construct containing a gata2 minimal promoter-eGFP-polyA cassette, and recombined using LR Clonase into a destination vector with a Gateway R4-R2 cassette flanked by Tol2 recombination sites to produce the LjPax6β-CNE-gata2-eGFP reporter construct. The minimal gata2 promoter-eGFP reporter cassette has been used to report on the tissue-specific expression patterns driven by a wide variety of linked enhancers and does not produce reporter expression without the presence of linked enhancer elements57.
Generation of transgenic zebrafish
Maintenance of zebrafish and the generation of transgenic fish were done according to previously described procedures57. LjPax6β-CNE-gata2-eGFP reporter plasmid DNA was isolated using a Qiagen miniprep kit and further cleaned via a Qiagen PCR purification column. Tol2 transposase RNA was synthesized with the SP6 mMessage mMachine kit (Ambion) from a NotI-linearized pCS2-TP plasmid58. An injection mix containing 25 ng/μl each of the reporter plasmid DNA and transposase RNA was micro-injected into the cytoplasm of ~200 embryos at the 1- to 2-cell stage. Embryos showing mosaic fluorescence at 1–5 days post-fertilization (dpf) were raised to adulthood and used to establish lines.
Imaging of transgenic embryos was performed as previously described57. Embryos, treated with 0.003% PTU (1-phenyl2-thio-urea) from 24 hpf to repress pigmentation, were anaesthetised with tricaine and mounted in 1% low-melting agarose for imaging on a Nikon A1R confocal microscope.
Ethics statement
The zebrafish experiments were approved by the University of Edinburgh Ethical Committee and performed under UK Home Office license number PIL PA3527EC3; PPL IFC719EAD. Extraction of DNA and RNA from lamprey tissues was approved by the Institutional Animal Care and Use Committee of the Biological Resource Centre, Agency for Science, Technology and Research (A*STAR), Singapore.
Data availability
Sequences for the three Japanese lamprey Pax6 loci generated in this study have been submitted to GenBank with accession numbers MH778922-MH778924.
References
Simpson, T. I. & Price, D. J. Pax6; a pleiotropic player in development. BioEssays: news and reviews in molecular, cellular and developmental biology 24, 1041–1051 (2002).
Osumi, N., Shinohara, H., Numayama-Tsuruta, K. & Maekawa, M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26, 1663–1672 (2008).
Cvekl, A. & Callaerts, P. PAX6: 25th anniversary and more to learn. Experimental eye research 156, 10–21 (2017).
Nakayama, T. et al. Xenopus pax6 mutants affect eye development and other organ systems, and have phenotypic similarities to human aniridia patients. Developmental biology 408, 328–344 (2015).
St-Onge, L., Sosa-Pineda, B., Chowdhury, K., Mansouri, A. & Gruss, P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 387, 406–409 (1997).
Dohrmann, C., Gruss, P. & Lemaire, L. Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mechanisms of development 92, 47–54 (2000).
Gosmain, Y. et al. Pax6 is crucial for beta-cell function, insulin biosynthesis, and glucose-induced insulin secretion. Mol Endocrinol 26, 696–709 (2012).
Hart, A. W., Mella, S., Mendrychowski, J., van Heyningen, V. & Kleinjan, D. A. The developmental regulator Pax6 is essential for maintenance of islet cell function in the adult mouse pancreas. PloS one 8, e54173 (2013).
Hill, R. E. et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354, 522–525 (1991).
Ashery-Padan, R., Marquardt, T., Zhou, X. & Gruss, P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes & development 14, 2701–2711 (2000).
Marquardt, T. et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43–55 (2001).
Klimova, L. & Kozmik, Z. Stage-dependent requirement of neuroretinal Pax6 for lens and retina development. Development 141, 1292–1302 (2014).
Ravi, V. et al. Sequencing of Pax6 loci from the elephant shark reveals a family of Pax6 genes in vertebrate genomes, forged by ancient duplications and divergences. PLoS genetics 9, e1003177 (2013).
Feiner, N., Meyer, A. & Kuraku, S. Evolution of the vertebrate Pax4/6 class of genes with focus on its novel member, the Pax10 gene. Genome biology and evolution 6, 1635–1651 (2014).
Dehal, P. & Boore, J. L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3, e314 (2005).
Putnam, N. H. et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008).
Kleinjan, D. A. et al. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Human molecular genetics 10, 2049–2059 (2001).
Bhatia, S. et al. A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos. Developmental biology 387, 214–228 (2014).
Miyashita, T. et al. Hagfish from the Cretaceous Tethys Sea and a reconciliation of the morphological-molecular conflict in early vertebrate phylogeny. Proceedings of the National Academy of Sciences of the United States of America 116, 2146–2151 (2019).
Mehta, T. K. et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proceedings of the National Academy of Sciences of the United States of America 110, 16044–16049 (2013).
Smith, J. J. & Keinath, M. C. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res 25, 1081–1090 (2015).
Smith, J. J. et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nature genetics 50, 270–277 (2018).
Lamb, T. D., Collin, S. P. & Pugh, E. N. Jr. Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nature reviews. Neuroscience 8, 960–976 (2007).
Youson, J. H. & Al-Mahrouki, A. A. Ontogenetic and phylogenetic development of the endocrine pancreas (islet organ) in fish. General and comparative endocrinology 116, 303–335 (1999).
Murakami, Y. et al. Identification and expression of the lamprey Pax6 gene: evolutionary origin of the segmented brain of vertebrates. Development 128, 3521–3531 (2001).
