Abstract
Infection of humans with Chlamydia trachomatis, a bacterial pathogen with a unique intracellular replication cycle, may cause a variety of clinical manifestations. These are linked to various serovars of the pathogen; trachoma to serovars A-C, oculogenital infections to serovars D-K, and lymphogranuloma venereum to serovars L1-L3. Nineteen serovars are known as human pathogens. The aim of the study was to determine the serovars of 401 C. trachomatis DNA positive extracts from original clinical specimens of patients in Austria including cervical and urethral swabs, urine, genital secretions and conjunctival swabs - collected from 2014 to 2017. Sequence analysis of the omp1 gene, encoding major outer-membrane protein was performed on each sample. In 50.1% of samples serovar E was identified and serovars F, D/Da and G/Ga were found in 16.2%, 9.7% and 9.0%, respectively. Remaining serovars were J (6.0%), K (4.7%), H (2.7%), B/Ba (1.0%), and I/Ia (0.5%). In 19 patients follow up samples could be tested. The majority of C. trachomatis serovars were associated with urogenital tract infections (D-K), however, one of them – serovar B/Ba - is linked to both, ocular and genital tract infection.
Similar content being viewed by others
Introduction
Sexually transmitted infections in the industrialised world are most commonly caused by Chlamydia trachomatis. According to WHO, about 130 million new cases occur each year worldwide1. Chlamydial urogenital infection mainly affects sexually active young people between the ages of 15 and 24 years2. Genital chlamydial infections are asymptomatic in a higher proportion in women than in men; thus, in women, chlamydiae can affect the upper genital tract causing inflammation and scarring, possibly followed by infertility, ectopic pregnancy and pre-term delivery3. Epididymitis, urethral obstructions and decreased fertility may occur in men. Infection with C. trachomatis also facilitates the transmission of HIV and is associated with cervical cancer4.
Currently, 19 serovars of C. trachomatis are recognised (A, B/Ba, C, D/Da, E, F, G/Ga, H, I/Ia, J, K, L1, L2, L2a and L3) according to specific epitopes of the major outer membrane protein (MOMP) encoded by the gene omp1. This gene encodes highly conserved protein structures that contain four evenly spaced domains whose sequences vary among the different serovars5. Classification of C. trachomatis isolates on the basis of the MOMP variable domains correlates with immunotyping using MOMP-specific monoclonal antibodies6. Serovars A, B, Ba and C are recognised as the aetiological agents of trachoma, serovars D–K are linked to oculogenital infections, and serovars L1–L3 are commonly associated with lymphogranuloma venereum.
Diagnosis of C. trachomatis infection is well established in Austria; however, little is reported on the occurrence and distribution of the serovars. We therefore analysed C. trachomatis serovars in DNA extracts from clinical samples submitted to our Institute by Austrian laboratories.
The study was approved by the Ethics Committee of the Medical University of Vienna, (EK-Number 1557/2017). Because of the retrospective design of the study, the Ethics Committee waived the need for informed consent.
Results
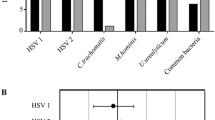
Sequence analysis of the omp1 gene fragments showed that among the 401 C. trachomatis samples, serovar E was the type most frequently identified (201/401, 50.1%), followed by serovars F (65/401, 16.2%), D/Da (39/401, 9.7%), G/Ga (36/401, 9.0%), J (24/401, 6.0%), K (19/401, 4.7%), H (11/401, 2.7%), B/Ba (4/401, 1.0%) and I/Ia (2/401, 0.5%). Information on the serovars and corresponding clinical samples are given in the Table 1. We identified nine different serovars in samples from female patients and five from male patients. The majority of samples were obtained from the urogenital tract and included samples of urine. Ten samples were derived from ocular swabs, including one from a new born infant. C. trachomatis was repeatedly detected in 19 patients, the time intervals between the first and the follow-up examinations ranging from 19 to 919 days. Sixteen patients had one follow-up visit, two patients had three and one patient had four. In 17 patients the same C. trachomatis serovar was identified in the first and follow-up sample: serovar E was detected the most frequently (8 patients), followed by serovar D/Da (3 patients) and J (2 patients); serovars B/Ba, F, G/Ga and I/Ia were found repeatedly in one patient each. In two patients different serovars were found in the follow-up samples; in one patient serovar E followed serovar K, and in the other serovar H followed serovar E. With regard to patient age, the frequency of C. trachomatis infection was highest in patients aged 21–25 years (143 samples), followed by those aged 15–20 (95 samples) and 26–30 (84 samples) (Fig. 1). Serovar E was the most common in all the age groups, followed by serovars F and G/Ga. There were only two positive samples from patients aged below 15 years: one ocular infection in a newborn (serovar E), one genital infection in a teenager (serovar B/Ba).
Discussion
In 401 clinical specimens from Austrian patients who tested positive for C. trachomatis, omp1 PCR identified serovar E as the most common type (50.1%), followed by serovars F, D/Da, G/Ga, J, K, H, B/Ba and I/Ia. The serovars D–F are recognised as the most prevalent worldwide2,3,7,8. Serovar B/Ba, detected in a genital sample, was the only serovar from the ocular group of C. trachomatis; this serovar has already been described as causing ocular and genital tract disease9. The majority of samples tested were derived from female patients (95%), where the most frequent serovars were E and F; these were also prominent in the small number of samples from male patients. Serovar E was also detected in an ocular swab of a newborn.
We did not find serovars L1-L3 in our mainly female study population.
