Abstract
Inherited pathogenic variants in genes that confer moderate to high risk of breast cancer may explain up to 50% of familial breast cancer. This study aimed at identifying inherited pathogenic variants in breast cancer cases from Puerto Rico that were not linked to BRCA1 or BRCA2. Forty-eight breast cancer patients that met the clinical criteria for BRCA testing but had received a negative BRCA1/2 result were recruited. Fifty-three genes previously implicated in hereditary cancer predisposition were captured using the BROCA Agilent cancer risk panel followed by massively parallel sequencing. Missense variants of uncertain clinical significance in CHEK2 were evaluated using an in vitro kinase assays to determine their impact on function. Pathogenic variants were identified in CHEK2, MUTYH, and RAD51B in four breast cancer patients, which represented 8.3% of the cohort. We identified three rare missense variants of uncertain significance in CHEK2 and two variants (p.Pro484Leu and p.Glu239Lys) showed markedly decreased kinase activity in vitro comparable to a known pathogenic variant. Interestingly, the local ancestry at the RAD51B locus in the carrier of p.Arg47* was predicted to be of African origin. In this cohort, 12.5% of the BRCA-negative breast cancer patients were found to carry a known pathogenic variant or a variant affecting protein activity. This study reveals an unmet clinical need of genetic testing that could benefit a significant proportion of at-risk Latinas. It also highlights the complexity of Hispanic populations as pathogenic factors may originate from any of the ancestral populations that make up their genetic backgrounds.
Similar content being viewed by others
Introduction
Women with a pathogenic variant in BRCA1 or BRCA2 are at substantially higher risk of developing breast, ovarian and other cancers1. For these women, risk reduction options include increased surveillance, chemoprevention and prophylactic surgery2. While the identification of a BRCA pathogenic variant constitutes a clear benefit for probands and their relatives3,4, a proportion of at-risk families do not present pathogenic variants in BRCA1 or BRCA2 and their genetic risk of breast cancer remains unexplained.
Significant progress has been made in the identification of inherited genetic factors underlying hereditary cancers. Pathogenic variants in PTEN, TP53, CHEK2, ATM, NBS1, RAD50, BRIP1 and PALB2, amongst others, have also been shown to confer moderate to high risk of breast cancer5,6. Recently, several commercial laboratories have replaced BRCA testing with extended panels of cancer susceptibility genes7. Yet, for many of the genes tested, there are no approved guidelines dictating the optimal prevention or treatment strategy should a loss of function variant be identified8.
The Hispanic/Latino population is the fastest growing ethnic minority in the US and is predicted to account for 35% by 20509. Although breast cancer (BC) incidence and mortality are lower compared to most ethnic groups in the US, the outcome and prognosis in Hispanic women are worse10. The prevalence and spectrum of BRCA variants varies significantly within Latin America and the Caribbean, with little overlap in the pathogenic variants most frequently reported in each country11. In Puerto Rico, our previous work estimated that a majority of the BRCA cases are explained by few recurrent founder mutations12,13. This study aimed at identifying hereditary cancer variants in BRCA-negative breast cancer cases from Puerto Rico using the BROCA panel of cancer predisposition genes14.
Methods
Study participants
The study protocol was approved by the Institutional Review Board of Ponce Research Institute (#120829-JD) and Moffitt Cancer Center (#MCC17404) and all methods were performed in accordance with relevant guidelines and regulations. Participants were recruited between September 2012 and September 2014 from a private surgery practice located in San Juan, Puerto Rico. Evaluation of the potential study participants was conducted by a single surgeon. All women were diagnosed with breast cancer and met the NCCN (National Comprehensive Cancer Network current at the time of the study design, version 1.2012) guidelines for BRCA testing, but had received a negative BRCA clinical test result. All study participants provided informed consent and received an orientation about genetics and hereditary cancers. Trained study personnel gathered a three-generation pedigree depicting family history of cancer. Participants whose grandparents had not been born in Puerto Rico were excluded. A complete surgery clinical pathology report was obtained for the tumor(s) of each participant. Not all tumors were evaluated by the same pathologist. A total of 97 women were interviewed, 56 (57.7%) met the inclusion criteria and agreed to participate. In addition, a sample containing a known BRCA2 c.3922G > T (p.Glu1308Ter) pathogenic variant was included as a blinded control. A cohort of 500 women from Puerto Rico that do not have a cancer diagnosis and that received a negative breast cancer screening mammography within 6 months of enrollment was used to assess frequency of variants of interests in the population of Puerto Rico.
BRCA1/2 large rearrangement screening
To ensure that all women included were true negative for BRCA1/2 pathogenic variants, comprehensive screening for large rearrangements was performed by multiplex ligation-dependent probe amplification (MLPA). The MLPA kits P002-D1 (BRCA1 NM_007294.3) and P045-B3 (BRCA2 NM_000059.3) and the protocol for the final assessment of allele dosage were used as described by the manufacturer (MRC-Holland, Amsterdam, the Netherlands)15.
