Abstract
Alongside various clinical prognostic factors for diffuse large B-cell lymphoma (DLBCL) such as the international prognostic index (IPI) components (ie, age, tumor stage, performance status, serum lactate dehydrogenase concentration, and number of extranodal sites), prognostic gene signatures have recently shown promising efficacy. However, previously developed signatures for DLBCL suffer from many major inadequacies such as lack of reproducibility in external datasets, high number of members (genes) in a signature, and inconsistent association with the survival time in various datasets. Accordingly, we sought to find a reproducible prognostic gene signature with a minimal number of genes. Seven datasets—namely GSE10856 (420 samples), GSE31312 (470 samples), GSE69051 (157 samples), GSE32918 (172 samples), GSE4475 (123 samples), GSE11318 (203 samples), and GSE34171 (91 samples)—were employed. The datasets were randomly categorized into training (1219 samples comprising GSE10856, GSE31312, GSE69051, and GSE32918) and validation (417 samples consisting of GSE4475, GSE11318, and GSE34171) groups. Through the univariate Cox proportional hazards analysis, common genes associated with the overall survival time with a P value less than 0.001 and a false discovery rate less than 5% were identified in 1219 patients included in the 4 training datasets. Thereafter, the common genes were entered into a multivariate Cox proportional hazards analysis encompassing the common genes and the international prognostic index (IPI) factors as covariates, and then only common genes with a significant level of difference (P < 0.01 and z-score >2 or <−2) were selected to reconstruct the prognostic signature. After the analyses, a 7-gene prognostic signature was developed, which efficiently predicted the survival time in the training dataset (Ps < 0.0001). Subsequently, this signature was tested in 3 validation datasets. Our signature was able to strongly predict clinical outcomes in the validation datasets (Ps < 0.0001). In the multivariate Cox analysis, our outcome predictor was independent of the routine IPI components in both training datasets (Ps < 0.0001). Furthermore, our outcome predictor was the most powerful independent prognostic variable (Ps < 0.0001). We developed a potential reproducible prognostic gene signature which was able to robustly discriminate low-risk patients with DLBCL from high-risk ones.
Similar content being viewed by others
Introduction
Diffuse large B-cell lymphoma (DLBCL) as the most common type of lymphoma in adults accounts for approximately 30% of all cases of lymphoma1,2.
Development of prognostic gene signatures was started in a study conducted by Alizadeh et al.3, who proposed 2 distinct subtypes of DLBCL (ie, germinal center B cell-like [GCB] and activated B cell-like [ABC]) based on gene expression profiling. The authors indicated that the overall survival (OS) time was significantly higher in patients with GCB-DLBCL than in those with ABC-DLBCL. In a study by Rosenwald et al.4, another molecular subtype of DLBCL (type 3), which did not express the gene characteristics of either GCB or ABC DLBCL, was added to the previous subtypes. In addition, the authors proposed a 17-gene signature which could predict OS after chemotherapy. Via gene expression profiling and supervised machine learning, a 13-gene predictive model was reconstructed in 58 patients with DLBCL5. Surprisingly, their results revealed that the clinical outcome was not significantly different between 2 groups of patients based on the 90-gene model proposed by Alizadeh et al.3. Via a statistical method based on Bayes’ rule, a classifier comprising 27 genes was developed to subtly assign patients with DLBCL to ABC and GCB subgroups and a concluding 14-gene model was proposed as the final subgroup predictor6. Lossos et al.2, seeking to develop a predictive model using prognostic genes previously identified as single prognostic genes or as a member of prognostic signatures, suggested a 6-gene model among 36 genes as the final prognostic signature. Finally, a 108-gene model was created using a combination of 3 gene-expression signatures—namely “germinal-center B-cell,” “stromal-1,” and “stromal-2”—by Lenz et al.7. This large signature could predict survival in CHOP-treated or R-CHOP treated patients.
Despite the introduction of various prognostic gene signatures, there are still many disadvantages curtailing the clinical use of these signatures. Indeed, the most salient disadvantage of the previously developed signatures is lack of reproducibility in various datasets, with many of the genes in the proposed prognostic signatures failing to show a significant association with survival in external validation analyses (See the Results.) Furthermore, our analysis showed that many of these genes failed to exhibit a consistent prognostic pattern in different datasets as some genes with positive associations with the survival time in a dataset were negatively associated with survival in another dataset (See the Results). In addition, some of these signatures are considerably large and contain large numbers of genes (90 genes, 27 genes, and 180 genes in signatures developed by Alizadeh et al.3, Wright et al.6, and Lenz et al.8, respectively), rendering the clinical application of such large signatures difficult or impossible. Moreover, these developed signatures have shown minimal common genes with each other. For example, there were no common genes in the models derived by Shipp et al. (2003) and Rosenwald et al.4. Similarly, BCL6 is the only common gene between signatures developed by Losses et al.2, Rosenwald et al.4, and Wright et al.6.
As another disadvantage, some of these studies used old microarray platforms, which might not be compatible with new platforms. For instance, some genes in Lymphochip-spotted cDNA microarrays3,4 cannot be found in new Affymetrix arrays.
Given all the above mentioned problems, we endeavored to develop a reproducible prognostic gene signature with a minimal number of genes using a strict pipeline in patients with DLBCL. Accordingly, using 4 training datasets, we identified common genes associated with the OS time in 1219 patients through stringent criteria. We reconstructed a prognostic signature with the extracted common genes and validated it externally in 417 patients included in 3 validation datasets. Finally, we produced a reproducible 7-gene signature, which was significantly associated with the survival time in both training and validation datasets and was by far the most powerful independent prognostic factor in comparison with the prognostic components of the IPI.
