Abstract
Murashige & Skoog (MS) and Hoagland’s media were previously used for in vitro culture of Landoltia punctata. During subsequent ex vitro culture, the use of MS medium resulted in a higher growth rate, compared to Hoagland’s medium. Thus, a higher starch content of L. punctata in MS medium was previously hypothesized. Here, L. punctata strain 5632 was isolated and characterized using morphological characteristics and the atpF-atpH intergenic region. During early cultivation stage, fresh weight and relative growth rate in MS medium were lower than Hoagland’s medium. Conversely, starch content in MS medium was considerably higher than in Hoagland’s medium. Medium effects on expression of genes coding for starch-biosynthesis ADP-glucosepyrophosphorylase (AGPase) were determined. Genomic fragments of small (LeAPS) and large (LeAPL1) AGPase subunits were characterized. Differential expression between each AGPase subunit genes was observed in both media. Additionally, in MS medium, the highest correlation coefficients between starch content and gene expression was found with LeAPS (0.81) and followed by LeAPL3 (0.67), LeAPL2 (0.65) and LeAPL1 (0.28). In Hoagland’s medium, the coefficients of LeAPL3 (0.83) and LeAPL2 (0.62) were higher than LeAPS (0.18) and LeAPL1 (−0.62). This suggested different levels of contributions of these genes in starch biosynthesis in both media.
Similar content being viewed by others
Introduction
Starch functions as an important energy reserve in plants1. During photosynthesis, carbon compounds are generated and converted into glucose that serves as the precursor for starch formation1. There are three committed steps in the process1. The first one is the generation of ADP-glucose. Secondly, the glucosyl moiety of ADP-glucose is linked to an existing glucan chain through the formation of the α(1-4) linkage, resulting in an extension of the starch chain. Finally, branching of the chain is formed through the formation of the α(1-6) linkage between glucosyl moieties and the chain. ADP-glucose pyrophophorylase (AGPase) is responsible for the formation of ADP-glucose that was proposed as the rate-limiting step2. In higher plants, AGPase is heterotetrameric and consists of two small and two large subunits3,4. Small subunits play the catalytic role, while large subunits mainly function in regulating the enzyme activity5,6. Genes that code for large and small subunits were previously identified in various dicotyledonous and monocotyledonous plants, e.g., Arabidopsis7, potato8,9, chickpea10, maize11,12 and wheat13,14. Additionally, overexpression of AGPase large subunit genes increased starch content in maize and wheat grains12,15. Another study showed that a transgenic wheat line, overexpressing an AGPase small subunit gene, accumulated starch at a level substantially higher than the wild-type cultivar6.
Duckweeds are small aquatic plants that belong to the family Lemnaceae16. Thus far, 37 duckweed species were affiliated with the family and classified into five genera, including Spirodela, Landoltia, Lemna, Wolffiella and Wolffia16,17. Fronds are leaf-like and function as both vegetative and reproductive organs18. L. punctata has gained the interest in research communities, because of its high starch content that could be used in bioethanol production19,20,21. Several advantages of L. punctata over other energy crops were also cited22. These included rapid growth rate, high starch content and low levels of fiber and lignin content22. Additionally, the use of L. punctata for the energy industry did not compete for land with food crops23. Various external and internal factors affected starch biosynthesis of L. punctata. Nutrient starvation induced starch accumulation4, while phytohormones were internal regulators of starch biosynthesis24,25. Expression of L. punctata AGPase subunit genes, including LeAPS, LeAPL1, LeAPL2 and LeAPL3, were also affected by growth conditions4,25. Expression of LeAPL2 under nitrogen starvation was relatively lower than that under phosphorus starvation4. In contrast, nitrogen deficiency induced an early response of LeAPL3, while phosphorus deprivation resulted in an upregulation of LeAPL3 at a later phase4. A transcriptomic study showed that, when L. punctata was treated with uniconazole, expression of two AGPase large subunit genes and starch content were concurrently increased25.
Synthetic media were generally used for culturing duckweeds under controlled environments26,27. Medium compositions had various physiological impacts on duckweeds. In Spirodela polyrhiza, exogenous addition of abscisic acid (ABA) in culture medium induced a morphological transition of fronds to turions, a starch accumulating structure28. SpAPL2 and SpAPL3, encoding two different AGPase large subunits, were also upregulated under such conditions, whereas SpAPL1 expression was not significantly increased28. For L. punctata, biomass production was higher when plants were cultivated in Hoagland’s medium for 13 days, compared to MS medium26. However, after transferred to nutrient-limited pond water for ex vitro culture, L. punctata obtained from MS medium grew 17.1% faster than those from Hoagland’s medium. It was previously hypothesized that cultivation in MS medium resulted in higher starch accumulation that subsequently promoted ex vitro growth26. This hypothesis was tested here. L. punctata strain 5632 was newly isolated and characterized, based on its morphologies and the atpF-atpH intergenic region sequence. Strain 5632 was long-term (35 days) cultured in MS and Hoagland’s media to observe its biomass production and starch accumulation. The cDNA fragments of LeAPS and LeAPL1, coding for AGPase large and small subunits, respectively, were cloned and compared with the reference sequences of L. punctata strain 02024. Genomic fragments of both genes were also cloned, and their introns and exons were characterized. Expression levels of four AGPase subunit genes, including LeAPS, LeAPL1, LeAPL2 and LeAPL3, in both media were analysed. Correlation coefficients, between AGPase gene expression levels and starch content, were calculated to determine the contribution of each gene in starch biosynthesis, during the cultivation in MS and Hoagland’s media.