Sequencing
Genomic DNA was extracted using Paxgene blood DNA Kit (Qiagen, Valencia, CA) and used for the preparation of paired-end libraries with 200 base pairs (bp) inserts. An oligonucleotide pool targeting exons, non-repeating introns and selected promoter regions of the genes of interest was used for enrichment (SureSelect). Following capture, samples were sequenced on an Illumina MiSeq. The total genomic region covered was 1.1 million bp (Mbp). The 53 genes of the BROCA (http://tests.labmed.washington.edu/BROCA_versions) panel are listed in Supplementary Fig. 1. This panel was developed for the evaluation of patients at risk of hereditary cancers, it is not restricted to BC susceptibility genes. Forty-eight samples achieved proper QA/QC standards and remained for further analyses.
Alignment, variation detection, annotation
Sequence reads were aligned to the reference human genome (hs37d5) with the Burrows-Wheeler Aligner (BWA)16, and duplicate identification, insertion/deletion realignment, quality score recalibration, and variant calling were performed with PICARD (http://picard.sourceforge.net/), the Genome Analysis ToolKit (GATK)17. Sequence variants were annotated using ANNOVAR18. Additional contextual information was incorporated, including allele frequency in other studies such as 1000 Genomes (Phase 3 20130502, downloaded 11/2015), NHLBI Exome Sequence Project (ESP6500 v2, downloaded 8/2013), and the Exome Aggregation Consortium (release 0.2, downloaded 1/2015). In silico functional impact predictions, and observed impacts were assessed from databases like ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) (release 9/2016), the Catalog Of Somatic Mutations In Cancer (COSMIC) (v68), RegulomeDB19, and FunSeq20. Analyses were performed using custom bash, Perl, R scripts, and VarSifter21. Variants distribution plots were realized using Lollipops (https://zenodo.org/badge/latestdoi/20224/pbnjay/lollipops). We obtained an average of 2.7 million reads per sample, 99% aligning to the human genome and an average depth of coverage of 83.9 (Supplementary Fig. 1). After filtering for genotype quality, read depth and 1 kG population frequencies, we identified a total of 2,270 single nucleotide variants and 490 indels with allelic frequency of less than 1% in the 1 kG Admixed American reference populations (Supplementary Fig. 2).
Variant classification
Each non-synonymous coding variant was assessed individually for classification into the following categories: (1) likely benign or benign, (2) likely pathogenic or pathogenic, (3) variant of unknown significance (VUS). This classification did not distinguish variants of high penetrance from those of moderate penetrance sometimes refered to as ‘risk factor’ in public databases. First, 16 variants that have been classified as benign or likely benign by expert panels (ENIGMA for BRCA1 and BRCA222 and Insight for Lynch Syndrome genes23) were not further investigated (n = 17). For the remaining variants, those that had a frequency >0.5% in any of the gnomAD24 populations (African, East Asian, non-Finnish European, Latino, South Asian) were filtered from the analysis unless they had been reported as likely pathogenic or pathogenic on ClinVar25.Finally, the remaining variants were classified based on the ACMG criteria26. The frequency of likely pathogenic/pathogenic variants and VUS identified in NCCN clinically actionable gene was assessed in a cohort of 500 women that do not have cancer history by multiplex PCR using a Sequenom analyzer. In Silico predictions of functional significance was obtained for each non-synonymous variant using the following algorithms: fathmm27, MutationAssessor28, PolyPhen-229 and SIFT30.
Ancestry
Global ancestry proportions were estimated using a panel of 106 ancestry informative SNPs that can discriminate indigenous American, African, and European ancestry31. Genotyping was done by multiplex PCR using a Sequenom analyzer as previously described31. For each individual, respective proportions of European, African and Native American ancestry were estimated using ADMIXTURE32. In pathogenic variant carriers, local ancestry at the site of the genes of interest was extracted from Affymetrix Axiom UK array genotypes that had been phased using Shape-it33 and for which locus-specific ancestry was determined using RFMix34.
CHK2 kinase assays
To determine the impact of CHEK2 missense variants we inserted p.Glu239Lys, p.Pro484Leu, and pArg519Leu variants in a wild type human CHEK2 sequence in pCMV2-FLAG by site directed mutagenesis using QuikChange II kit (Agilent, Santa Clara, CA) (primers available upon request). We also constructed two known pathogenic variants, p.His143Tyr and p.Ile157Thr, to serve as negative controls. Constructs were confirmed by Sanger sequencing.
Human embryonic kidney 293FT cells were transfected with empty pCMV2-FLAG vectors, or with expression vectors containing wild-type CHK2 or variants using Fugene HD (Promega, Madison, WI). After ~48 h cells were exposed to 6 Gy of ionizing radiation (IR) and lysates were collected after 1 h in RIPA buffer (150 mM NaCl, 10 mM Tris-Cl [pH 7.4], 5 mM EDTA, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 0.1% sodium deoxycholate). Levels of total and phosphorylated CHK2 in lysates were analyzed by immunoblotting on a 10% SDS-PAGE gel. The following antibodies were used: anti-phospho-Serine 516 CHK2 (Cell signaling), anti-phospho-Threonine 68 CHK2 (Cell signaling), anti-CHK2 (EMD Millipore) and anti-β-actin (Santa Cruz Biotechnology).