Our aim was to get a first impression about C. trachomatis serovars obtained from clinical samples of Austrian patients. We identified eight different serovars of the oculogenital group, together with serovar B/Ba, one of the trachoma group and known as an agent of genital tract infection. We considered sequencing of the omp1 gene fragment by using Sanger sequencing an efficient method for this purpose. Multilocus sequence typing and next generation sequencing of C. trachomatis would be the next step to analyse the strains in more detail in order to compare their relation to other isolates and in respect to clinical manifestations.
Our findings might serve as an incentive for future surveillance of C. trachomatis infection in Austria. Further, knowledge of the C. trachomatis serovar is relevant for deciding about the length of treatment of a C. trachomatis infection; e.g. 21 days for C. trachomatis serovars L1-L310. Finally, it might have an impact on the selection of suitable immunogenic antigens for vaccine development.
Materials and Methods
A total of 401 clinical specimens were analysed, originating from 378 patients (359 females and 19 males). In 19 patients (18 females and 1 male) repeated samples were taken - between 2 and 4 times per patient. Most of the specimens were genital/urethral samples. Under inclusion of retested patients, 371 samples were from female patients with a median age of 26 years (range 15–65 years), and 13 samples from male patients with a median age 36 years (range 23–42 years). Seven specimens were urine samples from 2 female patients (median age 24 years, range 22–26 years), 5 from male patients (median age 27 years, range 22–37 years), and 10 were ocular swabs derived from 7 female patients (median age 20 years, range 0.1–32 years) and 3 male patients (median age 32 years, range 28–39 years).
The PEQGoldTissue DNA Mini Kit (Peqlab, VWR, Darmstadt, Germany) was used for extraction of C. trachomatis DNA from the clinical samples. The DNA was stored at −20 °C until serovar determination.
The omp1 gene fragment (ca. 1130 bp) was amplified in nested PCR according to a method described by Hsu et al.11. Briefly, the outer primer pair NLO/NRO was used for primary amplification and the inner primer pair MOMP87/C214 for the second run, to obtain the 584 bp products. The final PCR products were analysed on 1% agarose gel using GelRed® Nucleic Acid Gel Stain (Biotium, Germany). It should be noted that serovar subtypes Da, Ba, Ga and Ia cannot be discriminated using this method.
The omp1 fragments were purified from the gel using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). DNA concentration was then measured (DeNovix Spectrophotometer/Fluorometer), followed by bi-directional sequencing of the samples (Microsynth, Vienna, Austria) using the primer set MOMP87/C214. Consensus sequences were compared with sequences of known C. trachomatis strains by BLAST search provided by NCBI (www.ncbi.nlm.nih.gov/GenBank).
Data availability
All data generated or analysed during this study are included in this published article.
References
Newman, L. et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systemic review and global reporting. PLoS One. 10, 12, https://doi.org/10.1371/journal.pone.0143304 (2015).
European Center for Disease Prevention and Control. Sexually transmitted infections in Europe 2013. Stockholm: ECDC, htpp://ecdc.europa.eu/en/publications/Publications/sexual-transmitted-infections-europe-surveillance-report-2013.pd (2015)
CDC. Sexually Transmitted Disease Surveillance 2012. Atlanta: U.S. Department of Health and Human Science, htpp://www.cdc.gov/sTD/stats12/Surv2012.pdf (2013)
Malhotra, M., Sood, S., Mukherjee, A., Muralidhar, S. & Bala, M. Genital Chlamydia trachomatis: an update. Indian J Med Res. 138, 303–16 (2013).
Schachter J. Infection and disease epidemiology. In: Stephens RS, ed. Chlamydia. Washington, DC: ASM Press 139–69 (1999)
Wang, S. P., Kuo, C. C., Barnes, R. C., Stephens, R. S. & Grayston, J. T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis. 152, 791–800 (1985).
Lee, G. et al. ompA genotyping of Chlamydia trachomatis from Korean female sex workers. J Infect. 52, 451–454 (2006).
Machado, A. C. et al. Distribution of Chlamydia trachomatis genovars among youths and adults in Brazil. J Med Microbiol. 60, 472–476 (2011).
Wang, S. P. & Grayston, J. T. Immunologic relationship between genital TRIC, lymphogranuloma venereum, and related organisms in a new microtiter indirect immunofluorescence test. Am J Ophthalmol. 70, 367–374 (1970).
de Vries H. et al. 2019 European guideline on the management of lymphogranuloma venereum. J Eur Acad Dermatol Venereol., https://doi.org/10.1111/jdv.15729 (2019)
Hsu, Mc et al. Genotyping of Chlamydia trachomatis from clinical specimens in Taiwan. J Med Microbiol. 55, 301–8 (2006).
Author information
Authors and Affiliations
Contributions
Iwona Lesiak-Markowicz and Anna-Margarita-Schötta conceived of the study. Iwona Lesiak-Markowicz performed all experiments, prepared table and figure, and wrote the manuscript. Anna-Margarita Schötta, Hannes Stockinger, Gerold Stanek und Mateusz Markowicz provided critical feedback and helped shape the research, analysis and manuscript. All authors contributed to the final version and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lesiak-Markowicz, I., Schötta, AM., Stockinger, H. et al. Chlamydia trachomatis serovars in urogenital and ocular samples collected 2014–2017 from Austrian patients. Sci Rep 9, 18327 (2019). https://doi.org/10.1038/s41598-019-54886-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54886-5
This article is cited by
-
Feasibility of using volatile urine fingerprints for the differentiation of sexually transmitted infections
Applied Microbiology and Biotechnology (2023)
-
Prevalence of selected sexually transmitted infectious agents in a cohort of asymptomatic soldiers in Austria
Parasites & Vectors (2022)
-
A Chlamydia trachomatis VD1-MOMP vaccine elicits cross-neutralizing and protective antibodies against C/C-related complex serovars
npj Vaccines (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.