Abstract
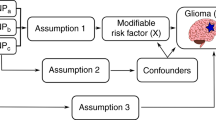
Genetic risk score (GRS) is used to demonstrate the genetic variants contributing to the polygenic architecture of complex diseases. By using a GRS, we have investigated the additive impact of the known adult glioma susceptibility loci on the pediatric brain tumor (PBT) risk and assessed the proportion of PBT heritability attributable to these susceptibility loci. A GRS was generated for PBTs based on the alleles and associated effect sizes derived from a previously published genome-wide association study on adult glioma. The GRS was calculated in CEFALO, a population-based case-control study of brain tumors in children and adolescents including saliva DNA of 245 cases and 489 controls. The unconditional logistic regression model was used to investigate the association between standardized GRS and risk of PBTs. To measure the variance explained by the effect of GRS, Nagelkerke pseudo-R2 was calculated. The GRS for adult brain tumors was associated with an increased risk of PBTs (OR 1.25 [95% CI 1.06–1.49], p = 0.009) and 0.3% of the variance in PBTs could be explained by the effect of GRS on the liability scale. This study provides evidence that heritable risks of PBTs are in-part attributable to some common genetic variants associated with adult glioma.
Similar content being viewed by others
Introduction
Genetic risk score (GRS), or polygenetic risk score, calculations are used to analyze the oligo- or polygenic architecture underlying complex genetic disorders. In such calculations, first, the alleles which are associated with a trait at a certain threshold and their effect sizes are identified in a discovery sample. Then, a GRS, being a composite score of the alleles identified in the discovery sample and weighted by the effect size (e.g. log odds ratio (OR)), is calculated for each individual in an independent target sample. Later, the regression model is applied to assess the association between the GRS and the phenotype in the target sample adjusting for covariates1.
The current study aimed to examine whether the genetic risk score for adult brain tumors, taking into account the combined effect of five susceptibility loci, is associated with risk of pediatric brain tumors and to assess the proportion of pediatric brain tumor heritability attributable to these known adult glioma susceptibility loci.
Methods
We generated the GRS for pediatric brain tumors based on the alleles and associated effect sizes derived from a previously published genome-wide association study (GWAS) on adult glioma2, which in turn was based on the meta-analysis of six previously published GWAS and also two new GWAS including in total 12496 cases and 18190 controls. Quality control procedures and the GWAS results were described in details in the original paper2. The CEFALO study was employed as the target sample. CEFALO is a population-based case-control study of brain tumors in children and adolescents aged 7–19 years conducted in Sweden, Denmark, Norway, and Switzerland. Details of the study methods, single nucleotide polymorphism (SNP) selection, genotyping, and the quality control procedures have been described previously3,4,5. Briefly, saliva DNA of 245 cases and 489 controls was included in the study and was satisfactorily genotyped for 92 SNPs. The study was approved by the national data protection boards and ethical committees in all participating countries, all research was performed in accordance with relevant guidelines/regulations and written informed consent was obtained from all participants and/or their parents. Table 1 summarizes demographic characteristics of included cases and controls and the distributions of diagnostic pediatric brain tumor types. We selected the discovery sample for these analyses based on its similarity with the target sample for ethnicity, as well as its large sample size leading to the more accurate reported estimates.
SNPs which were associated with the adult glioma risk at the genome-wide significance level in all the discovery, validation and combined results of either all glioma or non-glioblastoma glioma in the discovery sample were selected for the GRS analyses. Thus, the GRS in the CEFALO study was based on 5 SNPs (rs6010620, rs2736100, rs4977756, rs498872, rs2297440) and corresponding to the number of the risk alleles weighted by the logarithm of the ORs from the discovery sample across this set of SNPs (Table 2). PLINK was used to perform the analyses6.
The GRS was subsequently standardized using its mean value and standard deviation (SD); for which the OR associated with it can then be interpreted as the increased (or decreased) odds of pediatric brain tumors for a one SD change in GRS. The unconditional logistic regression model was applied to evaluate whether the standardized GRS is associated with the risk of pediatric brain tumors adjusted for age, sex and country as covariates (full model); unmatched analyses have been performed since not all CEFALO participants provided saliva sample. To measure the variance explained by the effect of GRS, Nagelkerke pseudo-R2 was calculated as the difference of R2 in the full model compared to the reduced model including the covariates but not the GRS. The analyses were conducted using R7.
The study was approved by the Ethical Review Board in Stockholm, Sweden.
Results
The results indicated that the standardized genetic risk score for adult brain tumors was associated with increased risk of pediatric brain tumors (OR 1.25 [95% CI 1.06–1.49], p = 0.009). The identified OR of 1.25 can be interpreted as: one SD increase in GRS is associated with a 25% increased odds of pediatric brain tumors. Moreover, 1.2% of the variance in pediatric brain tumors could be explained by the effect of the GRS (R2 = 0.012). The estimated disease prevalence used to transform the estimated heritability to the liability scale was 0.04%. The SNP-heritability on the liability scale was estimated to be 0.003.
The minimum detectable ORs for each individual SNPs rs6010620 (G), rs2736100 (C), rs4977756 (G), rs498872 (A), and rs2297440 (C) in this dataset of 245 cases and 489 controls were 1.61, 1.56, 1.56, 1.58, and 1.62, respectively.
Discussion
This study was performed based on the hitherto largest series of pediatric brain tumor cases with the purpose to investigate the additive impact of the known adult glioma susceptibility loci on pediatric brain tumor risk and for the first time to investigate the similarity of SNP-heritability between adult and pediatric brain tumors.