For immunoprecipitation and kinase assays cell lysates were incubated at 4 °C for 2 h with anti-FLAG M2 affinity gel. Immunoprecipitates were washed three times with RIPA buffer and used for immunoblotting with anti-phospho-CHK2 and anti-CHK2. For kinase assay, the beads were incubated with Kinase Buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM PMSF, 40 μM cold ATP, and 13 μCi [γ-32P] ATP) and bacterially expressed GST-CDC25C peptide (aa 200–256; containing the phosphorylation site for CHK2) as an exogenous substrate. Kinase reactions were incubated for 30 min at 30 °C. Samples were resolved on a 12% SDS-PAGE gel.
Statistical analyses
Descriptive and statistical analyses were conducted in R Studio35.
Results
Characteristics of the study cohort
The description of demographic, hormonal and clinical characteristics of the study cohort is presented in Supplementary Table 1. Over half of the study participants (56.2%) had their breast cancer diagnosis before the age 50. The majority of the tumors were invasive ductal carcinomas (65.9%), of lower stages; grade I and II represented 77.4% of the invasive tumors, size below 2 cm (76.9%) and luminal A or B subtype (88.2%) (Supplementary Table 1).
Variants
There were 158 coding variants: 2 truncating, 2 deletions, 106 missense and 48 synonymous variants. Each affected proband had on average 0.8 non-synonymous coding VUS, ranging from 0 to 4.
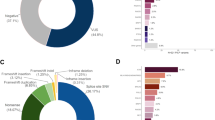
The study participants were classified according to their carrier status of a variant affecting the genes coding regions (truncating, nonsense and missense variants): Group I (n = 5; 10.2%), patients harboring at least one coding variant classified as likely pathogenic or pathogenic (L/Pathogenic) in any gene. Group II (n = 12, 24.5%), patients with VUS in breast cancer susceptibility genes classified as “clinically actionable” by NCCN27. Group III (n = 11, 22.4%), patients with variants in any other gene in the panel. Group IV (n = 21, 42.9%), patients with no pathogenic variant or VUS found in the coding sequences (Fig. 1).
(a) Proportion of unrelated BC patients by carrier status and (b) distribution after grouping by age of onset and family history characteristics. Carriers categories are: harboring at least one coding variant classified as likely pathogenic or pathogenic (L/Pathogenic), VUS in NCCN clinically actionable genes, VUS in other genes, or no L/Pathogenic variant or VUS identified. Early onset breast cancer refers to individuals for which diagnosis was at 45 years of age or less. Positive family history includes only individuals for which a first, or second degree relative was diagnosed with an early onset breast cancer. NCCN clinically actionable BC genes are ATM, BRCA1, BRCA2, CDH1, CHEK2, NBN, NF1, PALB2, PTEN, STK11, and TP53 (NCCN Guidelines for Genetic/Familial High-Risk assessment v3.201925). The complete list of genes included in the BROCA panel is available in the methods section. Coding variants included truncating, missense, deletions and duplications. Synonymous variants were excluded from this analysis. BC breast cancer; VUS variant of uncertain significance.
To assess the prior probability of being carrier, each proband was grouped according to age of onset and/or family history of early onset breast cancer (at or before age 45). There was no significant difference in the distribution of carrier status after grouping probands by age of onset and/or family history of early onset breast cancer (P = 0.8, Fig. 1). It is noteworthy however that 3 out of 4 carriers of a L/pathogenic variant had a breast cancer diagnosis at or before 50 years of age.
Pathogenic variants
After excluding the BRCA2 p.Glu1308Ter carrier (blind control), which was properly identified, four probands in 48 (8.3%) were found to carry a pathogenic variant in a non-BRCA breast cancer gene (Table 1). CHEK2 p.Ile157Thr was observed in two unrelated individuals (Fig. 2). Family history for the first patient was characterized by multiple first-degree relatives with late onset breast cancer, prostate and gastric cancer. The other carrier was diagnosed with breast cancer at the age of 44 years with no known family history of cancer.
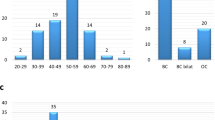
Family history of cancer (site and age of diagnosis, dx) in relatives of carriers of a known or predicted pathogenic variant in CHEK2 (a), MUTYH (b), and RAD51B (c). Sites of carcinoma are Br breast cancer, Col colorectal cancer, Gas gastric cancer, MM multiple myeloma, Pr prostate cancer. Probands are designated by a red arrow.
