Abstract
Gymnema sylvestre is a highly valuable medicinal plant in traditional Indian system of medicine and used in many polyherbal formulations especially in treating diabetes. However, the lack of genomic resources has impeded its research at molecular level. The present study investigated functional gene profile of G. sylvestre via RNA sequencing technology. The de novo assembly of 88.9 million high quality reads yielded 23,126 unigenes, of which 18116 were annotated against databases such as NCBI nr database, gene ontology (GO), KEGG, Pfam, CDD, PlantTFcat, UniProt & GreeNC. Total 808 unigenes mapped to 78 different Transcription Factor families, whereas 39 unigenes assigned to CYP450 and 111 unigenes coding for enzymes involved in the biosynthesis of terpenoids including transcripts for synthesis of important compounds like Vitamin E, beta-amyrin and squalene. Among them, presence of six important enzyme coding transcripts were validated using qRT-PCR, which showed high expression of enzymes involved in methyl-erythritol phosphate (MEP) pathway. This study also revealed 1428 simple sequence repeats (SSRs), which may aid in molecular breeding studies. Besides this, 8 putative long non-coding RNAs (lncRNAs) were predicted from un-annotated sequences, which may hold key role in regulation of essential biological processes in G. sylvestre. The study provides an opportunity for future functional genomic studies and to uncover functions of the lncRNAs in G. sylvestre.
Similar content being viewed by others
Introduction
Gymnema sylvestre R.Br (Family, Asclepidaceae), also known as gurmar or Madhunashini, is a woody climber and a well-known highly valued medicinal plant used to treat diabetes in India since ages1. The leaves of G. sylvestre contain triterpene saponins belonging to oleanane and dammarane classes. The major constituents like gymnemic acids and gymnemasaponins are members of oleanane type of saponins while gymnemasides are dammarane saponins2. They are known for their antidiabetic, hypolipidemic3, stomachic, diuretic, refrigerant and astringent properties1. In addition, it is also known to exhibit anticancer activity4. Most importantly, gymnemic acids stimulate an antihyperglycemic response by regeneration of pancreatic cells, causing insulin release and inhibition of glucose absorption5. It is also known that G. sylvestre leaves not only produce blood glucose homeostasis but also increase uptake and activities of enzymes like phosphorylase, gluconeogenic enzymes and sorbitol dehydrogenase that are involved in glucose utilization via insulin dependent pathways6. In recent year’s, genomic profiling technologies such as RNA sequencing have emerged as effective tool in understanding functional genomics profile of non-model plants. RNA-seq has been used by scientific community in the identification of functional genes involved in the biosynthesis of active compounds and metabolic engineering of important pathways in plants7,8,9. Recent studies with Curcuma longa10, Withania somnifera11, Camelina sativa12, Andrographis paniculata13, Solanum trilobatum14, Foeniculum vulgare15 and Arisaema heterophyllum16 have demonstrated the effectiveness of de novo assembly of transcriptomes. Despite the importance of this plant, there is a dearth of data relating to its functional genomics profile except for a recent report describing polyoxypregnane glycoside biosynthesis pathway17. However, a detailed description of the expressed transcripts of G. sylvestre and putative genes involved in terpenoid biosynthesis pathways is still not known. In the present study, de novo transcriptome sequencing of G. sylvestre leaf was performed using Ion-Proton platform and analysis using various bioinformatics tools. The study will serve as a road to discover and decipher expression information like biosynthetic pathways and putative candidates of important secondary metabolites, which may be further used for the scale up production of bioactive compound.
Materials and Methods
Plant material and RNA isolation
Young and fully expanded leaves were collected in biological triplicates from disease-free one year old plants Gymnema sylvestre, grown at State Medicinal and Aromatic Plants garden, Gandhinagar, Gujarat, India and identified by State Medicinal Plant Board, Gujarat. The leaves were snap chilled immediately by dipping in liquid nitrogen and ground into fine powder using mortar and pestle. Further, total RNA was isolated according to manufacturer’s protocol using Qiagen Plant RNeasy isolation kit and RNase-free DNase I treatment was given in order to remove any traces of genomic DNA. For quality check, QIAxpert and QIAxcel were used for determining RNA integrity, while quantification was done using Qubit4.