Results and Discussion
Morphological and molecular characterization
Duckweed samples were collected, and an axenic culture on Hoagland’s medium was established from a single mother frond to ensure the genetic similarity among samples throughout the study. The strain was designated as strain 5632. It was important to note here that the nutrient compositions of Hoagland’s medium were somewhat different from the original publication29. Originally, two different macronutrient solutions were described for the preparation of Hoagland’s medium29. The main difference between them was the nitrogen source. While one solution contained only NO3−, the other one was supplemented with both NO3− and NH4+ 29. Hoagland’s solution, used in this study, had nutrient compositions (Table 1) that were more similar to the latter one. However, there was a distinction in the NO3− concentration in the original solution (1 mM)29 and this study (6 mM). Additionally, iron was supplied as iron tartate in the original publication29, whereas it was given here as chelated iron. Basic morphological characteristics of strain 5632 were examined (Fig. 1). Fronds were inflated and oval-shaped. The upper surface was dark green. The lower surface was reddish, suggesting the accumulation of anthocyanins in the tissues. Several roots were present on the lower side of the fronds, indicating that strain 5632 was likely a member of the genus Landoltia. The presence and the number of roots were distinguishing morphological characteristics among the five genera of the family Lemnaceae30. For example, a single root was present on each frond of Lemna spp., whereas plants in genera Wolffia and Wolffiella did not produce roots. In contrast, several roots were present on each frond of members in genera Spirodela and Landoltia31. The number of roots of the genus Spirodela ranged from 7 to 21, while that of the genus Landoltia ranged from 2 to 732. Additional characteristics, e.g., a medial series of papillae on the upper surface, frond prophyllum, frond nerves and external anther locules, could be used to differentiate Landoltia from Spirodela32. However, because of their small and highly reduced structures, identification of duckweeds at the species levels could be challenging without a special expertise in the Lemnaceae family33. Biochemical and DNA sequence data were previously combined with morphological and anatomical markers to study the phylogeny of the family Lemnaceae34. The phylogenetic tree confirmed the presence of the paraphyletic subfamily Lemnoideae, consisting of Spirodela, Landoltia, and Lemna, and the monophyletic subfamily Wolffioideae, comprising of Wolffia and Wolffiella. Additionally, a DNA barcode, based on the atpF-atpH intergenic region, was developed to aid the characterization of lemnaceous plants33. For identification of strain 5632, the atpF-atpH intergenic region was amplified from plastid DNA and sequenced. Using the blastn analysis, the highest identity level (100%) was found between the sequences of strain 5632 and L. punctata strains DW2701-1 (accession number: KJ630554) and DW2701-4 (KJ630555). To understand the phylogenetic relationship between strain 5632 and other L. punctata strains, the phylogenetic tree was reconstructed based on the atpF-atpH intergenic sequences. It showed that strain 5632 was closely related to L. punctata strains DW2701-1, DW2701-4 and 20d (Fig. 2). This was supported by the 100% confidence level. Consistent with its morphologies, the molecular characterization indicated that strain 5632 was a member of the species L. punctata.
Phylogenetic relationship between strain 5632 and selected members in the family Lemnaceae. Reconstruction of the phylogenetic tree was based on the sequence of the atpF-atpH intergenic region, using the UPGMA method. Bootstrap values are shown at tree nodes as the percentage of 1,000 replicates. Only values equal to or higher than 70% are shown. Bar indicated the evolutionary distance as the number of base substitutions per site.
Growth and starch accumulation of L. punctata 5632 in synthetic media
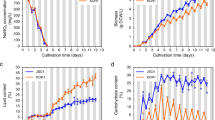
A previous study showed that the growth rate of L. punctata in MS medium was lower than that in Hoagland’s medium26. However, after transferred to nutrient-limited water, L. punctata obtained from MS medium grew at a higher rate than those from Hoagland’s medium26. This suggested a higher starch content of L. punctata in MS medium that could be used to sustain subsequent ex vitro growth. To test this hypothesis, strain 5632 was grown in MS and Hoagland’s liquid media for 35 days, and fresh weight was determined every seven days (Fig. 3a). It was important to note that there were some differences, e.g. types of some micronutrient salts and some nutrient concentrations, between the compositions of Hoagland’s solution used here (Table 1) and in the previous study26. However, during the first 14 days of cultivation, the fresh weight of strain 5632 in Hoagland’s solution was higher than in MS medium. This was consistent with the previous study26. This continued until day 21 of cultivation. Conversely, on day 28, the fresh weight in MS medium (3.14 ± 0.04 g) became significantly higher (P = 0.038) than that in Hoagland’s medium (3.04 ± 0.04 g). On day 35, it became 1.2 times of that in Hoagland’s medium.
Fresh weight (a), relative growth rate (b) and starch content (c) of L. punctata strain 5632 grown in MS (gray bars) and Hoagland’s (white bars) media. Standard deviation (n = 3) is shown by vertical bars. Asterisks indicate statistically significant differences (P < 0.05) between the parameters in MS and Hoagland’s media on each day.