The patient harboring a pathogenic variant in MUTYH (p.Gly396Asp, PRI-117 III-1) was diagnosed with breast cancer at the age of 50 and had a brother with early onset colorectal cancer. A truncating variant, p.Arg47* was identified in RAD51B predicted to result in a protein lacking ~90% of the coding region. This patient (PRI-136 III-1) was diagnosed with an invasive ductal carcinoma at age 33 and family history included early onset breast and colorectal cancer. Pedigrees representing family histories of patients carrying known pathogenic variants are depicted in Fig. 2.
Variants of uncertain clinical significance
The location of coding variants in each gene is depicted in Supplementary Fig. 3. Tables 1 and 2 present all the non-synonymous variants classified as L/Pathogenic and VUS by ClinVar25 and/or using ACMG26 criteria. The highest number of VUS was observed in ATM (n = 6) (Table 1), a clinically actionable breast cancer gene. Despite having received a negative BRCA1/BRCA2 test result, two missense VUS were identified in BRCA2.
All of the non-synonymous variants identified have previously been reported either in ClinVar25 or in the gnomAD24 databases. Despite a low observed prevalence in gnomAD, BLM p.Arg1407Lys and CDH1 p.Tyr523Cys were each present in two unrelated individuals in this cohort. Variants ATM p.Val2937Ala and BLM p.Ser854Asn, have been observed in the gnomAD Latino population exclusively (Supplementary Table 2). For variants that have been reported in more than one gnomAD populations, some exhibited significant differences in frequencies, notably BLM p.Arg1407Lys and CDH1 p.Tyr523Cys, which were 32 and 14 times more frequent in gnomAD Latinos than in non-Finnish European, respectively.
In Silico prediction analysis of functional significance led to conflicting interpretation for most of the non-synonymous variants, with the exception of ATM p.Arg2227His, MUTYH p.Glu396Asp, and SDHB p.Ser239Cys which were predicted damaging by all four models tested (Supplementary Table 3).
In 27.1% of the cohort (n = 13), no known pathogenic variant or coding VUS were identified. Among those, PRI-185 stands out for having strong family history of cancer including early onset breast cancer, ovarian cancer and colorectal cancer (data not shown). For this individual, the prior risk of being a BRCA carrier was estimated at 14.7% using the BRCAPRO model (data not shown).
Functional impact of the CHEK2 variants
In addition to the pathogenic CHK2 variant found in two unrelated probands (Table 1), we identified three rare missense variants of uncertain significance (p.Glu239Lys, p.Pro484Leu, and p.Arg519Leu) (Table 2). To assess their impact on CHK2 stability and activity we expressed epitope-tagged CHK2 constructs containing the variants in 293T HEK cells and compared them to the wild-type CHK2 construct (positive control) and with known CHK2 pathogenic variants p.His143Tyr and p.Ile157Thr.
Variant p.Pro484Leu, located at the end of the kinase domain showed dramatically decreased levels of expression when compared with the wild type (Fig. 3C,D). Levels of expression for p.Pro484Leu were lower than for both pathogenic variants p.His143Tyr and p.Ile157Thr, but differently from p.His143Tyr it still retains the ability to auto-phosphorylate at Serine 516 and to be phosphorylated by ATM at Threonine 68 (Fig. 3C,D). Variants p.Glu239Lys and p.Arg519Leu showed levels comparable to the wild type CHK2 (Fig. 3C,D).
(A) Diagram human CHK2 protein domains and variants tested in kinase assays in the current study (purple) or known pathogenic variants (red). (B) Ribbon and Surface view of human CHK2 dimer (aa 89–501) with variants tested in this study (purple) and known pathogenic variants (red) (PDB ID: 3I6U). (C) Western blot analysis of CHK2 expression and activation in HEK293T expressing wild-type CHK2 or the indicated missense variants. Cells were exposed to 6 Gy IR and WCL (whole cell lysates) were extracted 1 h post-exposure. (D) Immunoprecipitated FLAG-CHK2 complexes from HEK293T lysates were immunoblotted for CHK2 and phospho-CHK2. E. In vitro kinase assays from HEK293T cell lysates. FLAG-CHK2 immunoprecipitates were incubated with a purified GST-CDC25C (aa 200–256) peptide. Both auto-phosphorylation of CHK2 and phosphorylation of CDC25C were used as a measure of CHK2 kinase activity.
In an in vitro kinase assay, p.Glu239Lys and p.Pro484Leu showed markedly decreased kinase activity comparable to the known pathogenic variant while variant p.Arg519Leu resulted in substrate phosphorylation levels similar to wild type CHK2 (Fig. 3E). In conclusion, functional analysis suggest that two CHEK2 variants found in our cohort (p.Glu239Lys and p.Pro484Leu) are likely to affect CHK2 activity in vivo and be associated with increased risk of breast cancer. While the evidence is less clear for p.Arg519Leu, this variant in the nuclear localization sequence and proximal to the Serine 516 site seems to auto-phosphorylate less efficiently (Fig. 3C,D).