Generation of cDNA library and sequencing of transcriptome
Samples with RNA Integrity Score (RIS) values greater than 8.0 were further processed for preparing cDNA library. Ribosomal RNA depletion was carried out using RiboMinus RNA plant kit for RNA-Seq (Life Technologies, CA). The whole transcriptome cDNA library was prepared using Ion Total RNA-Seq kit V2 (Life Technologies Corporation, CA). Double stranded cDNA was ligated to barcoded adapters, loaded onto the Ion PI™ Chip (Ion torrent, Life technologies, CA) and sequenced in triplicate according to the standard protocol using Ion Proton System (Ion torrent, Life technologies, CA).
De novo assembly of transcriptome
Sequencing data was collected and further, the raw reads were subjected to stringent filtering conditions for the removal of reads with adaptors, reads with ambiguous bases and reads with low quality using FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). High quality (HQ) reads (i.e., each base having ≥20 phred score) were considered for assembling transcriptome. Primary assembly was carried out by merging the HQ reads using “Trinity” assembler18 with a minimum contig length of 200 bases and k-mer size of 25 bp. A minimum count of 2 k-mers were assembled by Inchworm algorithm and a minimum number of 5 reads were used to glue two Inchworm contigs together. In order to cluster contigs originating from the same gene or protein, a secondary assembly was carried out using CD-HIT EST (v4.6.1) tool19. Homologous contigs with 80% identity were clustered to generate full length transcripts. In order to determine the percentage of reads mapped to assembled transcriptome, we mapped the assembled transcriptome onto the processed reads using Bowtie220. The resulting file was further used as input for eXpress tool to determine the FPKM and TPM values for the reads (https://pachterlab.github.io/eXpress/index.html).
The sequence data generated in this study have been deposited at NCBI in the Short Read Archive database under the accession number SRR7876667, SRR9644907, SRR9644908.
Functional annotation and classification of transcripts
Assembled transcript sequences were functionally annotated using public databases. Sequence similarity search was performed using a BLASTX against the Uniprot and Swissprot databases and Pfam database using the Trinotate pipeline18. The Trinotate annotation pipeline includes several software packages such as BLASTX, BLASTP and PFAM search that are essential in transcriptome functional annotation. All analyses were performed in parallel using assembled FASTA sequences. Gene Ontology (GO) and Conserved Domain Database (CDD) were used to annotate the transcripts based on similarity. The GO analysis helps us in specifying all the annotated sequences comprising of GO functional group such as Biological Process, Molecular Function and Cellular Component21. Translated peptides were generated using the Transdecoder program embedded in the Trinity assembly pipeline for protein-based analysis using Eukaryotic Orthologous Group (KOG) classification. All results were deposited into Trinotate-provided SQLite database template and a spreadsheet summary report was generated from Trinotate using BLASTX E-value cutoff of 1e-5. Pathway assignment for the annotated transcripts was carried out using KEGG mapping (http://www.genome.ad.jp/kegg/). KEGG orthologs were identified using the KEGG Automated Annotation Server (KAAS) with default parameters. Transcripts were also annotated simultaneously using Function Annotator for transcriptome data22. FunctionAnnotator includes scripts and annotation tools, including LAST, BLAST2GO, PSORT, TMHMM, etc. for annotating GO terms, enzyme and domain identification, predictions for subcellular localization, lipoproteins, secretory proteins and transmembrane proteins, etc. FDR corrected GO terms were filtered and comparison with the closely related species were performed with similarity search E-value 10e-5.
Identification of transcription factor families
Transcription factors (TFs) were identified using genome-scale protein and nucleic acid sequences by analyzing InterProScan domain patterns in protein sequences with high coverage and sensitivity using PlantTFcat analysis tool (http://plantgrn.noble.org/PlantTFcat/)23.
Identification of simple sequence repeats (SSRs)
Simple sequence repeats were identified using MIcroSAtellite identification tool v1.0 (MISA) (http://pgrc.ipk-gatersleben.de/misa/). Unit size cut-off of six was used to consider a di-nucleotide repeat and 5 for SSRs of 3, 4, 5, and 6-nucleotide repeats. Maximum of 100 interrupting bases were allowed between two SSRS in a compound microsatellite.