Relative growth rate of strain 5632 in both media were calculated and found relatively consistent with fresh weight analysis (Fig. 3b). During the first 14 days of cultivation, strain 5632 in MS medium exhibited lower relative growth rate, compared to Hoagland’s medium. In contrast, on day 28 and 35 of cultivation, the relative growth rate in MS medium became substantially higher than in Hoagland’s medium. This indicated a more rapid reduction in growth of strain 5632 in Hoagland’s medium. A discrepancy between fresh weight and relative growth rate was observed on day 21. While the fresh weight in MS medium was lower than Hoagland’s medium (Fig. 3a), the relative growth rate became considerably higher than Hoagland’s medium (Fig. 3b). This was because of the differential increase between the fresh weight in both media. From day 14 to day 21, the fresh weight in MS medium elevated from 0.53 ± 0.04 g to 1.71 ± 0.08 g, respectively. This yielded the relative growth rate at 0.17 ± 0.004 day−1. During the same period, the fresh weight in Hoagland’s medium rose from 1.02 ± 0.07 g to 2.23 ± 0.12 g, and yielded relative growth rate at 0.11 ± 0.002 day−1.
Starch content of strain 5632 in MS and Hoagland’s media was also determined throughout the cultivation period (Fig. 3c). In MS medium, it continuously increased and reached the level of 3.31 ± 0.28% on day 28. This was followed by a decrease to 1.95 ± 0.25% on day 35. In contrast, starch content of strain 5632 in Hoagland’s medium increased relatively slowly during the first three weeks of cultivation. Subsequently, it reached the highest level (4.01 ± 0.24%) on day 28 and slightly decreased to 3.91 ± 0.18% on day 35. Comparison between the fresh weight and starch content in both media revealed an inverse correlation. From day 7 to 21, the fresh weight of strain 5632 in MS medium was lower than Hoagland’s medium, whereas starch content in MS medium was higher than the other medium. The opposite was found on day 28 and 35 of cultivation.
Growth and starch content of strain 5632 were likely affected by the differences between the concentrations of both NH4+ and NO3− in MS and Hoagland’s media (Table 1). During the early phase of cultivation, the slower growth in MS medium was likely caused by the high concentration of NH4+ in MS medium (20.6 mM). A previous study showed that NH4+ was preferable over NO3− for the uptake by L. punctata35. Thus, cultivation of strain 5632 in the medium likely caused excessive accumulation of NH4+ in strain 5632. This resulted in lower biomass production, compared to the use of Hoagland’s medium, that was supplemented with 1 mM NH4+. When present at a high concentration, NH4+ was known to negatively affect plant growth36. This was also demonstrated in other duckweed species. Relative growth rate of S. polyrhiza decreased upon the increase of NH4+ concentration37. Growth inhibition and frond chlorosis were also found in Lemna minor, grown in Hoagland’s medium supplemented with 280 and 840 mg l−1 NH4+-N, which equaled to 20 and 60 mM NH4+, respectively38. During the later cultivation period, growth of strain 5632 in Hoagland’s medium declined more rapidly, compared to that in MS medium. This may be attributed to the NO3− concentration in Hoagland’s medium (10 mM) that was only 25.4% of that in MS medium (39.4 mM). The result suggested that this level of NO3−, in Hoagland’s medium, was insufficient to sustain growth of strain 5632 for the entire cultivation period. Additionally, another nutrient that may cause the distinction in growth of strain 5632 in the two media was iron. It is an important microelement in various plant metabolic processes39. Generally, FeSO4 was added together with Na2EDTA to generate chelated iron that ensured its transport into plant cells40. Here, the concentrations of FeSO4 and Na2EDTA, in Hoagland’s medium, were approximately 9% of those in MS medium and may be insufficient to sustain growth during the extended cultivation period. On the other hand, starch biosynthesis was previously known to be upregulated under nutrient starvation conditions, which also resulted in growth retardation4,5,22,41,42. This was somewhat consistent with the increase in starch content of strain 5632 in MS medium, where growth was relatively lower than in Hoagland’s medium. However, as mentioned above, both NH4+ and NO3− in MS medium were supplied at much higher levels than Hoagland’s medium. This suggested that differential starch accumulation of strain 5632 in the two media was likely a secondary effect that resulted from growth modulation by the nitrogen sources.