Non coding variants
To determine whether our patients harbored potentially functional variants in addition to protein alteration, we examined variants with the two highest functional score using two annotation tools: RegulomeDB (classes: 1a/f, 2a/b/c) and FunSeq (classes: 5, 4). Of the six variants most likely to be functional by both methods, two were near CTNNA1: one was considered to be in an ultra-sensitive region by FunSeq (11 probands), and the other was predicted to be motif breaking for HDAC2 (3 probands). An MSH6 intronic variant was predicted to be motif breaking for ETS1, GABPA, and EGR1 (3 probands). A variant in the upstream promoter region of STK11 was predicted to be motif breaking for SP1 (1 proband). The promoter region of ATM harbored a variant predicted to be motif breaking for BCL11A and BCL3 (1 proband). Finally, a PALB2 promoter region variant is predicted to be motif breaking for MAX, MYC, and USF1 (1 proband).
Global and local ancestry in carriers of pathogenic variants
In the study cohort, the contribution of each ancestral population to the genome was 74.3% (SD 16.2), 14.9% (SD 12.2) and 10.8% (SD 7.4) for European, African and Native American ancestry, respectively (Supplementary Fig. 4,A). Carriers of a pathogenic variant were distributed throughout the spectrum of global ancestries. On the CHEK2 locus, both carriers of the p.Ile157Thr pathogenic variant were homozygous for European ancestry (Supplementary Fig. 4,B). The MUTYH p.Gly396Asp pathogenic variant was also located in a European ancestral context (Supplementary Fig. 4,C). In contrast, the RAD51B p.Arg47* variant was located on a chromosome segment of African origin (Supplementary Fig. 4,D).
Discussion
This study aimed at identifying inherited pathogenic variants in BRCA-negative breast cancer cases from Puerto Rico. Pathogenic variants were identified in CHEK2, MUTYH, and RAD51B in four cancer patients, which represented 8.3% of the cohort. In addition, ~30% of the VUS identified were found in NCCN actionable genes. Our study reveals an unmet clinical need of genetic testing that could benefit a significant proportion of at-risk women.
We identified four carriers with pathogenic or likely pathogenic CHEK2 variants (p.Ile157Thr in two unrelated individuals, p.Glu239Lys and p.Pro484Leu), which represents over 8% of our cohort. In ClinVar, the p.Ile157Thr variant (VCV000005591.3) currently has conflicting interpretations with most submissions representing Likely pathogenic (8) or Pathogenic (17/20). The Ile157 residue is central in the FHA-kinase domain interface and functionally analyses of this variant by several independent groups indicates that the substitution generates a partially defective protein36,37,38,39,40. The p.Ile157Thr variant may confer a modest increase in unselected and familial BC risk (OR ~ 1.5) and a high risk (OR > 4) for lobular breast cancer, although risk estimates are not available for Puerto Rican women41.
Two missense VUS (p.Glu239Lys and p.Pro484Leu), located in the N- and C-terminal lobes of the kinase domain36,38, respectively, were shown to have marked decreases in kinase activity, and may constitute pathogenic variants. Although not central to its stabilization, the p.Glu239Lys is also located at the FHA-Kinase dimerization interface36. A change from a negatively to a positively charged amino acid might have an impact on CHK2 dimerization. Pro484 faces away from the dimerization axes but replacing proline may be reflected in a less rigid positioning of the alpha helix adjacent to the C-terminal end of the kinase domain36. Of note, although p.Arg519Leu, located in the NLS region, only displayed mild effects, our assay did not evaluate its cellular distribution. It is currently a variant of uncertain significance.
Significant differences in the prevalence of CHEK2 variants across ethnic groups have been observed in the literature42. The cancer spectrum in families of carriers of pathogenic or likely pathogenic CHEK2 variants based on functional assays, was consistent with the literature including breast, early onset breast, prostate, and colorectal cancers. Remarkably, two out of four families reported multiple cases of second primary or bilateral breast cancers (PRI-177 III-2 and III-3; PRI-193 III-1 and IV-1), consistent with published meta-analysis reporting the association of CHEK2 with second primary breast cancers43. Potentially pathogenic CHEK2 variants were overrepresented in this cohort, suggesting that CHEK2 variants may affect the Puerto Rico population disproportionately. We have previously described a founder effect for a recurrent BRCA2 variant in Puerto Rico (p.Glu1308Ter)13, a phenomenon that is likely contributing to increased variant frequencies in this population.
One patient was heterozygous for a known pathogenic variant in MUTYH, with a family history of early onset colorectal cancer consistent with the reported cancer risk for this gene. MUTYH-associated polyposis is inherited as an autosomal recessive trait44, with bi-allelic carriers demonstrating up to 28-fold increase in colorectal cancer risk45. Mono-allelic carriers have modest increases in risk of colorectal cancer, which was shown to differ in magnitude based on the variant46,47 and family history of colorectal cancer47. There is also evidence supporting a role of MUTYH in extra colonic tumor risk48, including emerging associations with the risk of BC, as two missense variants in MUTYH were shown to confer a significant increase in breast cancer risk in Sephardic Jews, a population in which the MUTYH mutation prevalence was high49. As of now, the data supporting the association between monoallelic MUTYH pathogenic variants and BC risk remains sporadic and additional evidence will be required before management guidelines are modified for this gene.
