Abstract
Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) is an accurate and fast method to measure gene expression. Reproducibility of the analyses is the main limitation of RT-qPCR experiments. Galaxy is an open, web-based, genomic workbench for a reproducible, transparent, and accessible science. Our aim was developing a new Galaxy tool for the analysis of RT-qPCR expression data. Our tool was developed using Galaxy workbench version 19.01 and functions implemented in several R packages. We developed PIPE-T, a new Galaxy tool implementing a workflow, which offers several options for parsing, filtering, normalizing, imputing, and analyzing RT-qPCR data. PIPE-T requires two input files and returns seven output files. We tested the ability of PIPE-T to analyze RT-qPCR data on two example datasets available in the gene expression omnibus repository. In both cases, our tool successfully completed execution returning expected results. PIPE-T can be easily installed from the Galaxy main tool shed or from Docker. Source code, step-by-step instructions, and example files are available on GitHub to assist new users to install, execute, and test PIPE-T. PIPE-T is a new tool suitable for the reproducible, transparent, and accessible analysis of RT-qPCR expression data.
Similar content being viewed by others
Introduction
Quantitative real-time polymerase chain reaction (qPCR) is a routinely used technique for the detection of specific nucleic acids, RNA expression profiling, quantification of DNA and DNA methylation, and validation of microarray hybridization data1. Reverse transcription qPCR (RT-qPCR) is an accurate, sensitive, and fast method to quantify gene expression from qPCR experiments2, and is widely accepted as the Golden Standard for the analysis of gene expression1,3. Briefly, RT-qPCR measures the expression of a set of target RNAs through repeated cycles of sequence-specific amplification followed by expression measurements4. The cycle at which the observed expression first exceeds a user-specified threshold is commonly called the threshold cycle (Ct) or quantification cycle. The Ct values of the target RNAs represent a quantitative assessment of gene expression and are often treated as the raw data for subsequent analyses4. Two methods can be used to quantify gene expression from the Ct value: the absolute and the relative quantification3. In the absolute quantification, a standard curve is used as reference calibrator. In the relative quantification, the signal is related to the expression of a user-specified group3. Therefore, the difference between the two approaches depends on the data used as reference calibrator to which relating the signal.
In many RT-qPCR experiments not all Ct values can be numerically defined. For example, when the starting RNA abundance is too low, or an off-target product is amplified, or no reliable Ct can be determined, the corresponding Ct value cannot be quantified numerically and is flagged as missing value5. Handling missing data is a crucial step in the analysis of RT-qPCR experiments because procedures used in the subsequent analyses of these data are based on statistics that are unable to handle both numeric and missing values4. Imputation is an established technique to solve the problem6. Imputation substitutes a missing value with a rationally selected numeric value4. K-nearest neighbors (KNN)6, maximum Ct plus one cycle (Mestdagh)7, and cubic spline interpolation (Cubic)1 are known methods to impute missing values in RT-qPCR data5,6.
Another key step in the analysis of RT-qPCR data is the assessment of true biological changes associated with the phenomenon or disease of interest. In fact, biological changes are often masked by nonspecific technical variability introduced in the data during the experimental procedure6. Data normalization is expected to reduce/eliminate any technical variability without affecting the true biological results6. Global mean8, DeltaCt based on universal normalizers9, Modified global mean10, Quantile9, and Rank Invariant9 are among the most accepted methods used for RT-qPCR data normalization5.
RT-qPCR experiments allow measuring the expression of several transcripts in parallel using high-density plates9. Plates have been used in several explorative studies to find novel biomarkers from the analysis of different diseases, tissues, experimental conditions, and cell types3,5,6. The large number of studies published in the literature stimulated companies to develop commercial technologies to perform RT-qPCR experiments3. For each experiment, these technologies generate textual reports summarizing a number of experimental parameters and data such as feature name, quality control flags, and Ct values. Different technologies generate reports that can be of different format. According to our experience, SDS, EDS, and OpenArray are among the most used file formats for reporting results of RT-qPCR experiments.
Although the computational procedures and technologies for analyzing RT-qPCR data are well established, the heterogeneity of the assays employed in RT-qPCR experiments and the lack of a consensus on the best normalization system and on the missing values imputation approach to adopt makes it hard to set up a standardized analysis procedure6. Furthermore producing high quality publications and reproducible data are among the most critical pitfalls of qPCR experiments11.