Nucleotide variations in full-length cDNA sequences of LeAPL1 and LeAPS
Full-length LeAPS and LeAPL1 cDNA were previously sequenced in L. punctata strain 0202 that was collected in China4. However, strain 5632 was obtained from a local pond in Thailand. Thus, cDNA sequences of both genes were determined to investigate nucleotide variations that may exist between the two genes of these strains. Full-length LeAPS (1,578 bp) and LeAPL1 (1,554 bp) cDNA of strain 5632 were sequenced and compared with their corresponding sequences of strain 0202 (LeAPS: KJ603243; LeAPL1: KJ603244)4. For LeAPS, three nucleotide variations were found at positions 27, 575 and 769 (Table 2). Two of these resulted in changes of the deduced amino acid residues. For LeAPL1, there were three nucleotide variations at positions 563, 680 and 754, all of which resulted in changes of corresponding amino acid residues (Table 2). To investigate whether these amino acid variations were inherent to strain 5632, the deduced LeAPL1 and LeAPS protein sequences of strain 5632 were used for multiple alignment analysis with those of strain 0202 and their orthologs from other plant species. The analysis indicated that amino acid positions 9, 192 and 257 of LeAPS (Fig. 4a) and 188 and 227 of LeAPL1 (Fig. 4b) of strain 5632 were more similar to the orthologs than strain 0202. These differences between the deduced amino acid sequences of strains 5632 and 0202 were possibly derived from DNA amplification errors during the cloning step. LeAPS and LeAPL1 cDNA of strain 0202 were amplified, using rTaq DNA polymerase4. In contrast, the cloned sequences of strain 5632 were obtained with Q5 high-fidelity DNA polymerase, of which the error rate was described as 280 times lower than regular Taq DNA polymerase (www.neb.com). Thus, these amino acids of strain 5632 likely represented the actual residues of LeAPS and LeAPL1. The only exception was the amino acid position 252 of LeAPL1 that was derived from its corresponding codon containing the nucleotide variant position 754. The amino acid sequences of strain 5632 and other orthologs carried methionine at this position, as opposed to valine found in L. punctata 0202 and S. tuberosum sequences (Fig. 4b). Thus, this nucleotide position may represent a genotypic variation between LeAPL1 of strains 5632 and 0202.
Organization of genomic LeAPS and LeAPL1
Although their cDNA sequences were previously characterized4, the genomic sequences of LeAPS and LeAPL1 were still unavailable. Here, amplification of genomic LeAPS with the CLeAPS-F and LeAPS-R primers was successful. The length of the amplified fragment was 2,621 bp. It was important to note that the fragment was only partial and did not cover the remaining 128 nucleotides on the 3′ region of its cDNA. Eight exons and seven introns were identified in the sequence (Fig. 5a). The numbers of exons and introns found in genes coding for AGPase small subunits were variable among different plant species. A study on the genomic structure of maize Bt2 showed that it consisted of ten exons and nine introns11. In contrast, sAGP of potato43 and ibAGP1 and ibAGP2 of sweet potato44 were similarly organized into nine exons and eight introns. On the other hand, the genomic fragment of LeAPL1 (3,161 bp) was obtained using the CLeAPL1-F and CLeAPL1-R primers. The genomic sequence consisted of 14 exons and 13 introns (Fig. 5b). The average length of the exons was 111 bp. Exon 1 was the longest (210 bp), while exon 13 was the shortest (61 bp). The organization of exons and introns in LeAPL1 was different from its ortholog, SpAPL1 of S. polyrhiza, where 15 exons and 14 introns were characterized28.
Expression of AGPase subunit genes and correlation with starch accumulation
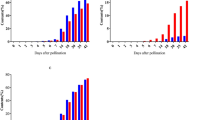
Expression of AGPase subunit genes, including LeAPS, LeAPL1, LeAPL2 and LeAPL3, was analysed, in order to understand the molecular responses towards nutrient compositions in MS and Hoagland’s media. Based on their genomic sequences, new primers were designed and used to quantify expression of LeAPS and LeAPL1. Unlike the previously described primers4, they spanned over intron regions to ensure the amplified products were derived from cDNA of processed mRNA molecules. For LeAPL2 and LeAPL3, the primers used in the analysis were from the previous study4, because their genomic sequences were unavailable. Additionally, the expression analysis here covered the cultivation period of five weeks. This was much longer than other previous studies where it ranged from seven to ten days4,25,42 and enabled the monitoring of the long-term gene regulation in both media. Expression levels of each gene during the cultivation in both media are shown in Fig. 6. In MS medium, the highest expression levels of LeAPS (0.65 ± 0.07), LeAPL2 (1.92 ± 0.88) and LeAPL3 (1.48 ± 0.77) were observed on day 21 and approximately 8.2 times of the level on day 0. In contrast, LeAPL1 expression increased and reached the maximum level (0.66 ± 0.14) on day 14, which was approximately 2.6 times of that on day 0. Similar to LeAPS, the highest levels of LeAPL2 and LeAPL3 expression occurred on day 21 and were 4.0 and 8.1 folds of day 0, respectively. In Hoagland’s medium, expression of LeAPS (0.72 ± 0.10) and LeAPL1 (1.02 ± 0.15) similarly peaked on day 14 and were 10.3 and 4.2 folds of day 0, respectively. LeAPL2 expression became the highest on day 28 (2.38 ± 0.80) and was 9.5 times of day 0. In contrast, expression of LeAPL3 continuously increased throughout the cultivation period and was the highest on day 35 (1.48 ± 0.61) and 3.6 folds of day 0. Significant differences were also observed between expression levels of some AGPase subunit genes in both media. On day 21, LeAPS in MS medium was expressed at a level significantly higher (P = 0.049) than in Hoagland’s medium. On the other hand, LeAPL1 in MS medium was expressed at levels significantly lower than in Hoagland’s medium on day 14 (P = 0.039) and 21 (P = 0.029). However, its expression in MS medium became significantly higher than in Hoagland’s medium on day 28 (P = 0.03) and 35 (P = 0.014). In contrast, differences in LeAPL2 and LeAPL3 expression levels observed in MS and Hoagland’s media were not substantial throughout the cultivation period. The result observed here indicated differential regulation of each AGPase subunit gene. This was also previously found with L. punctata strain 0202, that was grown under nitrogen and phosphorus starvation conditions4. In response to nitrogen starvation, upregulation of LeAPL1 and LeAPL3 was observed earlier than LeAPL24. Under phosphorus deprivation, a dramatic increase in LeAPL1 and LeAPL2 expression occurred more rapidly than LeAPL24. In contrast, LeAPS was highly expressed throughout the nitrogen starvation period and during the early stage of phosphorus starvation4. ABA may play a role in regulating expression of AGPase genes in this study, since its application on L. punctata was previously shown to increase starch content and AGPase activity20,45. Consistently, a transcriptomic study demonstrated that both ABA content and expression levels of AGPase large subunit genes were elevated, when L. punctata was treated with uniconazole24,25. Reports in S. polyrhiza28 and rice cell culture46 suggested that each AGPase subunit gene may respond to ABA differently. Expression of SpAPL2 and SpAPL3 was upregulated by the ABA treatment28. In contrast, the expression level of SpAPL1 remained relatively unchanged and was much lower than SpAPL2 and SpAPL3. On the other hand, application of ABA to rice cell culture resulted in elevated expression of OsAPL3 and OsAPS1, while its effects on other rice AGPase subunit genes were much less pronounced46. These previous observations suggested that ABA effects on expression of AGPase genes may also vary among different plant species.