A RAD51B truncating variant p.Arg47* was observed in a family characterized by early onset breast cancer and colorectal cancer. Although a segregation analysis would be required to assess the carrier status of relatives with colorectal cancer, to our knowledge, there has been no report of the association of RAD51B variant with the familial risk of colorectal cancer. In a French study, RAD51B likely deleterious variants were observed in both breast-ovarian and breast-only cancer families50. GWAS have consistently identified the 14q24.1 genomic interval locus as a breast cancer susceptibility locus, which can be broken down into 2 signals mapping within or close to RAD51B. The first has been shown to be associated with triple negative phenotype and mammographic density51, and the second with male breast cancer risk52. When combined with GWAS evidence, our results suggest that RAD51B may be a candidate that warrants reconsideration for inclusion in breast cancer susceptibility gene panels. The local ancestry at the RAD51B locus in the carrier of p.Arg47* was predicted to be of African origin, which highlights the complexity of Hispanic populations in studying genetic basis to disease as pathogenic factor may originate from any of the ancestral populations that make up their genetic backgrounds. The identification of pathogenic variants in genes not commonly included in hereditary cancer testing is a reminder that negative results, especially in the context of strong family history, should be interpreted with care.
In the current study, over 46% of the study participants were found to carry a non-synonymous coding VUS. In the BRCA1/2 genes, previous studies have reported higher rates of novel mutations or VUS in historically understudied populations53,54,55. In the clinic, VUS are associated with additional challenges. In U.S community-based practices, clinicians reported less confidence in interpreting and counseling multigene panel testing for cancer risk assessment when compared to single-gene testing56. In a survey among members of the National Society of Genetic Counselors, only 63.2% felt their patients understood the meaning of a VUS result57. For counselees, VUS results are less understood58 and have been shown to be associated with psychological distress59. The BROCA panel was developed to provide comprehensive testing for hereditary cancers, and includes several genes that have not been associated with BC risk. While standardized panel simplifies genetic testing, it also results in a higher probability of identifying VUS, which remain challenging in the clinic.
In 42.9% of the cohort, no potential causative variant was identified. Familial aggregation and twin studies have shown that even in non-hereditary breast cancer cases, a significant proportion of risk is inherited60,61. To date, genome-wide association studies (GWAS) have identified over 100 loci associated with breast cancer risk62, accounting for up to 14% of the familial risk attributed to common variants63. Although the risk associated with individual GWAS loci is not elevated enough to inform clinical decisions, polygenic risk scores were proposed as risk stratification tools in population screening programs and targeted prevention64,65,66,67. It is possible that high-risk families for which no potential causative variant is identified share an excess of low penetrance variants.
Although this is the first study addressing specifically hereditary cancer risks in an entirely Puerto Rican population, the sample size was limited. In addition, because the inclusion criteria required a BRCA1/2 negative clinical test result, patients of higher socio-economic status with better access to health care are likely to be overrepresented in this cohort. While the clinic where recruitment took place serves patients from all over the island, its location in the San Juan metropolitan area probably favored the recruitment of patients from the metropolitan area. The European contribution to the global genetic ancestry of the cohort was estimated at 74.3%, which is higher than the previously reported average of 63.7% for the Island in a census based population68. European ancestry in Puerto Rico has been associated with higher socio-economic status68. Consequently, having access to BRCA genetic testing, which was an inclusion criterion in this study, is likely to be correlated with a higher socio-economic status and might explain this enrichment in European ancestry.
Our understanding of the genetic factors driving disease risk is biased towards populations of European descent69. This is especially concerning, as it has been documented that rarer variants are more likely to show geographic clustering. In this study, we provide the first description of the pathogenic variants underlying BC genetic risk in Puerto Rican Hispanic women who tested negative for a BRCA mutation. Over 12% of the study participants had a known pathogenic variant or a variant that was predicted pathogenic by functional assay. For a large proportion of the genes included in hereditary cancer gene panels, clinical guidelines for management of risk have yet to be established pending the availability of accurate estimates of their respective cancer spectrum and the magnitude of associated risk.
References
Kuchenbaecker, K. B. et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017).
Metcalfe, K. A., Kim Sing, C., Ghadirian, P. & Narod, S. A. Health care provider recommendations for reducing cancer risks among women with a BRCA1 or BRCA2 mutation. Clin. Genet. 85, 21–30 (2014).
Grann, V. R. et al. Effect of prevention strategies on survival and quality-adjusted survival of women with BRCA1/2 mutations: an updated decision analysis. J. Clin. Oncol. 20, 2520–2529 (2002).