Several open-access software packages, tools, and web applications, such as R packages, have been proposed in the last years for the analysis of RT-qPCR data1. HTqPCR is a well-known open source R\Bioconductor package for the high-throughput analysis of RT-qPCR data9. It provides several functions and parameter options for assessing the quality of the experiment, filtering unreliable data, normalizing raw data, finding potential candidate biomarkers, and visualizing RT-qPCR data9. However, R-based analysis suffers from some known limitations. First of all, analysis procedures are implemented in several packages lacking a unified framework. Second, users with biological background who want to use the functionalities of R packages need non-trivial coding skills. Furthermore, the lack of a simple framework for reusing, sharing, and communicating experimental procedures and results limits reproducibility, transparency, and accessibility of R-based analysis12.
Galaxy is an open, collaborative, web-based, genomic workbench for a reproducible, transparent, and accessible science12. Galaxy provides a very active developer community. More than 6746 public tools and workflows are freely available in the Galaxy Tool Shed repositories12. New tools and workflows are easily deployable in the Galaxy repositories. To this purpose, Galaxy offers fresh installations of R and Python environments, a fast dependency resolver, a step-by-step documentation, a simple graphical interface, and GitHub integration13. However, to the best of our knowledge, no Galaxy tool or workflow has been reported to date for analyzing RT-qPCR data.
In the present work, we developed pipette (PIPE-T), a new tool for analyzing RT-qPCR expression data integrating the functionalities implemented in various R packages into one unified, reusable, transparent, accessible, and easy to use Galaxy wrapper.
Methods
Overview of the main procedures implemented by PIPE-T
PIPE-T implements the relative quantification method using the R language and computing environment14.
To start a PIPE-T analysis, users must upload two input files:
-
A List collection of tab-separated text files for all samples generated as report of the RT-qPCR experiment (ListOfFile).
-
A tab-separated text file associating each filename in ListOfFile with a treatment group (FileTreatment).
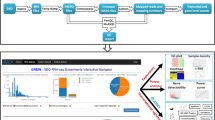
Five distinct computational procedures are implemented in PIPE-T. Procedures are summarized in Fig. 1 and a detailed description of each procedure is provided in the following sections.
Schematic representation of the analysis procedures implemented in PIPE-T. Input files are uploaded and parsed for initiating the analysis. Transcripts are categorized according to a user-defined range of values and\or quality control flag to label unreliable Ct values. Ct values are normalized to reduce\remove technical variability in the data. Transcripts are filtered out according to a user-specified maximum number of missing values to maintain the bias as low as possible. Imputation is applied to handle missing values. Transcripts discriminating between two treatments are identified for subsequent analyses.
The execution of PIPE-T outputs the following output files:
-
A tab-separated text file containing the raw Ct values for every sample and transcript
-
A PNG file showing the distribution of the Ct values of every samples obtained after the Ct filtering and categorization step visualized as sequence of boxplots.
-
A tab-separated text file containing the normalized Ct values
-
A PNG file showing the cumulative distribution plot before and after data normalization of the coefficient of variation of every transcript.
-
A PNG file showing the distribution of the normalized Ct values visualized as sequence of boxplots.
-
A tab-separated text file containing data after imputation
-
A tab-separated text file containing the results of the differential expression analysis.
File uploading and parsing
Heterogeneity of assays quantifying RT-qPCR gene expression is often associated with heterogeneity of the file formats reporting data summarizing the results of the RT-qPCR experiment. Hence, it is crucial that the user uploads files whose content is compliant with the file format parsable by PIPE-T before running any PIPE-T analysis.
“Upload File from your computer” is a Galaxy tool that allows uploading files into Galaxy. This tool is available on any fresh Galaxy instance or on the main Tool Shed repository15.
PIPE-T processes tab-separated text files containing a dot as decimal separator uploaded with “Upload File from your computer” tool. The formats supported by PIPE-T are:
-
Applied Biosystems Sequence Detection Systems (SDS)
-
ThermoFisher Experiment Detection Systems (EDS)
-
Applied Biosystems OpenArray (OpenArray)
-
Roche LightCycler (LightCycler)
-
Bio-Rad CFX (CFX)
-
Fluidigm Biomark Table format (BioMark)
-
User-formatted plain text (Plain)
SDS, OpenArray, LightCycler, CFX, BioMark, and Plain are HTqPCR R package9 parsable file formats. We updated the parsing procedure to adapt it working with R 3.5.0 and tab-separated text files. We extended the list of the parsable file formats including the possibility of processing EDS format, which is one of the most used by Thermo Fisher Scientific real-time qPCR instruments.