Prediction of long non-coding RNA (lncRNAs)
The non-coding DNA sequences (CDS) of G. sylvestre were used as the starting point for the prediction of lncRNAs. The CDS with length greater than 200 nucleotides24 were retained. The coding potential for the sequences were checked by Coding Potential Calculator (CPC), developed on support vector machine25. Based on CPC score (S), sequences were classified into non-coding (S ≤ −0.5), neutral (−0.5 < S < 1.0) and coding (S ≥ 1.0). The sequences were further searched using BLASTX against the SWISS-PROT database with an e-value cut-off of 0.001 in order to be sure that the sequences were non-protein coding. A database of lncRNAs was created using 45 plant species from the GreeNC26 and Blastn was performed. The sequences with more than 90% identity were predicted to be lncRNAs.
Quantitative reverse transcription PCR (qRT-PCR) of selected secondary metabolite biosynthetic pathway genes in G. sylvestre leaf sample
qRT-PCR enables the detection and identification of target mRNA transcripts. Hence, to validate our dataset, some of the assembled G. sylvestre unitranscripts involved in Terpenoid biosynthetic pathway were selected for performing qRT-PCR. Total RNA from the leaves of G. sylvestre in biological triplicates was isolated from using Plant RNeasy isolation kit according to manufacturer’s protocol. cDNA was synthesized using Oligo(dT) and SuperScript III Reverse Transcriptase. Transcripts encoding squalene monooxygenase (SQLE), farnesyl-diphosphate farnesyltransferase (FDFT1) involved in Sesquiterpenoid and triterpenoid biosynthesis and 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (ispF), (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ispG), farnesyl diphosphate synthase (FDPS2) & diphosphomevalonate decarboxylase (MVD) involved in Terpenoid backbone biosynthesis were validated against house-keeping transcript Actin B, GAPDH, Beta-tubulin and Ubiquitin C as reference. Transcript specific primers were designed and PCR based expression profiling was carried out for each transcript in triplicates.
Results
Sequencing of cDNA library and de novo assembly of transcriptome
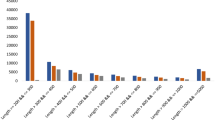
Sequencing of cDNA library using the Ion Proton generated millions of reads with an average length of 200 bp after the removal of adapter sequences and low quality reads with Phred score <20. 88.9 million of high quality data reads were obtained representing 85% of the transcriptome. Currently no reference genome is available for G. sylvestre therefore the Trinity assembler20 was used for de novo assembly of the high quality reads. Assembly of high quality reads using Trinity assembler produced a total of 23126 unigenes post removal of redundant transcripts using CD-HIT. Transcript length ranged from 200–7200 bp with an average of 369 bp and N50 of 372 bp was obtained. De novo assembly of transcriptome revealed 42.69% of GC content. The raw reads were mapped on to the assembly using Bowtie2 and 85% alignment was observed indicating good quality assembly.
Functional annotation and classification of the clustered transcripts
Extensive functional annotation was performed in order to decipher the profile and information regarding molecular functions, SSRs, transcription factors as well as signal peptides. Additionally, lncRNA were also predicted via in silico approach. Total 18116 unigenes were functionally annotated, whereas 5010 did not show similarity to any proteins or domains. Corresponding GO IDs were classified into biological functions, cellular components and molecular functions. Functional annotations of the assembled transcripts revealed that almost 52% of them showed homology to 11 other species. While the majority of them (35.73%) were homologous to the species Coffea canephora lowest homology was found with Populus trichocarpa (1.5%) (Fig. 1). The un-annotated unigenes show that there may be genus specific or species-specific functions.
Annotation with gene ontology (GO)
Out of 18116 annotated unigenes, 14987 were found to be associated with gene ontology terms. A total of 13085 unigenes were found in the category of biological processes with majority belonging to oxidation-reduction processes, metabolic processes, protein phosphorylation, response to cadmium ion, and regulation of transcription etc. (Fig. 2). About 12349 transcripts mapped to different molecular functions with majority of them belonging to ATP binding followed by zinc ion binding, DNA binding, protein serine/threonine kinase activity etc and 12690 transcripts mapped to cellular components belonging to membranes of nucleus, plasma membrane, cytosol, plasmodesma etc.