Weitzel, J. N. et al. Effect of genetic cancer risk assessment on surgical decisions at breast cancer diagnosis. Arch. Surg. 138, 1323–1329 (2003).
Walsh, T. & King, M. C. Ten genes for inherited breast cancer. Cancer Cell 11, 103–105 (2007).
Easton, D. F. et al. Gene-panel sequencing and the prediction of breast cancer risk. N. Engl. J. Med. 372, 2243–2257 (2015).
Desmond, A. et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 1, 943–951 (2015).
Eccles, D. M. et al. & ENIGMA Clinical Working Group. BRCA1 and BRCA2 genetic testing-pitfalls and recommendations for managing variants of uncertain clinical significance. Ann. Oncol. 26, 2057–2065 (2015).
Penedo, F. J. et al. Self-reported cancer prevalence among Hispanics in the US: results from the Hispanic Community Health Study/Study of Latinos. PLoS ONE 11, e0146268 (2016).
Iqbal, J., Ginsburg, O., Rochon, P. A., Sun, P. & Narod, S. A. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA 313, 165–173 (2015).
Dutil, J. et al. The spectrum of BRCA1 and BRCA2 alleles in Latin America and the Caribbean: a clinical perspective. Breast Cancer Res. Treat. 154, 441–453 (2015).
Dutil, J., Colon-Colon, J. L., Matta, J. L., Sutphen, R. & Echenique, M. Identification of the prevalent BRCA1 and BRCA2 mutations in the female population of Puerto Rico. Cancer Genet. 205, 242–248 (2012).
Diaz-Zabala, H. J. et al. A recurrent BRCA2 mutation explains the majority of hereditary breast and ovarian cancer syndrome cases in Puerto Rico. Cancers 10, E419 (2018).
Walsh, T. et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. USA 107, 12629–12633 (2010).
Hogervorst, F. B. et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 63, 1449–53 (2003).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res. 38, e164–e164 (2010).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Khurana, E. et al. 1000 Genomes Project Consortium, Dermitzakis, E.T., Yu, H., Rubin, M.A., Tyler-Smith, C. & Gerstein, M. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 342, 1235587–1235587 (2013).
Teer, J. K., Green, E. D., Mullikin, J. C. & Biesecker, L. G. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics 28, 599–600 (2012).
Spurdle, A. B. et al. & ENIGMA. ENIGMA–evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum. Mutat. 33, 2–7 (2012).
Plazzer, J. P. et al. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam. Cancer 12, 175–80 (2013).
gnomAD genome aggregation database, https://gnomad.broadinstitute.org/ (accessed on August 2019).
National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Genetic/Familial High-Risk Assessment: Breast and Ovarian, http://www.nccn.org/ (accessed August 2019).
Richards, S. et al. & ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–24 (2015).
Shihab, H. A. et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34, 57–65 (2013).
Reva, B., Antipin, Y. & Sander, C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 8, R232 (2007).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 76, 7.20.1–7.20.41 (2013).
Vaser, R., Adusumalli, S., Leng, S. N., Sikic, M. & Ng, P. C. SIFT missense predictions for genomes. Nat. Protoc. 11, 1–9 (2016).
Fejerman, L. et al. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res. 68, 9723–9728 (2008).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655–1664 (2009).
Delaneau, O., Coulonges, C. & Zagury, J.-F. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics 9, 540 (2008).
Maples, B. K., Gravel, S., Kenny, E. E. & Bustamante, C. D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013).
R Studio Team. RStudio: Integrated Development for R. RStudio, Inc, Boston, MA, http://www.rstudio.com/ (accessed August 2017).
Cai, Z., Chehab, N. H. & Pavletich, N. P. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol. Cell. 35, 818–29 (2009).
Falck, J., Mailand, N., Syljuåsen, R. G., Bartek, J. & Lukas, J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410, 842–7 (2001).
Li, J. et al. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 9, 1045–54 (2002).
Chrisanthar, R. et al. CHEK2 mutations affecting kinase activity together with mutations in TP53 indicate a functional pathway associated with resistance to epirubicin in primary breast cancer. PLoS One 3, e3062 (2008).
Kleiblova, P. et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int. J. Cancer 145, 1782–1797 (2019).
Liu, C., Wang, Y., Wang, Q. S. & Wang, Y. J. The CHEK2 I157T variant and breast cancer susceptibility: a systematic review and meta-analysis. Asian Pac. J. Cancer Prev. 13, 1355–60 (2012).
Bell, D. W. et al. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int. J. Cancer 121, 2661–2667 (2007).
Weischer, M. et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J. Clin. Oncol. 30, 4308–4316 (2012).
Sampson, J. R. & Jones, N. MUTYH-associated polyposis. Best Pract. Res. Clin. Gastroenterol. 23, 209–218 (2009).
Theodoratou, E. et al. A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br. J. Cancer. 103, 1875–84 (2010).
Ma, X., Zhang, B. & Zheng, W. Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut 63, 326–36 (2014).