FileTreatment should have only two columns named SampleName and Treatment. The column named SampleName lists the name and the extension of the files uploaded into the ListOfFile collection. The column named Treatment associates each sampleName with an experimental condition or group of interest. Group specification is necessary since PIPE-T implements the relative quantification method to analyze data from RT-qPCR experiments. PIPE-T admits the specification of two treatment groups. In the GitHub documentation we provided a checklist of recommendations to help users formatting their input files and checking that these files contain sufficient data to run PIPE-T without errors.
If file format is correct, PIPE-T populates a qPCRset object containing the following data for each transcript and sample:
-
Raw Ct\Cq value,
-
Value of the internal quality control flag,
-
Transcript and sample names,
-
FeatureCategory
Data parsing and qPCRset object generation are carried out using the readCtData function of the HTqPCR R package9.
Ct filtering and categorization
Feature categorization is a procedure for describing the level of reliability of a transcript and can be used to filter out features whose expression is not sufficiently reliable9. HTqPCR package defines three possible categories: “Undertermined”, “Unreliable”, and “OK”9. “Undetermined” is used to flag Ct values above a user-defined threshold, and “Unreliable” indicates Ct values that are so low as to be estimated by the user to be problematic9.
By default, only Ct values labeled as “undetermined” in the input data files are placed into the “Undetermined” category, and the rest are classified as “OK”9.
The FeatureCategory for a transcript can be altered on the basis of two criteria9:
-
Range of Ct values. Some Ct values might be too high or too low to be considered a reliable measure of gene expression in the sample and, therefore, should not be marked as “OK”.
-
Flags. Depending on the qPCR input, the values might have associated flags, such as “Passed” or “Failed”, which are used for assigning categories.
PIPE-T implements the two criteria allowing users to set up a range of Ct values and a List button. Any Ct value exceeding the user-defined range is categorized as “Unreliable”. Users can force PIPE-T to check internal control flag status. In this case, the FeatureCategory for a transcript is replaced by an “Undetermined” if the transcript did not pass internal quality control.
PIPE-T uses FeatureCategory labels to replace any Ct values corresponding to “Undertermined” and “Unreliable” with a not accessible value (NA).
These operations are carried out using setCategory and filterCategory functions of HTqPCR package9.
Normalization
Data normalization allows to minimize unwanted systematic technical and experimental variation in the data for better appreciating true biological changes16.
PIPE-T offers six different normalization options that are listed below:
Global mean, quantile, norm rank invariant, and scale rank invariant were already implemented in HTqPCR R package9. However, as Norm Rank Invariant and Scale rank invariant worked only if missing values were absent, we extended the procedure substituting any missing value with a numeric value using the na.spline function implemented in the zoo R package17. D’haene and collegues showed the benefits of using the geometric mean for the normalization of microRNA expression data by introducing the so-called modified global mean method10. For these reasons, we integrated the modified global mean method in PIPE-T.
PIPE-T supports the deltaCt method. Housekeeping genes can be specified by the user or can be estimated by the geNorm or NormFinder methods implemented in the NormqPCR R\Bioconductor package18. When geNorm is selected, PIPE-T identifies candidate normalizers taking those transcripts whose stability was greater than 1.5 as reported by Vandesompele and collegues19.
Newly implemented normalization methods have been integrated in PIPE-T as an updated version of the function normalizeCtData of the HTqPCR R package9.
Transcript filtering and imputation
High-throughput data may often contain missing values. For this reason, handling missing values is a crucial step of any RT-qPCR analysis5,6. The simplest solution for handling missing values would be to exclude from the analysis any transcript with at least one missing value. In such a case, missing values do not represent a problem anymore because they are removed from the analysis. However, this approach could filter out a considerable number of potential useful transcripts. Another solution would be to take every transcript no matter of the number of missing values. In such a case, all potential useful transcripts are taken into account for subsequent analysis, but the probability of making an error increases with the number of missing values6. In the literature, there is a wide accepted approach that consists in keeping transcripts with a reasonable number of missing values and filtering out those exceeding this threshold6. Transcripts that do not exceed the threshold are imputed using a suitable method. In the literature, several imputation methods have been proposed20.