To our knowledge, to date, two studies have assessed the heritability of adult glioma based on the known glioma susceptibility loci. Sampson et al. assessed the adult glioma heritability based on 10 loci (rs1412829, rs2157719, rs2736100, rs2853676, rs4295627, rs4809324, rs4977756, rs498872, rs6010620, rs891835) and reported the estimated SNP-heritability of 1.7% (h2 = 0.017)8. Moreover, Kinnersley et al. investigated the proportion of glioma heritability attributable to known glioma susceptibility loci (rs2736100, rs11979158, rs2252586, rs4295627, rs4977756, rs498872, rs6010620) and the estimated proportion of genetic variance of adult glioma explained by these seven risk loci was 1.6%9.
In the present study, we estimated that 0.3% of the variance in the pediatric brain tumors could be explained by the effect of the genetic risk score containing five known adult glioma susceptibility loci. This provides evidence that the heritable risks of pediatric brain tumors are in-part attributable to some common genetic variants associated with adult glioma. Also, previously, based on CEFALO study, we could show that adult and pediatric brain tumors have some genetic risk factors in common4,5. However, more polygenic analyses based on large sample sizes and big genotyping data for both adult and pediatric brain tumors are required to determine the similarity of genetic architecture in adult and pediatric brain tumors which is clinically relevant from a risk prediction and treatment perspective. In addition, this line of research provides the basis to develop comprehensive risk prediction models including both genetic and environmental factors for brain tumors that eventually may lead to brain tumor prediction and prevention. This can be achieved by incorporating large collaborative genetic association studies and high quality registration data and could be leveraged by refinement of molecular classification and development of somatic characteristics of brain tumors10.
References
Wray, N. R. et al. Research Review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psyc 55, 1068–1087, https://doi.org/10.1111/jcpp.12295 (2014).
Melin, B. S. et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nature genetics 49, 789–794, https://doi.org/10.1038/ng.3823 (2017).
Aydin, D. et al. Mobile phone use and brain tumors in children and adolescents: a multicenter case-control study. Journal of the National Cancer Institute 103, 1264–1276, https://doi.org/10.1093/jnci/djr244 (2011).
Adel Fahmideh, M. et al. CCDC26, CDKN2BAS, RTEL1 and TERT Polymorphisms in pediatric brain tumor susceptibility. Carcinogenesis, https://doi.org/10.1093/carcin/bgv074 (2015).
Adel Fahmideh, M. et al. Common genetic variations in cell cycle and DNA repair pathways associated with pediatric brain tumor susceptibility. Oncotarget 7, 63640–63650, https://doi.org/10.18632/oncotarget.11575 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81, 559–575, https://doi.org/10.1086/519795 (2007).
R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL, http://www.R-project.org/ (2013).
Sampson, J. N. et al. Analysis of Heritability and Shared Heritability Based on Genome-Wide Association Studies for Thirteen Cancer Types. Journal of the National Cancer Institute 107, djv279, https://doi.org/10.1093/jnci/djv279 (2015).
Kinnersley, B. et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci Rep 5, 17267, https://doi.org/10.1038/srep17267 (2015).
Ostrom, Q. T. et al. Risk factors for childhood and adult primary brain tumors. Neuro-oncology 21, 1357–1375, https://doi.org/10.1093/neuonc/noz123 (2019).
Acknowledgements
The Swedish part of the CEFALO study was supported by grants from the Swedish Council for Working Life and Social Research [grant numbers 2004-0504 and 2007-0224]; the Swedish Research Council [grant number K2008-70X-15366-04-3]; the Swedish Cancer Society [grant number 09 0666]; the Swedish Childhood Cancer Society [grant numbers PROJ06/050 and PROJ09/086]; and the Swedish Radiation Protection Authority [grant number SSI P 1572]. The Danish CEFALO study was supported by the Danish Strategic Research Council [grant numbers 2103-05-0006 and 2064-04-0010]. The Swiss part of the CEFALO study was supported by the Swiss Federal Office of Public Health [grant number 05.001626]; the Swiss Research Foundation on Mobile Communication [grant number A2006.18]; and the Swiss National Science Foundation [grant number PDFMP3_122873]. The Norwegian CEFALO study was supported by the Research Council of Norway [grant number 175163/V40]. M.A.F. was partly supported by a Research Training Award for Cancer Prevention Post-Graduate Training Program in Integrative Epidemiology from the Cancer Prevention and Research Institute of Texas (grant number RP160097, PI: M. Spitz).
Author information
Authors and Affiliations
Contributions
M.F., J.S., M.R., K.K., M.A.G. and C.E.K. designed the study and organized patient recruitment and control selection in their respective countries. M.A.F. and M.F. oversaw the genotyping. M.A.F. performed statistical and bioinformatic analyses and drafted the manuscript, with contributions from C.L. and M.F.. G.T. and H.D. contributed in statistical analyses. B.L., L.S.S. and C.J. managed recruitment of patients and controls and organized collection of saliva. All authors critically reviewed the manuscript and approved the final version. M.F. and M.A.F. have had full access to all the data in the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adel Fahmideh, M., Lavebratt, C., Tettamanti, G. et al. A Weighted Genetic Risk Score of Adult Glioma Susceptibility Loci Associated with Pediatric Brain Tumor Risk. Sci Rep 9, 18142 (2019). https://doi.org/10.1038/s41598-019-54701-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54701-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.