Metabolic pathway analysis by KEGG
Present study annotated the leaf transcriptome of G. sylvestre and focused primarily on the terpenoid pathway unigenes. Identification of candidate genes and key enzymes are crucial in understanding the biosynthetic pathways of functional terpenoids in G. sylvestre. As pharmaceutical properties of G. sylvestre is largely dependent on its terpenoid profile, the present study was mainly focused on the identification of transcripts involved in terpenoid biosynthesis. The KEGG predictions of the present study mapped 111 transcripts encoding for various enzymes involved in the biosynthesis of different isoprenoids such as mono-terpenes, di-terpenes, tri-terpenes, and ubiquinones (Fig. 3). Analysis of transcripts involved in the terpenoid and diterpenoid biosynthetic pathways identified majority of them being involved in terpenoid biosynthesis followed by ubiquinone and other terpenoid-quinone biosynthesis (Figs 4–6). It was observed that the transcripts involved in Vitamin E synthesis, beta-amyrin synthesis and squalene synthesis were also mapped on the pathway as evident from Figs 5 and 6. Pathway analysis also showed 32 transcripts involved in the flavonoid biosynthesis pathway such as chalcone synthase, naringenin 3-dioxygenase, flavonoid 3′-monooxygenase, shikimate O-hydroxycinnamoyltransferase etc as depicted in Fig. 7.
Transcripts mapped on the Terpenoid Biosynthetic pathway (Enzymes highlighted in one colour code for one enzyme. Green colour depicts different enzyme code). KEGG pathway map 00900 is mined here from http://www.kegg.jp/kegg/kegg1.html. The KEGG database has been reported previously55,56,57.
Transcripts mapped on ubiquinone and other terpenoid-quinone biosynthesis (Enzymes highlighted in one colour code for one enzyme. Green colour depicts different enzyme code). KEGG pathway map 00130 is mined here from http://www.kegg.jp/kegg/kegg1.html. The KEGG database has been reported previously55,56,57.
Transcripts mapped on biosynthesis of sesquiterpenoid and triterpenoid biosynthesis pathway (Enzymes highlighted in one colour code for one enzyme. Green colour depicts different enzyme code). KEGG pathway map 00909 is mined here from http://www.kegg.jp/kegg/kegg1.html. The KEGG database has been reported previously55,56,57.
Transcripts mapped on the flavonoid biosynthetic pathway (Enzymes highlighted in one colour code for one enzyme. Green colour depicts different enzyme code). Green colour depicts different enzyme code). KEGG pathway map 00941 is mined here from http://www.kegg.jp/kegg/kegg1.html. The KEGG database has been reported previously55,56,57.
Identification of Transmembrane proteins, signal peptides, subcellular localization and CYP450
Analysis of the transcript sequences revealed 293 transcripts to have at least one enzyme hit. Total 4075 transcripts were identified to have at least one domain with >50% coverage (Supplementary Fig. S1). Total 2163 transcripts were predicted to have at least 1 transmembrane domain, whereas 541 transcripts were predicted to have signal peptides (Supplementary Table 1). In our present study total 39 transcripts which showed homology to CYP450 sequences.
Transcription factor (TF) analysis and identification of SSRs
The analysis of transcripts revealed 809 unique transcripts belonging to 78 transcription factor families (Fig. 8). Among the identified unigenes, most of them represent WD40 family followed by C2H2, CCHC(Zn), Hap3/NF-YB, PHD etc. MISA analysis of 23126-clustered transcripts revealed a total number of 1428 SSRs in 1262 transcripts. More than 1 SSR was found in 135 transcripts including 96 transcripts with compound SSRs. A maximum number of SSRs were identified as di-nucleotide repeats followed by tri-nucleotide, mono-nucleotide, tetra-nucleotide, and penta-nucleotide repeats.
Identification of long non-coding RNA (lncRNAs)
The 5010 un-annotated sequences were considered for predicting lncRNA. The coding potential of non-coding transcripts was determined using Coding Potential Calculator (Supplementary Table 2). Coding potential calculator provides coding probability, isoelectric points and fickett scores for the transcripts and gives a probability whether or not the transcript may be coding or non-coding. Transcripts having CPC score <0.2 were considered as non-coding. A database of lncRNAs was created using 45 plant species from the GREENC and Blastn was performed. Total 8 putative lncRNA were predicted from 2 plant species, in 10 Gymnema sylvestre transcripts (Table 1). Majority of predicted lncRNA were from Arabidopsis lyrata (932008, 932001, 931993, 484743, 932003, 930998) followed by from Ananas comosus species. These candidate sequences were searched in the Phytozome database27 and PANTHER database28 to determine whether any functional role was reported for the homologous sequences.