PIPE-T offers a slider that the user can move to specify the maximum percentage of missing values admissible for a specific transcript. PIPE-T allows filtering transcripts using a user-defined percentage of missing values and/or a user-defined list of transcripts to be removed by using the filterCtData function of the HTqPCR package9.
In addition, PIPE-T gives the possibility of selecting one of three well-known imputation methods. These methods are:
-
KNN
-
Mestdagh
-
Cubic
KNN and Cubic imputation methods were already implemented in the impute and zoo R packages.
Mestdagh is an imputation method that substitutes a missing Ct value with a numeric value obtained adding one cycle to the highest Ct value across samples7. This method has already been described in other reports5. This method assumes that missing values depends on the low or null abundance of the transcript in the sample.
Differential expression analysis
Differential expression is a very popular analysis for identifying candidate transcripts whose expression can discriminate between two predefined conditions. Among the methods eligible for a differential expression analysis21, PIPE-T offers the possibility of choosing between three approaches:
T-test and two sample Wilcoxon test are among the most used statistical tests to perform a differential expression analysis21. Tests are implemented by ttestCtData and mannwhitneyCtData functions of the HTqPCR R package9. For the t-test and the two sample Wilcoxon test, PIPE-T offers the possibility of setting up six distinct parameters, which include: the types of alternative hypothesis to assess significance, the choice of a paired or an unpaired analysis, the presence in the data of replicated transcripts, the choice of a more or less stringent analysis, and the choice of the method for adjusting p-values in case of multiple hypothesis testing.
Rank Product is a popular method originating from a biological reasoning22. Rank Product is carried out using RP function of RankProd R package23.
If users do not specify any differential expression analysis method, PIPE-T allows them to select an option named NONE. In this case, no differential expression analysis is performed on the data.
Data visualization and outputting
Quality assessment of RT-qPCR data is crucial for enhancing the accuracy of the results and the reliability of the conclusions2. HTqPCR provides several visualization options for assessing the quality of qPCR data, which include histograms, boxplots, density distributions, and scatter plots9. PIPE-T uses two boxplot visualizations showing the distribution of the expression values across all samples. The boxplots show the distribution of expression values before and after data normalization, respectively. The visual inspection of the two boxplots is used as qualitative assessment of the normalization procedure because boxplots show the noise reduction comparing the data before and after data normalization8. Empirical Cumulative Distribution Function (ECDF) is also used in the literature for measuring noise reduction as an effect of data normalization8,10. PIPE-T computes and plots ECDF before and after data normalization by using ecdf function of the stats R package14. The significance of the difference between the two ECDF curves is estimated by Kolmogorov-Smirnov test and p-value is reported on top of the figure and in the standard output.
Tabular output files include raw data, filtered data, imputed data and statistics to assess differential expression. A detailed description of the row and column names can be found in HTqPCR and RankProd R packages documentation. A detailed description of visualization, sharing, and workflow integration using Galaxy graphical interface can be found in the Galaxy documentation.
Results
We tested the ability of PIPE-T of analyzing RT-qPCR data using two example datasets whose tab-separated text files were available in the Gene Expression Omnibus (GEO) with accession identifiers GSE25552 and GSE43000. Datasets were relative to two published studies on various metastatic tumors24 and non-small cell lung (NSCL) cancers25. The first study reported the results of the analysis of sixteen different tumors including Lung, Renal, Colon, Sarcoma, Ovarian, and Head and neck squamous cell carcinoma24. The second study reported the results of the analysis of forty-four NSCL tumor samples25. We carried out PIPE-T analysis of both datasets on a test Galaxy instance version 19.01, installed in a local Linux machine. Parameter settings for the two analyses have been taken from the original publications when available. When the parameters were not specified we selected them arbitrarily.