Win, A. K. et al. Cancer risks for monoallelic MUTYH mutation carriers with a family history of colorectal cancer. Int. J. Cancer 129, 2256–62 (2011).
Vogt, S. et al. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology 137, 1976–1985.e10 (2009).
Rennert, G. et al. MUTYH mutation carriers have increased breast cancer risk. Cancer 118, 1989–1993 (2012).
Golmard, L. et al. Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer 13, 484 (2013).
Vachon, C. M. et al. Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 21, 1156–1166 (2012).
Orr, N. et al. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nature Genet. 44, 1182–1184 (2012).
Solano, A. R. et al. BRCA1 and BRCA2 analysis of Argentinean breast/ovarian cancer patients selected for age and family history highlights a role for novel mutations of putative south-American origin. Springerplus 1, 20 (2012).
Calderón-Garcidueñas, A. L., Ruiz-Flores, P., Cerda-Flores, R. M. & Barrera-Saldaña, H. A. Clinical follow up of Mexican women with early onset of breast cancer and mutations in the BRCA1 and BRCA2 genes. Salud Pública Méx. 47, 110–115 (2005).
Hall, M. J. et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast‐ovarian cancer. Cancer 115, 2222–2233 (2009).
Blazer, K. R. et al. Next-generation testing for cancer risk: perceptions, experiences, and needs among early adopters in community healthcare settings. Genet. Test. Mol. Biomarkers 19, 657–665 (2015).
Petrucelli, N., Lazebnik, N., Huelsman, K. M. & Lazebnik, R. S. Clinical interpretation and recommendations for patients with a variant of uncertain significance in BRCA1 or BRCA2: a survey of genetic counseling practice. Genet. Test. 6, 107–113 (2002).
Richter, S. et al. Variants of unknown significance in BRCA testing: impact on risk perception, worry, prevention and counseling. Ann. Oncol. 2013 24(Suppl 8), viii69–viii74 (2013).
O’Neill, S. C. et al. Distress among women receiving uninformative BRCA1/2 results: 12‐month outcomes. Psycho‐Oncology 18, 1088–1096 (2009).
Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer — analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85 (2000).
Peto, J. & Mack, T. M. High constant incidence in twins and other relatives of women with breast cancer. Nat. Genet. 26, 411–414 (2000).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361 (2013).
Pharoah, P. D. P., Antoniou, A. C., Easton, D. F. & Ponder, B. A. J. Polygenes, risk prediction, and targeted prevention of breast cancer. N. Engl. J. Med. 358, 2796–2803 (2009).
Mavaddat, N. et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 107, djv036 (2015).
Burton, H., Chowdhury, S., Dent, T. & Hall, A. Public health implications from COGS and potential for risk stratification and screening. Nat. Genet. 45, 349–51 (2013).
Garcia-Closas, M., Gunsoy, N. B. & Chatterjee, N. Combined associations of genetic and environmental risk factors: implications for prevention of breast cancer. J. Natl. Cancer Inst. 106, dju305 (2015).
Via, M. et al. History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS ONE 6, e16513 (2011).
Popejoy, A. B. & Fullerton, S. M. Genomics is failing on diversity. Nature 538, 161–164 (2016).
Acknowledgements
The authors are grateful to all the women who participated in this study. We also want to thank the clinical collaborators, Wanda Vargas at Ponce Research Institute and Terangelie Rivera at Auxilio Mutuo Hospital. This work was supported by grants of the National Institute of Health (NIH) National Cancer Institute (NCI) (U54 CA163071 and U54 CA163068 for BROCA analysis and 1SC1CA182845 for ancestry analyses). This work was supported in part by the Molecular Genomics Core Facilities at the Moffitt Cancer Center through its NCI CCSG grant (P30-CA76292).
Author information
Authors and Affiliations
Contributions
Idea conception, planning and funding acquisition: J.D., J.K.T., J.L.M., A.N.M.; Acquisition of data: J.D., V.G., S.Y., N.A., R.K., M.E., A.N.M.; Data analysis and interpretation: J.D., J.K.T., V.G., W.L.T., R.K., A.N.M.; Writing of the original draft: J.D., J.K.T., A.N.M.; Manuscript review and editing: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dutil, J., Teer, J.K., Golubeva, V. et al. Germline variants in cancer genes in high-risk non-BRCA patients from Puerto Rico. Sci Rep 9, 17769 (2019). https://doi.org/10.1038/s41598-019-54170-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54170-6
This article is cited by
-
Genetic and chemotherapeutic influences on germline hypermutation
Nature (2022)
-
Germline mutational spectrum in Armenian breast cancer patients suspected of hereditary breast and ovarian cancer
Human Genome Variation (2021)
-
Prevalence of germline variants in consensus moderate-to-high-risk predisposition genes to hereditary breast and ovarian cancer in BRCA1/2-negative Brazilian patients
Breast Cancer Research and Treatment (2021)
-
New germline mutations in non-BRCA genes among breast cancer women of Mongoloid origin
Molecular Biology Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.