Expression levels of LeAPS (a), LeAPL1 (b), LeAPL2 (c) and LeAPL3 (d) of L. punctata strain 5632, grown in MS (gray bars) and Hoagland’s (white bars) media. Standard deviation (n = 3) is shown by vertical bars. Asterisks indicate statistically significant differences (P < 0.05) between expression levels of each AGPase subunit genes in MS and Hoagland’s media on each day.
Because of their differential expression, each AGPase subunit genes may contribute to starch accumulation of strain 5632 in MS and Hoagland’s media differently. Correlation coefficients between the expression levels of AGPase subunit genes and the starch content were determined. In MS medium, LeAPS had the highest correlation coefficient (0.81) and was followed by LeAPL3 (0.67) and LeAPL2 (0.65). A small correlation (0.28) was observed with LeAPL1. In Hoagland’s medium, expression of LeAPL3 was the most correlated (0.83) with starch content. An intermediate level of correlation was found with LeAPL2 (0.62). In contrast, the correlation coefficient of LeAPS was only 0.18, while LeAPL1 was negatively correlated (−0.62). The intermediate and high correlation coefficients of LeAPL2 and LeAPL3 were consistent with their expression levels that were found comparable in both media (Fig. 6c,d). This suggested the roles of LeAPL2 and LeAPL3 in starch biosynthesis may be non-specific to the media. In contrast, LeAPS contribution may be more pronounced in MS medium. On the other hand, the correlation coefficients of LeAPL1 in the two media were highly distinctive and ranged from low to negatively intermediate levels. This indicated its input in starch content was likely dependent on medium compositions. This was also reflected on the differences between its expression levels in both media (Fig. 6b). The correlation coefficients observed here were consistent with a previous study, where L. punctata strain 0202 was grown in Hoagland’s medium under nitrogen- and phosphorus-deficient conditions for seven days4. The correlations between LeAPL2 and LeAPL3 expression and starch content were either intermediate or high under both conditions. LeAPS was found correlated to starch accumulation specifically under phosphorus starvation. In contrast, low or negative correlations were also found with LeAPL1 under nitrogen and phosphorus deprivation.
Conclusions
Long-term (35 days) physiological and molecular responses were demonstrated in L. punctata strain 5632 cultured in MS and Hoagland’s media. During the early stage, the use of MS medium resulted in lower growth and higher starch content, compared to Hoagland’s medium. The situation was reverse during the last 14 days of the experiment. Expression of AGPase subunit genes was differentially regulated in both media. Based on the correlation coefficients between starch content and gene expression, LeAPL2 and LeAPL3 were likely important for starch biosynthesis in both media, while LeAPS was more specific to MS medium. In contrast, low or negative correlation coefficients were associated with LeAPL1.
Methods
Plant materials and cultivation
L. punctata (G. Mey.) Les & D. J. Crawford was collected from a local pond in Bangkok, Thailand. To establish in vitro culture, fronds were surface-sterilized, using 10% (v/v) NaClO solution and a few drops of Tween-20 for two minutes with vigorous shaking. They were washed in sterilized distilled water three times. Surface-sterilized fronds were grown on Hoagland’s solid medium (PhytoTechnology Laboratories, USA), containing 2% (w/v) sucrose and 0.7% (w/v) agar, pH 5.7. Growth conditions were 25 ± 2 °C and 16-hour-light/8-hour-dark photoperiods. Photosynthetically active radiation (PAR) flux was 65 µmol m−2 s−1. Daughter fronds derived from a single mother frond were obtained. Morphological characteristics were examined. This duckweed strain was registered to the Rutgers Duckweed Stock Cooperative as strain 5632.