Various metastatic cancers
We downloaded input tab-delimited files from GEO and we added a SDS version 2.4 format header to each of these files because it lacked. Input files contained experimental data for 384 microRNAs. We coupled RT-qPCR data with information about tumor status, which was oligometastatic (OLIGO) for ten out of sixteen patients and polymetastatic (POLY) for the remaining six patients. File names and tumor status were organized into a tab-delimited text file. The newly created file and the sixteen tab-separated text files were uploaded in Galaxy as fileTreatment and ListOfFile through “Upload File from your computer” tool. Analysis was carried out with parameters settings reported in Fig. 2.
Our tool successfully completed the execution, returning seven output files (see Tables S1–S4 and Figs S1–S3). Boxplots and EDCF before and after data normalization as well as the significant genes and statistics reported by the differential expression analysis procedure are depicted in Figs 3, 4, and Table 1, respectively.
Quantitative assessment of the noise reduction for metastatic cancer data. ECDFs (y axis) and coefficient of variation (CV) is displayed for the metastatic cancer samples after Ct filtering and categorization (blue line) and after normalization (green line) procedures. Kolmogorov-Smirnov test assessing the significance of the separation between the curves and p value is reported on top of the plot.
We found 12 significantly upregulated and 11 downregulated microRNAs in polymetastatic tumors (p value < 0.05 and FC > 2 or FC < 0.5; Table 1).
Interestingly, among the significantly modulated microRNAs reported in the Lussier and coworkers manuscript24, 11 out of 12 microRNAs were consistently up regulated in polymetastatic tumors and 8 out of 11 microRNAs were consistently upregulated in oligometastatic tumors. Any difference between our findings and those reported by Lussier and collegues24 are probably due to the different approaches used in the experiments to filter and handle missing values. Lussier and collegues did not report any information about filtering based on the percentage of missing values or the application of any method for handling missing or unreliable Ct values. These results provide the first evidence that PIPE-T is able to correctly analyze RT-qPCR expression data.
Non-small cell lung cancer
NSCL input files were compliant with SDS format version 2.3 and reported experimental data for 381 microRNAs. Since the downloaded files used a comma as decimal separator, each comma was replaced with a dot before running PIPE-T. RT-qPCR data were coupled with histological data provided in the original publication25, which refer to twenty lung adenocarcinoma (LA) and twenty-four squamous cell lung cancer (SCLC). File names and tumor subtypes were organized into a text file. We uploaded the newly created file as fileTreatment, and the forty-four tab-separated text files as ListOfFile. Analysis was carried out with the parameter settings reported in Fig. 5.
Our tool successfully completed the execution returning seven output files (see Tables S5–S8 and Figs S4–S6). Boxplots and EDCF before and after normalization, as well as the significant microRNAs identified by the differential expression analysis procedure, are depicted in Figs 6, 7, and Table 2, respectively.
Quantitative assessment of the noise reduction for NSLC data. ECDFs (y axis) and coefficient of variation (CV) is displayed for the NSLC samples after Ct filtering and categorization (blue line) and after normalization (Green line) procedures. Kolmogorov-Smirnov test assessing the significance of the separation between curves and p value is reported on top of the plot.
We found 16 significantly modulated microRNAs (p value < 0.05 and FC > 2 or FC < 0.5; Table 2). Interestingly, miR-205, miR-149, miR-422a, and miR-708 were significantly upregulated in SCLC and miR-375 was significantly upregulated in LA in accordance with the results of the original manuscript25. Any difference of fold change or p-value between our study and that by Molina-Pinelo and collegues25 can be explained by the different handling of missing values. Authors did not report their approach to missing or unreliable Ct values. In spite of three small differences, our results provide evidences that PIPE-T is able to correctly analyze RT-qPCR expression data.
Conclusions
We developed PIPE-T, a new Galaxy tool that offers several state-of-the-art options for parsing, filtering, normalizing, imputing, and analyzing RT-qPCR expression data. Integration of PIPE-T into Galaxy allows researchers with strong bioinformatic background, as well as those without any programming expertise, to perform complex analysis in a simple to use, transparent, accessible, reproducible, and user-friendly environment.
Availability of Supporting Source Code and Requirements
Project name: Pipe-t
Project home page: https://github.com/igg-molecular-biology-lab/pipe-t (2019)26
Operating system(s): Linux (Galaxy), and platform independent
Programming language: R
Other requirements: Galaxy
License: GNU GPL
PIPE-T is available on the Main Tool Shed15 at the link27, on the Docker28 at the link29 and on the web30 at the link31. PIPE-T code is freely available on GitHub at the link https://github.com/igg-molecular-biology-lab/pipe-t (2019)26.