Derobert, Y., Baratte, B., Lepage, M. & Mazan, S. Pax6 expression patterns in Lampetra fluviatilis and Scyliorhinus canicula embryos suggest highly conserved roles in the early regionalization of the vertebrate brain. Brain research bulletin 57, 277–280 (2002).
Epstein, J. A. et al. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & development 8, 2022–2034 (1994).
Kozmik, Z., Czerny, T. & Busslinger, M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. The EMBO journal 16, 6793–6803 (1997).
Fabian, P., Kozmikova, I., Kozmik, Z. & Pantzartzi, C. N. Pax2/5/8 and Pax6 alternative splicing events in basal chordates and vertebrates: a focus on paired box domain. Frontiers in genetics 6, 228 (2015).
Kuraku, S. & Kuratani, S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoological science 23, 1053–1064 (2006).
Kammandel, B. et al. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Developmental biology 205, 79–97 (1999).
Zhang, H. et al. Lampreys, the jawless vertebrates, contain only two ParaHox gene clusters. Proceedings of the National Academy of Sciences of the United States of America 114, 9146–9151 (2017).
Qiu, H., Hildebrand, F., Kuraku, S. & Meyer, A. Unresolved orthology and peculiar coding sequence properties of lamprey genes: the KCNA gene family as test case. BMC genomics 12, 325 (2011).
Coffill, C. R. et al. The p53-Mdm2 interaction and the E3 ligase activity of Mdm2/Mdm4 are conserved from lampreys to humans. Genes & development 30, 281–292 (2016).
Nah, G. S., Tay, B. H., Brenner, S., Osato, M. & Venkatesh, B. Characterization of the Runx gene family in a jawless vertebrate, the Japanese lamprey (Lethenteron japonicum). PloS one 9, e113445 (2014).
Brudno, M. et al. Glocal alignment: finding rearrangements during alignment. Bioinformatics 19(Suppl 1), i54–62 (2003).
Plaza, S., Dozier, C., Langlois, M. C. & Saule, S. Identification and characterization of a neuroretina-specific enhancer element in the quail Pax-6 (Pax-QNR) gene. Molecular and cellular biology 15, 892–903 (1995).
Mulley, J. F., Hargreaves, A. D., Hegarty, M. J., Heller, R. S. & Swain, M. T. Transcriptomic analysis of the lesser spotted catshark (Scyliorhinus canicula) pancreas, liver and brain reveals molecular level conservation of vertebrate pancreas function. BMC genomics 15, 1074 (2014).
Hastie, N. D. Wilms’ tumour 1 (WT1) in development, homeostasis and disease. Development 144, 2862–2872 (2017).
McEwen, G. K. et al. Early evolution of conserved regulatory sequences associated with development in vertebrates. PLoS genetics 5, e1000762 (2009).
Xu, P. X. et al. Regulation of Pax6 expression is conserved between mice and flies. Development 126, 383–395 (1999).
Yue, J. X. et al. Conserved Noncoding Elements in the Most Distant Genera of Cephalochordates: The Goldilocks Principle. Genome biology and evolution 8, 2387–2405 (2016).
Marletaz, F. et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564, 64–70 (2018).
Antosova, B. et al. The Gene Regulatory Network of Lens Induction Is Wired through Meis-Dependent Shadow Enhancers of Pax6. PLoS genetics 12, e1006441 (2016).
Dimanlig, P. V., Faber, S. C., Auerbach, W., Makarenkova, H. P. & Lang, R. A. The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development 128, 4415–4424 (2001).
McBride, D. J., Buckle, A., van Heyningen, V. & Kleinjan, D. A. DNaseI hypersensitivity and ultraconservation reveal novel, interdependent long-range enhancers at the complex Pax6 cis-regulatory region. PloS one 6, e28616 (2011).
Oron-Karni, V. et al. Dual requirement for Pax6 in retinal progenitor cells. Development 135, 4037–4047 (2008).
Remez, L. A. et al. Pax6 is essential for the generation of late-born retinal neurons and for inhibition of photoreceptor-fate during late stages of retinogenesis. Developmental biology 432, 140–150 (2017).
Kim, Y. et al. The LIM protein complex establishes a retinal circuitry of visual adaptation by regulating Pax6 alpha-enhancer activity. eLife 6 (2017).
Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8, 175–185 (1998).
Gordon, D., Abajian, C. & Green, P. Consed: a graphical tool for sequence finishing. Genome Res 8, 195–202 (1998).
Hall, T. A. In Nucleic Acids Symposium Series, Vol. 41 95–981999).
Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Kohany, O., Gentles, A. J., Hankus, L. & Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC bioinformatics 7, 474 (2006).
Mayor, C. et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 (2000).
Bhatia, S. et al. Functional assessment of disease-associated regulatory variants in vivo using a versatile dual colour transgenesis strategy in zebrafish. PLoS genetics 11, e1005193 (2015).
Fisher, S. et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nature protocols 1, 1297–1305 (2006).
Acknowledgements
The work in the B.V. lab was supported by the Biomedical Research Council of A*STAR, Singapore. S.B. and D.A.K. were supported by core funding from the Medical Research Council UK to the Institute of Genetics and Molecular Medicine at the University of Edinburgh.
Author information
Authors and Affiliations
Contributions
B.V. and D.A.K. conceived the project; V.R., S.B., P.S. and B.-H.T. performed the experiments; V.R., S.B., B.V. and D.A.K. analysed the data; and B.V. and D.A.K. wrote the manuscript with inputs from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ravi, V., Bhatia, S., Shingate, P. et al. Lampreys, the jawless vertebrates, contain three Pax6 genes with distinct expression in eye, brain and pancreas. Sci Rep 9, 19559 (2019). https://doi.org/10.1038/s41598-019-56085-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-56085-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.