Determination of growth and starch content
Twenty colonies of strain 5632 were randomly selected and transferred to glass bottles containing 100 ml of either MS or Hoagland’s liquid media (Phytotechnology, USA), supplemented with 2% sucrose, pH 5.7. The cultures were grown for 35 days under 25 ± 2 °C and 16-hour-light/8-hour-dark photoperiods. PAR flux was 65 µmol m−2 s−1. Samples were collected on day 0, 7, 14, 21, 28 and 35 of cultivation and rinsed thoroughly with distilled water. Excessive water was removed by blotting on tissue paper. Total fresh weight was determined. Relative growth rate on day 7, 14, 21, 28 and 35 was calculated as follows:
where Wt = fresh weight on day 7, 14, 21, 28 and 35 of cultivation; Wt-1 = fresh weight of prior sample collection and time = the time interval (7 days).
One-hundred milligrams of fresh samples collected on each day were used for the analysis of starch content with Total Starch Assay Kit (Megazyme, Ireland), according to the manufacturer’s protocol. Water was used as the blank control, and 1 mg ml−1 D-glucose was used as the standard. The experiment was done in triplicate. Two-tailed student’s t-test was used to determine statistically significant differences (P < 0.05) between fresh weight, relative growth rate, and starch content, in MS and Hoagland’s media on each day.
Amplification and phylogenetic analysis of the atpF-atpH intergenic region
Fronds from axenic culture were used for DNA extraction with a FavorPrep Plant Genomic DNA Extraction Mini Kit (Favorgen, Taiwan), according to the manufacturer’s protocol. Amplification and sequencing of the atpF-atpH intergenic region were done with the atpF-atpH forward (5′-ACTCGCACACACTCCCTTTCC-3′) and reverse (5′-GCTTTTATGGAAGCTTTAACAAT-3′) primers, according to the previous study33. The sequence was analysed with blastn. Multiple alignment of the sequence with those of other duckweed strains in the GenBank database used the CLUSTAL W program version 1.8147. Gaps were manually removed and adjusted. A phylogenetic tree was reconstructed, using unweighted pair group method with arithmetic mean (UPGMA), in MEGA 7.048. Bootstrap analysis49 with 1,000 re-samplings determined the confidence level of each clade.
Cloning and sequencing of LeAPS and LeAPL1 cDNA
Total RNA was extracted from 100 mg of fronds, using a FavorPrep Plant Total RNA Mini Kit (Favorgen, Taiwan), according to the manufacturer’s protocol. Genomic DNA was removed with DNaseI (Promega, USA) by the on-column method, described in the protocol. cDNA of all samples was synthesized from 500 ng of total RNA, using iScript cDNA Synthesis Kit (Bio-Rad, USA). Oligo-dT supplied with the kit was used as the primer for the first-strand synthesis. CLeAPS-F (5′-ATGGCGGCGACGAGCTTC-3′) and CLeAPS-R (5′-TCATATGATGGTTCCGCTAGGG-3′) primers were used for amplification of LeAPS cDNA, while CLeAPL1-F (5′-ATGGCGCTGCGGATTGAG-3′) and CLeAPL1-R (5′-TCAGATGACAAGGCCATCCTT-3′) primers were used for LeAPL1 cDNA. The primer sequences and amplification cycles were according to the previous study4. Q5 High-Fidelity DNA polymerase (New England Biolabs, USA) was used for the amplification of full-length cDNA. The nucleotide sequences were compared with those of L. punctata strain 0202, using blastn. Multiple alignment analysis was performed between the deduced amino acid sequences of LeAPS and LeAPS and their homologs, using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Characterization of LeAPS and LeAPL1 genomic fragments
Q5 High-Fidelity DNA polymerase (New England Biolabs, USA) was used for the amplification of LeAPL1 and LeAPS genomic fragments. CLeAPS-F and LeAPS-R (5′-GCCCAATCCGAGCATTCT TAT-3′)4 primers were used for amplification of partial LeAPS genomic fragment, while CLeAPL1-F and CLeAPL1-R primers were for genomic LeAPL1. The amplified cycles were: 95 °C for 5 min; 40 cycles of 95 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 90 sec; 72 °C for 5 min. The amplified fragments were cut from an agarose gel and purified using FavorPrep GEL/PCR Purification Kit (Favorgen, Taiwan). Subsequently, the fragments were ligated with the pGEM-T vector (Promega, USA) and cloned in Escherichia coli strain DH5-α. Recombinant plasmids were extracted, using FavorPrep Plasmid DNA Extraction Mini Kit (Favorgen, Taiwan). Universal primers M13F (−20) and M13R (−40) were used for initial DNA sequencing. Additional primers were designed based on the sequences and used to complete the sequences. The genomic sequences of LeAPS (accession number: MK878513) and LeAPL1 (MK878512) were deposited in GenBank database.