Validation of transcripts using qRT-PCR
In order to validate the relative expression levels from the transcript abundance estimation, six important transcripts related to terpenoid, sesquiterpenoid biosynthetic pathways were chosen for RT-qPCR. The primer sequences for the transcripts designed as shown in Table 2. The nucleotide sequences for the same are provided in Supplementary Table S3. The relative expression of these transcripts was calculated using equation provided in Saussoy et al.29. Actin B, GAPDH, Ubiquitin C, beta-tubulin were chosen as reference for calculation of dCT. The expression of the transcripts have been provided in Supplementary Fig. S2. The expression of transcripts with individual housekeeping genes as reference are provided in Supplementary Fig. S3.
Discussion
In the present study, we performed leaf transcriptome sequencing and reported de novo assembly of Gymnema sylvestre. The denovo assembly of G. sylvestre resulted, 23,126 unigenes with an N50 of 372 bp and 42.69% of GC content. The quality of assembly based on N50 of unigenes, were near to the earlier transcriptome studies i.e. Camellia sinensis30 and Rubber tree31. These assembled unigenes and 85% alignment of reads onto assembled unigenes indicated good assembly quality. Functional analysis of the transcriptome annotated and classified 18116 unigenes into different biological processes, molecular functions and cellular components. The un-annotated unigenes show that there may be genus specific or species-specific functions.
Plant secondary metabolites have significant use in the food and pharmaceutical industries, which makes the study of biosynthesis, regulation and metabolic engineering of valuable secondary metabolites extremely useful32,33. Earlier report on G. sylvestre transcriptome indicated the synthesis of bioactive gymnemic acid takes place primarily in the leaf 34. Identification of candidate genes and key enzymes is crucial in understanding the biosynthetic pathways of functional terpenoids in G. sylvestre. As pharmaceutical properties of G. sylvestre largely depend on its terpenoid profile, the present study was mainly focused on the identification of transcripts involved in terpenoid biosynthesis. KEGG analysis mapped 111 transcripts encoding for various enzymes involved in the biosynthesis of different isoprenoids such as mono-terpenes, di-terpenes, tri-terpenes, and ubiquinones.
Precursor molecules for terpenoid biosynthesis are derived from the cytosolic mevalonate (MVA) and plastidial methyl-erythritol phosphate (MEP) pathways. Transcripts mapped on both MVA and MEP pathways which was evident from the data analysis. The results correlate with the hypothetical pathway provided by Tiwari et al.1 for gymnemic acid biosynthesis. We found many transcript genes related to isoprenoid biosynthesis from the MEP pathway including gene transcripts such as 1-deoxy-D-xylulose-5-phosphate synthase, 1-deoxy-D-xylulose-5-phosphate reductoisomerase, 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase, isopentenyl-diphosphate Delta-isomerase and geranyl-diphosphate synthase. These transcripts were also validated via qRT-PCR and represented positive involvement in terpenoid biosynthesis via the MEP pathway. G sylvestre is known to produce atleast 34 different compounds including Vitamin E, squalene, beta-amyrin and related glycosides35. Pathway analysis also showed transcripts involved in synthesis of Vitamin E which is considered as an important free radical scavenger involved in the prevention of prostate cancer35. Apart from Vitamin E, transcripts were also found for (3S)-2,3-Epoxy-2,3-dihydrosqualene also known as Beta-amyrin synthase which is an enzyme that catalyzes the reaction to form beta-amyrin. Beta-amyrin is known to exhibit anti-inflammatory, anti-microbial activities36. Besides this, transcripts involved in flavonoid synthesis pathway were also found. G. sylvestre is also known to exhibit wound healing properties which may be attributed to the free radical scavenging action and presence of flavonoids20,37.
Transcription factors (TFs) play a major role in plant development and their response to the environment. The Transcription factors identified in the present study showed presence of WD40 like and CCHC as major transcription factor families in the transcriptome. Members of WD40 superfamily are increasingly being recognized as key regulators of plant-specific developmental events38 whereas CCHC (Zn) also known as transcription factor interactor and regulator specifically interact with single-stranded DNA or RNA oligonucleotides carrying recognition sequences39. Another transcription factor, which was abundant, was the plant homeodomain (PHD) which has been termed as an epigenome reader. PHD zinc fingers are known to be conserved and modify chromatin as well as mediate molecular interactions in gene transcription40.