PIPE-T has the following dependencies:
<requirements>
<requirement type = “package” version = “3.5.0”>r-base</requirement>
<requirement type = “package” version = “7.2.0”>libgcc</requirement>
<requirement type = “package” version = “1.36.0”>bioconductor-htqpcr</requirement>
<requirement type = “package” version = “3.8.0”>bioconductor-rankprod</requirement>
<requirement type = “package” version = “1.56.0”>bioconductor-impute</requirement>
<requirement type = “package” version = “1.11.0”>r-bbmisc</requirement>
<requirement type = “package” version = “1.8.4”>r-psych</requirement>
<requirement type = “package” version = “1.8_3”>r-zoo</requirement>
</requirements>
If Conda32 is installed and enabled, Galaxy locates and resolves any tool dependencies automatically during tool installation.
Data availability
The tab-separated text files included in the ListOfFile collections of the two example applications are available in GEO repository with accession numbers: GSE25552 and GSE43000. A detailed documentation, step-by-step tool installation instructions, configuration, example applications are available on GitHub at the link https://github.com/igg-molecular-biology-lab/pipe-t (2019)26.
References
Pabinger, S., Rodiger, S., Kriegner, A., Vierlinger, K. & Weinhausel, A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol Detect Quantif 1, 23–33, https://doi.org/10.1016/j.bdq.2014.08.002 (2014).
Derveaux, S., Vandesompele, J. & Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230, https://doi.org/10.1016/j.ymeth.2009.11.001 (2010).
VanGuilder, H. D., Vrana, K. E. & Freeman, W. M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44, 619–626, https://doi.org/10.2144/000112776 (2008).
McCall, M. N., McMurray, H. R., Land, H. & Almudevar, A. On non-detects in qPCR data. Bioinformatics 30, 2310–2316, https://doi.org/10.1093/bioinformatics/btu239 (2014).
de Ronde, M. W. J., Ruijter, J. M., Moerland, P. D., Creemers, E. E. & Pinto-Sietsma, S. J. Study Design and qPCR Data Analysis Guidelines for Reliable Circulating miRNA Biomarker Experiments: A Review. Clin Chem 64, 1308–1318, https://doi.org/10.1373/clinchem.2017.285288 (2018).
Marabita, F. et al. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform 17, 204–212, https://doi.org/10.1093/bib/bbv056 (2016).
Mestdagh, P. et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11, 809–815, https://doi.org/10.1038/nmeth.3014 (2014).
Mestdagh, P. et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10, R64, https://doi.org/10.1186/gb-2009-10-6-r64 (2009).
Dvinge, H. & Bertone, P. HTqPCR: high-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics 25, 3325–3326, https://doi.org/10.1093/bioinformatics/btp578 (2009).
D’haene, B., Mestdagh, P., Hellemans, J. & Vandesompele, J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol 822, 261–272, https://doi.org/10.1007/978-1-61779-427-8_18 (2012).
Taylor, S. C. et al. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol 37, 761–774, https://doi.org/10.1016/j.tibtech.2018.12.002 (2019).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11, R86, https://doi.org/10.1186/gb-2010-11-8-r86 (2010).
Blankenberg, D. et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 19, 19.10.1–19.10.21, https://doi.org/10.1002/0471142727.mb1910s89 (2010).
R Core Team. R: A language and environment for statistical computing; Vienna, https://www.R-project.org (2019).
Blankenberg, D. et al. Dissemination of scientific software with Galaxy Tool Shed. Genome Biol 15, 403, https://doi.org/10.1186/gb4161 (2014).
Meyer, S. U., Pfaffl, M. W. & Ulbrich, S. E. Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnol Lett 32, 1777–1788, https://doi.org/10.1007/s10529-010-0380-z (2010).
Zeileis, A. & Grothendieck, G. Zoo: S3 Infrastructure for Regular and Irregular Time Series. In. Journal of Statistical Software 14, 1–27, https://doi.org/10.18637/jss.v014.i06 (2005).