Gene expression analysis
Total RNA was extracted from 100 mg of strain 5632 fronds, collected at each time point. The extraction and cDNA synthesis followed the methods mentioned above. Quantitative reverse transcription PCR was done using SensiFast Real-Time PCR kit (Bioline, USA), on CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA). New primers for quantification of LeAPS and LeAPL1 expression were designed. APSQ-F1 (5′-TCCCAGATTTCAGCTTCTATGATCGG-3′) and APSQ-R1 (5′ TGAATCTTGCAGTTCTTAATCACGC-3′) primers were for LeAPS, while APL1Q-F1 (5′-AGAACTCGAAGATCAGGAACTGC-3′) and APL1Q-R1 (5′-TCTTTCGGCTTCTTGGATTCCCTC-3′) primers were for LeAPL1. Previously described primers were used to quantify expression of LeAPL2 (LeAPL2-F/LeAPL2-R) and LeAPL3 (LeAPL3-F/LeAPL3-R)4. Gene expression levels were determined according to a previously described method28. Briefly, five μl of cDNA of all samples were combined. A standard dilution series of 1x, 10−1X, 10−2X and 10−3X concentrations were made. The quantitation cycle (Cq) values of the standards and all samples were determined. Standard curves of standard cDNA concentrations and Cq values were generated. Gene expression levels of all samples were determined by normalizing the sample Cq values against the standard curves. The experiment was performed in triplicate. Two-tailed student’s t-test was used to determine statistically significant differences (P < 0.05) between expression levels of each AGPase subunit genes in MS and Hoagland’s media on each day. Correlation coefficients between the averages of starch content and expression levels were determined for each gene.
References
Martin, C. & Smith, A. M. Starch biosynthesis. Plant Cell 7, 971–985 (1995).
Smidansky, E. D. et al. Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. USA 99, 1724–1729 (2002).
Ventriglia, T. et al. Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol. 148, 65–76 (2008).
Zhao, Z. et al. Effect of nitrogen and phosphorus deficiency on transcriptional regulation of genes encoding key enzymes of starch metabolism in duckweed (Landoltia punctata). Plant Physiol. Biochem. 86, 72–81 (2015).
Tao, X. et al. Comparative transcriptome analysis to investigate the high starch accumulation of duckweed (Landoltia punctata) under nutrient starvation. Biotechnol. Biofuels 6, 72 (2013).
Yang, Y. et al. Functional analysis of a wheat AGPase plastidial small subunit with a truncated transit peptide. Molecules 22, E386 (2017).
Villand, P., Olsen, O. A. & Kleczkowski, L. A. Molecular characterization of multiple cDNA clones for ADP-glucose pyrophosphorylase from Arabidopsis thaliana. Plant Mol. Biol. 23, 1279–1284 (1993).
La Cognata, U., Willmitzer, L. & Müller-Röber, B. Molecular cloning and characterization of novel isoforms of potato ADP-glucosepyrophosphorylase. MGG Mol. Gen. Genet. 246, 538–548 (1995).
Chen, X., Salamini, F. & Gebhardt, C. A potato molecular-function map for carbohydrate metabolism and transport. Theor. Appl. Genet. 102, 284–295 (2001).
Singh, S., Choi, S. B., Modi, M. K. & Okita, T. W. Isolation and characterization of cDNA clones encoding ADP-glucose pyrophosphorylase (AGPase) large and small subunits from chickpea (Cicer arietinum L.). Phytochemistry 59, 261–268 (2002).
Hannah, L. C. et al. Maize genes encoding the small subunit of ADP- glucose pyrophosphorylase. Plant Physiol. 127, 173–183 (2001).
Li, N., Zhang, S., Zhao, Y., Li, B. & Zhang, J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 233, 241–250 (2011).
Ainsworth, C., Tarvis, M. & Clark, J. Isolation and analysis of a cDNA clone encoding the small subunit of ADP-glucose pyrophosphorylase from wheat. Plant Mol. Biol. 23, 23–33 (1993).
Ainsworth, C. et al. Adenosine diphosphate glucose pyrophosphorylase genes in wheat: differential expression and gene mapping. Planta 197, 1–10 (1995).
Kang, G. et al. Increasing the starch content and grain weight of common wheat by overexpression of the cytosolic AGPase large subunit gene. Plant Physiol. Biochem. 73, 93–98 (2013).
Sree, K. S., Bog, M. & Appenroth, K. J. Taxonomy of duckweeds (Lemnaceae), potential new crop plants. Emirates J. Food Agric. 28, 291–302 (2016).
Appenroth, K. J., Borisjuk, N. & Lam, E. Telling duckweed apart: Genotyping technologies for the Lemnaceae. Chinese Journal of Applied and Environmental Biology 19, 1–10 (2013).
Lemon, G. D., Posluszny, U. & Husband, B. C. Potential and realized rates of vegetative reproduction in Spirodela polyrhiza, Lemna minor, and Wolffia borealis. Aquat. Bot. 70, 79–87 (2001).
Chen, Q. et al. Improving production of bioethanol from duckweed (Landoltia punctata) by pectinase pretreatment. Energies 5, 3019–3032 (2012).
Liu, Y. et al. Improving biomass and starch accumulation of bioenergy crop duckweed (Landoltia punctata) by abscisic acid application. Sci. Rep. https://doi.org/10.1038/s41598-018-27944-7 (2018).
Souto, L. R. F. et al. Effect of hydrothermal pre-treatment on duckweed (Landoltia punctata) biomass for simultaneous saccharification and fermentation process. Biomass and Bioenergy, https://doi.org/10.1016/j.biombioe.2019.105259 (2019).
Liu, Y. et al. The effects of photoperiod and nutrition on duckweed (Landoltia punctata) growth and starch accumulation. Ind. Crops Prod. 115, 243–249 (2018).
Huang, M. et al. Proteomic analysis to investigate the high starch accumulation of duckweed (Landoltia punctata) under nutrient starvation. Ind. Crops Prod. 59, 299–308 (2014).