Apart from gene discovery, transcriptome sequencing has also been proven to be an important tool for molecular marker development41. SSRs also known as microsatellites, are short repeating sequences with a unit size of mono-, di-, tri-, tetra-, or penta-nucleotides. In terms of the types of motifs found in SSR loci other than the mono- and large sized repeats, we found similar results as in previous report with plant microsatellites42. The most common tri-nucleotide repeats found were GAA/TTC, GAT/ATC, TCT/AGA and CAG/CTG. Interestingly, the proportions of di- and tri-nucleotide repeats were quite close (38.86% versus 34.87%) as reported earlier43.
In recent years, the functional characterization of one of the largest gene families, i.e. CYP450s, has created immense interest in the scientific community. They were known to catalyze the oxidative modification of various substrates using oxygen and NAD(P)H44. Many studies focusing on the transcriptome-wide identification of CYP450s for terpene biosynthesis have been reported45,46. An earlier research performed transcriptomic analyses based on 454 pyrosequencing data of Panax ginseng flowers, roots, stems, and leaves, which identified 326 potential CYP450s, including CYP716A47, which is related to the ginsenoside biosynthesis47. The current study identified 39 transcripts exhibiting homology to CYP450 sequences which may be of further interest to understand the involvement for the targeted biosynthetic pathway. Analysis of domains showed presence of a high number of transcripts for kinases like protein kinase-like domains, serine/threonine kinases, tyrosine-protein kinase etc. This suggests that most of the transcripts may be involved in signaling and regulatory processes, which correlates with our functional analysis.
With the advancements in sequencing technologies and high-throughput analysis tools the traditional view that protein-coding genes are the only effectors of gene function has been challenged. Micromolecules such as long noncoding RNAs (lncRNAs), miRNA etc. have been identified as key regulatory cascade of the eukaryotic transcriptomes, involved in the regulation of important biological processes in plants48,49,50 as well as in cross-kingdom gene regulation51,52,53. The study predicted 8 putative candidate lncRNA sequences using computational screening against database of 45 Plants species. Although some sequences showed annotation for the locus in PANTHER database28 no specific function was provided for these sequences, which may be due to the lag in lncRNA research in plants as compared to that in humans and animals54.
Conclusion
In summary, our findings give a molecular insight of the transcriptome profile of an important antidiabetic medicinal plant. Due to its bioactive principle and potential use in Indian system of medicine through many polyherbal formulations, our study will enrich the understanding of the biosynthesis of its active principle. Our data provides us a glimpse of the transcripts, involved in secondary metabolic pathways. The transcriptome profile reveals the terpenoid, flavonoid and other secondary metabolic pathway genes, which adds information to G. sylvestre dataset and may help in accelerating the design-build-develop approach in metabolite engineering. Further, qRT-PCR results confirmed expression of a few selected transcripts proving the reliability of our G. sylvestre transcriptome study. Additionally, identified putative lncRNAs in the present study may further be explored in future experimental studies to uncover their role in regulation of various biological process in G. sylvestre. Such study on non-model plants will be of great potential in scaling up the targeted metabolite for therapeutic purposes.
References
Tiwari, P., Mishra, B. N. & Sangwan, N. S. Phytochemical and pharmacological properties of Gymnema sylvestre: an important medicinal plant. BioMed. Research. International. 2014 (2014).
Khramov, V. A., Spasov, A. A. & Samokhina, M. P. Chemical composition of dry extracts of Gymnema sylvestre leaves. Pharm. Chem. J. 42, 29 (2008).
Kumar, H., Nagendra, N. I., Huilgol, S. V., Yendigeri, S. M. & Narendar, K. Antidiabetic and hypolipidemic activity of Gymnema sylvestre in dexamethasone induced insulin resistance in albino rats. International Journal of Medical Research and Health Sciences. 4, 639–645 (2015).
Arunachalam, K. D., Arun, L. B., Annamalai, S. K. & Arunachalam, A. M. Potential anticancer properties of bioactive compounds of Gymnema sylvestre and its biofunctionalized silver nanoparticles. Int. J. Nanomedicine. 10, 31 (2015).
Patel, D. K., Prasad, S. K., Kumar, R. & Hemalatha, S. An overview on antidiabetic medicinal plants having insulin mimetic property. Asian. Pac. J. Trop. Biomed. 2, 320 (2012).
Shanmugasundaram, K. R., Panneerselvam, C., Samudram, P. & Shanmugasundaram, E. R. B. Enzyme changes and glucose utilisation in diabetic rabbits: the effect of Gymnema sylvestre. J. Ethnopharmacol. 7, 205–234 (1983).