Perkins, J. R. et al. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics 13, 296, https://doi.org/10.1186/1471-2164-13-296 (2012).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034, https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Yadav, M. L. & Roychoudhury, B. Handling missing values: A study of popular imputation packages in R. In. Knowledge-Based Systems 160, 104–118, https://doi.org/10.1016/j.knosys.2018.06.012 (2018).
Andrew, H., Florence, G. & Kibria, G. B. Methods for identifying differentially expressed genes: An empirical comparison. Journal of Biometrics & Biostatistics 6, 1, https://doi.org/10.4172/2155-6180.1000265 (2015).
Breitling, R., Armengaud, P., Amtmann, A. & Herzyk, P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573, 83–92, https://doi.org/10.1016/j.febslet.2004.07.055 (2004).
Hong, F. et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827, https://doi.org/10.1093/bioinformatics/btl476 (2006).
Lussier, Y. A. et al. MicroRNA expression characterizes oligometastasis(es). PLoS One 6, e28650, https://doi.org/10.1371/journal.pone.0028650 (2011).
Molina-Pinelo, S. et al. MicroRNA-dependent regulation of transcription in non-small cell lung cancer. PLoS One 9, e90524, https://doi.org/10.1371/journal.pone.0090524 (2014).
GitHub, https://github.com/igg-molecular-biology-lab/pipe-t Accessed 20 May (2019).
Galaxy main tool shed repository, https://davidecangelosi@toolshed.g2.bx.psu.edu/repos/davidecangelosi/pipe_t Accessed 20 May (2019).
Merkel, D. Docker: lightweight linux containers for consistent development and deployment. Linux Journal 239, 2 (2014).
Docker, https://hub.docker.com/r/davidecangelosi/galaxy-pipe-t Accessed 20 May (2019).
Tangaro, M. A. et al. Laniakea: an open solution to provide “Galaxy on-demand” instances over heterogeneous cloud infrastructures. bioRxiv, 472464, https://doi.org/10.1101/472464 (2018).
Live Galaxy Instance website, http://igg.cloud.ba.infn.it/galaxy Accessed 22 October (2019).
Gruning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods 15, 475–476, https://doi.org/10.1038/s41592-018-0046-7 (2018).
Acknowledgements
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC 2015 Grant 17459 to L.V.), Italian Ministry of Health (Ricerca Corrente), Fondazione Italiana per la Lotta al Neuroblastoma (to L.V.), Fondo Aree Sottoutilizzate (FAS to L.V.), and Italian Ministry of Health (Ricerca Finalizzata, Project Code: RF-2016-02361048). Laniaka has been developed in the framework of the INDIGO-Datacloud project funded by the European Commision H2020 research and innovation program under grant agreement RIA 653549. Live Galaxy service is provided with the support of ELIXIR-IT and ReCaS-Bari. Funding institutions had no role in the design of the study, collection, analysis, and interpretation of data, decision to submit the article for publication, and in the writing of the manuscript. The authors would like to thank the Galaxy Help Community for invaluable support during tool implementation and Dr. Heidi Dvinge for interesting discussions about implementation of HTqPCR R package.
Author information
Authors and Affiliations
Contributions
N.Z. set up the GitHub project and the Docker image, installed and configured the Galaxy service in a Linux server and helped to write the documentation. M.M. helped in designing and testing the tool. M.A.T. and F.Z. provided technical support to create the live Galaxy instance. MCB interpreted the results. L.V. conceived the project, helped in designing and testing the original version of the tool, and provided the funding. A.E. supervised the project. D.C. conceived and implemented the Galaxy tool, performed literature search, tested the tool, wrote the documentation and wrote the manuscript. All authors read and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zanardi, N., Morini, M., Tangaro, M.A. et al. PIPE-T: a new Galaxy tool for the analysis of RT-qPCR expression data. Sci Rep 9, 17550 (2019). https://doi.org/10.1038/s41598-019-53155-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-53155-9
This article is cited by
-
deltaXpress (ΔXpress): a tool for mapping differentially correlated genes using single-cell qPCR data
BMC Bioinformatics (2023)
-
qRAT: an R-based stand-alone application for relative expression analysis of RT-qPCR data
BMC Bioinformatics (2022)
-
Laniakea@ReCaS: exploring the potential of customisable Galaxy on-demand instances as a cloud-based service
BMC Bioinformatics (2021)
-
Auto-qPCR; a python-based web app for automated and reproducible analysis of qPCR data
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.