Liu, Y. et al. Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) I: Transcriptome analysis of the effects of uniconazole on chlorophyll and endogenous hormone biosynthesis. Biotechnol. Biofuels, https://doi.org/10.1186/s13068-015-0246-7 (2015).
Liu, Y. et al. Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) II: Transcriptome alterations of pathways involved in carbohydrate metabolism and endogenous hormone crosstalk. Biotechnol. Biofuels 8, 64 (2015).
Kittiwongwattana, C. & Vuttipongchaikij, S. Effects of nutrient media on vegetative growth of Lemna minor and Landoltia punctata during in vitro and ex vitro cultivation. Maejo Int. J. Sci. Technol. 7, 60–69 (2013).
Moon, H. K. & Stomp, A. M. Effects of medium components and light on callus induction, growth, and frond regeneration in Lemna gibba (Duckweed). Vitr. Cell. Dev. Biol. - Plant 33, 20–25 (1997).
Wang, W. & Messing, J. Analysis of ADP-glucose pyrophosphorylase expression during turion formation induced by abscisic acid in Spirodela polyrhiza (greater duckweed). BMC Plant Biol. 12, 5 (2012).
Hoagland, D. & Arnon, D. I. The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Publ. 347, 36–39 (1938).
Les, D. H., Landolt, E. & Crawford, D. J. Systematics of the Lemnaceae (duckweeds): Inferences from micromolecular and morphological data. Plant Syst. Evol. 204, 161–177 (1997).
Landolt, E. Key to the determination of taxa within the family of Lemnaceae. Verӧff. Geobot. Inst. ETH, Stift. Rübel, Zürich 70, 13–21 (1980).
Les, D. H. & Crawford, D. J. Landoltia (Lemnaceae), a New Genus of Duckweeds. Novon 9, 530 (1999).
Wang, W. et al. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 10, 205 (2010).
Les, D. H., Crawford, D. J., Landolt, E., Gabel, J. D. & Kimbacll, R. T. Phylogeny and systematics of Lemnaceae, the duckweed family. Syst. Bot. 27, 221–240 (2002).
Fang, Y. Y., Babourina, O., Rengel, Z., Yang, X. E. & Pu, P. M. Ammonium and nitrate uptake by the floating plant Landoltia punctata. Ann. Bot. https://doi.org/10.1093/aob/mcl264 (2007).
Liu, Y. et al. Ammonium inhibits primary root growth by reducing the length of meristem and elongation zone and decreasing elemental expansion rate in the root apex in Arabidopsis thaliana. PLoS One. https://doi.org/10.1371/journal.pone.0061031 (2013).
Caicedo, J. R., Van Der Steen, N. P., Arce, O. & Gijzen, H. J. Effect of total ammonia nitrogen concentration and pH on growth rates of duckweed (Spirodela polyrrhiza). Water Res. https://doi.org/10.1016/S0043-1354(00)00128-7 (2000).
Wang, W. et al. Effects of high ammonium level on biomass accumulation of common duckweed Lemna minor L. Environ. Sci. Pollut. Res. https://doi.org/10.1007/s11356-014-3353-2 (2014).
Kobayashi, T. & Nishizawa, N. K. Iron Uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152 (2012).
Dalton, C. C., Iqbal, K. & Turner, D. A. Iron phosphate precipitation in Murashige and Skoog media. Physiol. Plant. 57, 472–476 (1983).
Cui, W. & Cheng, J. J. Growing duckweed for biofuel production: A review. Plant Biology 17, 16–23 (2015).
Yu, C. et al. Integrated analysis of transcriptome and metabolites reveals an essential role of metabolic flux in starch accumulation under nitrogen starvation in duckweed. Biotechnol. Biofuels 10, 167 (2017).
Nakata, P. A., Anderson, J. M., Okita, T. W. & Preiss, J. Structure and expression of the potato tuber ADP-glucose pyrophosphorylase small subunit. J. Biol. Chem. 269, 30798–30807 (1994).
Noh, S. A. et al. Genomic organizations of two small subunit ADP-glucose pyrophosphorylase genes from sweetpotato. Gene 339, 173–180 (2004).
Liu, Y. et al. The influence of different plant hormones on biomass and starch accumulation of duckweed: A renewable feedstock for bioethanol production. Renew. Energy. https://doi.org/10.1016/j.renene.2019.01.128 (2019).
Akihiro, T., Mizuno, K. & Fujimura, T. Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 46, 937–946 (2005).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Acknowledgements
This work was financially supported by the Research Grant of Faculty of Science, King Mongkut’s Institute of Technology Ladkrabang.
Author information
Authors and Affiliations
Contributions
C.K. designed and performed the experiments. C.K. analysed the results and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kittiwongwattana, C. Differential effects of synthetic media on long-term growth, starch accumulation and transcription of ADP-glucosepyrophosphorylase subunit genes in Landoltia punctata. Sci Rep 9, 15310 (2019). https://doi.org/10.1038/s41598-019-51677-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51677-w
This article is cited by
-
Identification of potential light deficiency response regulators in endangered species Magnolia sinostellata
Scientific Reports (2022)
-
Duckweeds: their utilization, metabolites and cultivation
Applied Biological Chemistry (2021)
-
Accumulation of starch in duckweeds (Lemnaceae), potential energy plants
Physiology and Molecular Biology of Plants (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.