Mata-Pérez, C. et al. Transcriptomic profiling of linolenic acid-responsive genes in ROS signaling from RNA-seq data in Arabidopsis. Front. Plant. Sci. 6, 122 (2015).
Wang, B. et al. Developing single nucleotide polymorphism (SNP) markers from transcriptome sequences for identification of longan (Dimocarpus longan) germplasm. Horticulture. Research. 2, 14065 (2015).
Miller, C. N. et al. Elucidation of the genetic basis of variation for stem strength characteristics in bread wheat by Associative Transcriptomics. BMC. Genomics. 17, 500 (2016).
Annadurai, R. S. et al. De Novo transcriptome assembly (NGS) of Curcuma longa L. rhizome reveals novel transcripts related to anticancer and antimalarial terpenoids. PLoS One. 8, e56217 (2013).
Dasgupta, M. G., George, B. S., Bhatia, A. & Sidhu, O. P. Characterization of Withania somnifera leaf transcriptome and expression analysis of pathogenesis–related genes during salicylic acid signaling. PLoS One. 9, e94803 (2014).
Mudalkar, S., Golla, R., Ghatty, S. & Reddy, A. R. De novo transcriptome analysis of an imminent biofuel crop, Camelina sativa L. using Illumina GAIIX sequencing platform and identification of SSR markers. Plant. Mol. Biol. 84, 159–171 (2014).
Cherukupalli, N., Divate, M., Mittapelli, S. R., Khareedu, V. R. & Vudem, D. R. De novo assembly of leaf transcriptome in the medicinal plant Andrographis paniculata. Front. Plant. Sci. 7, 1203 (2016).
Lateef, A., Prabhudas, S. K. & Natarajan, P. RNA sequencing and de novo assembly of Solanum trilobatum leaf transcriptome to identify putative transcripts for major metabolic pathways. Sci. Rep. 8, 15375 (2018).
Palumbo, F., Vannozzi, A., Vitulo, N., Lucchin, M. & Barcaccia, G. The leaf transcriptome of fennel (Foeniculum vulgare Mill.) enables characterization of the t-anethole pathway and the discovery of microsatellites and single-nucleotide variants. Sci. Rep. 8, 10459 (2018).
Wang, C. et al. De novo sequencing and transcriptome assembly of Arisaema heterophyllum Blume and identification of genes involved in isoflavonoid biosynthesis. Sci. Rep. 8, 17643 (2018).
Kalariya, K. A., Minipara, D. B. & Manivel, P. De novo transcriptome analysis deciphered polyoxypregnane glycoside biosynthesis pathway in Gymnema sylvestre. 3 Biotech. 8, 381 (2018).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature. Biotechnol. 29, 644 (2011).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22, 1658–1659 (2006).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie2. Nat. Methods. 9, 357 (2012).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome. Biol. 11, R14 (2010).
Chen, T. W. et al. FunctionAnnotator, a versatile and efficient web tool for non-model organism annotation. Sci. Rep. 7, 10430 (2017).
Dai, X., Sinharoy, S., Udvardi, M. & Zhao, P. X. PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinformatics. 14, 321 (2013).
Boerner, S. & McGinnis, K. M. Computational identification and functional predictions of long noncoding RNA in Zea mays. PloS One. 7, e43047 (2012).
Kong, L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic. Acids. Res. 35, W345–W349 (2007).
Paytuví Gallart, A., Hermoso Pulido, A., Anzar Martínez de Lagrán, I., Sanseverino, W. & Aiese Cigliano, R. GREENC: a Wiki-based database of plant lncRNAs. Nucleic. Acids. Res. 44, D1161–D1166 (2015).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic. Acids. Res. 40, D1178–D1186 (2011).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551 (2013).
Saussoy, P. et al. Differentiation of acute myeloid leukemia from B-and T-lineage acute lymphoid leukemias by real-time quantitative reverse transcription-PCR of lineage marker mRNAs. Clin. Chem. 50, 1165–73 (2004).
Yu, O. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 12, 131 (2011).
Li, D., Zhi, D., Bi, Q., Liu, X. & Men, Z. De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.). BMC Genomics. 13, 192 (2012).
Zhao, J., Davis, L. C. & Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 23, 283–333 (2005).
Tatsis, E. C. & O’Connor, S. E. New developments in engineering plant metabolic pathways. Curr. Opin. Biotechnol. 42, 126–132 (2016).
Stoecklin, W. Chemistry and physiological properties of gymnemic acid, the antisaccharine principle of the leaves of Gymnema sylvestre. J. Agric. Food. Chem. 17, 704–708 (1969).
Srinivasan, K. & Kumaravel, S. Unraveling the potential phytochemical compounds of Gymnema sylvestre through GC-MS study. Int J Pharm Pharm Sci 8, 450–453 (2015).
Kushiro, T., Shibuya, M. & Ebizuka, Y. β‐Amyrin synthase: cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. European Journal of Biochemistry 256, 238–244 (1998).
Malik, J. K., Manvi, F. V., Nanjware, B. R. & Sanjiv, S. Wound healing properties of alcoholic extract of Gymnema sylvestre R. Br. leaves in rats. Journal of Pharmacy Research. 2, 1029–1030 (2009).
Van Nocker, S. & Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics. 4, 50 (2003).
Klug, A. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293, 215–218 (1999).
Sanchez, R. & Zhou, M. M. The PHD finger: a versatile epigenome reader. Trends. Biochem. Sci. 36, 364–372 (2011).
Chen, H. et al. Transcriptome sequencing of mung bean (Vigna radiate L.) genes and the identification of EST-SSR markers. PLoS One. 10, e0120273 (2015).
La Rota, M., Kantety, R. V., Yu, J. K. & Sorrells, M. E. Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC. Genomics. 6, 23 (2005).
Huang, D. et al. Characterization and high cross‐species transferability of microsatellite markers from the floral transcriptome of Aspidistra saxicola (Asparagaceae). Mol. Ecol. Resour. 14, 569–577 (2014).
Chapple, C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant. Biol. 49, 311–343 (1998).
Banerjee, A. & Hamberger, B. P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem. Rev. 17, 81–111 (2018).
Liao, W. et al. Transcriptome Assembly and Systematic Identification of Novel Cytochrome P450s in Taxus chinensis. Front. Plant. Sci. 8, 1468 (2017).
Li, C. et al. Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng CA Meyer. BMC. Genomics. 14, 245 (2013).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome. Res. 22, 1775-1789 (2012).
Fabbri, M. & Calin, G. A. Beyond genomics: interpreting the 93% of the human genome that does not encode proteins. Curr. Opin. Drug. Discov. Devel. 13, 350–358 (2010).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Kumar, D. et al. Cross-Kingdom Regulation of Putative miRNAs derived from Happy Tree in Cancer Pathway: A Systems Biology Approach. Int. J. Mol. Sci. 18, 1191 (2017).
Mellis, D. & Caporali, A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem. Soc. Trans. 46, 11–21 (2018).
Yu, D., Tang, C., Liu, P., Qian, W. & Sheng, L. Targeting lncRNAs for cardiovascular therapeutics in coronary artery disease. Curr. Pharm. Des (2018).
Zhu, Q. H. & Wang, M. B. Molecular functions of long non-coding RNAs in plants. Genes. 3, 176–190 (2012).
Kanehisa, F. M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Acknowledgements
This work was supported by Gujarat State Biotechnology Mission (GSBTM) and Gujarat Biotechnology Research Centre (GBRC), Department of Science & Technology (DST), Government of Gujarat, India, Grant number – HLT-15.
Author information
Authors and Affiliations
Contributions
J.D., P.S2. and S.B.B. Conceived and designed the experiments, G.A., B.J., P.S1., P.S2. and L.S. performed the experiments, G.A. and I.S. analyzed the data, C.J., J.D. and S.B.B. Contributed reagents/materials/analysis tools, G.A., I.S., P.S2. and J.D. Wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ayachit, G., Shaikh, I., Sharma, P. et al. De novo transcriptome of Gymnema sylvestre identified putative lncRNA and genes regulating terpenoid biosynthesis pathway. Sci Rep 9, 14876 (2019). https://doi.org/10.1038/s41598-019-51355-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51355-x
This article is cited by
-
Multilayered regulation of secondary metabolism in medicinal plants
Molecular Horticulture (2023)
-
Effect of vitamin E supplementation in rheumatoid arthritis: a systematic review and meta-analysis
European Journal of Clinical Nutrition (2023)
-
Identification of a putative novel cholivirus in the transcriptome of Gymnema sylvestre R. Br
Archives of Microbiology (2023)
-
Characterization of squalene synthase gene from Gymnema sylvestre R. Br.
Beni-Suef University Journal of Basic and Applied Sciences (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.