Results
Extraction of the common genes associated with survival and the reconstruction of the prognostic signature
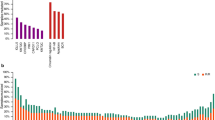
First, search was conducted to find the common genes associated with survival between the 4 training datasets, encompassing 1219 patients. Our analysis revealed that 12 genes consistently had significant associations with OS at a P value less than 0.001 and an FDR less than 5% in all the datasets (Supplementary Table 1). The common genes included APOC1, C5orf30, CALD1, CD84, CSF2RA, GPNMB, ITPKB, LPP, PDLIM4, PLAU, RTN1, and RGS3. These genes showed consistent expression patterns in the 4 datasets, with 11 out of the 12 genes being positively associated with survival and the remaining gene (C5orf30) being negatively associated with survival (Supplementary Table 1). These genes also emerged as members in the class predictors developed using 2 different algorithms, which revealed that their expressions were significantly different between the 2 classes (long survival vs. short survival).
More robust and reliable findings were obtained by entering the common genes into the multivariate Cox analysis, where various components of the IPI and the common genes were considered as covariates. In this stage, only genes which reached a significant level were retained. Hence, genes with a P value less than 0.01 and a z-score greater than 2 or below −2 were selected to reconstruct the prognostic signature. Our analysis retained 7 genes—namely APOC1, CALD1, CD84, GPNMB, ITPKB, PLAU, and RTN1—and excluded 5 genes—namely c5orf30, LPP, CSF2RA, PDLIM4, and RGS3 (Table 1). Although some genes such as LPP, CSF2RA, PDLIM4, and RGS3 passed the defined criteria in 1 dataset, they did not reach a significant level in another one (Table 1). Hence, they were excluded for subsequent analysis.
Finally selected 7 prognostic genes were used to reconstruct the prognostic gene signature as described in the method section. The patients in the training datasets were categorized into 2 groups based on this signature. As shown in Fig. 1, the survival time was significantly different between the low-risk and high-risk groups (P < 0.0001) in training datasets. In GSE10846, the rates of OS at 5 years in the low-risk and high-risk groups were 75% and 43%, respectively. Likewise, in GSE31312, the rates of OS at 5 years in the low-risk and high-risk groups were 75% and 48%, correspondingly. These value for low-risk and high-risk groups in GSE32918&69051 were 63% and 43%, respectively. The hazard ratio was significantly lower in the low-risk group than in the high-risk group in GSE10846 (HR = 0.39 [0.27–0.54]), GSE31312 (HR = 0.46 [0.33–0.63]) as well as in GSE32918&69051 (HR = 0.51 [0.35–0.75]) (Ps < 0.0001) (Table 2).
Further analysis revealed that our developed prognostic signature was independent of routine IPI components in both training datasets (GSE10846: HR = 0.39 [0.26–0.59], GSE31312: (HR = 0.49 [0.34–0.72]) (Ps < 0.0001). Our outcome predictor was the most powerful prognostic variable in the multivariate Cox proportional hazards analysis (Table 3). Among the various components of the IPI, only age was able to predict the outcome in both datasets (Ps < 0.01) (Table 3).
External validation of the prognostic gene signature
Next, the outcome predictor was checked to determine whether it could externally predict the outcome in the patients with DLBCL. Our results indicated that the developed signature was significantly associated with the clinical outcome of DLBCL in all the validation datasets, containing 417 patients, at a P value less than 0.0001 (Fig. 1). In GSE34171, our signature stratified the patients with distinct outcomes—with corresponding 5-year OS rates of 94% and 53% in the low-risk and high-risk groups, respectively. Additionally, in GSE4475, our signature divided the patients into 2 distinct outcomes—with corresponding 5-year OS rates of 60% and 20% in the low-risk and high-risk groups, respectively. In GSE11318, the rates of OS at 5 years in the low-risk and high-risk groups were 60% and 35%, correspondingly (Fig. 1). The hazard ratios for GSE4475, GSE11318, and GSE34171 were 0.32 (0.19–0.54), 0.51 (0.35–0.76), and 0.10 (0.02–0.45), respectively (Ps ≤ 0.001) (Table 2).
Final prognostic signature and subtype of diffuse large-B-cell lymphoma
Our findings revealed that the survival time was significantly different between the 2 risk groups, constituted based on our signature, when applied in all the molecular subtypes of DLBCL—namely ABC-like, GCB-like, and type 3 (Ps ≤ 0.001) (Fig. 2). Hence, this outcome predictor was able to subdivide the patients within each subgroup into distinct risk groups.
Our results also showed that the expressions of CALD1, ITPKB, PLAU, and RTN1 were significantly diminished in the subtype with inferior survival (ie, ABC-like) compared with the subtype with better survival (ie, GCB-like) in both datasets (ie, GSE31312 and GSE10846) (Ps < 0.05) (Fig. 3). In GSE31312, the expression of GPNMB was significantly lower in the ABC-like subtype than in the GCB-like subtype (Ps < 0.05) (Fig. 3).
Extraction of a revised prognostic gene signature from the final prognostic gene signature
The final signature was revised after the validation step. The goal of this step was to minimize the number of the genes to obtain a more practical signature which could be technically simple and applicable for routine clinical use. Our analysis showed that a combination of 3 genes—namely APOC1, RTN1, and PLAU—was able to divide the patients into high-risk and low-risk groups with distinct survival times in both training and validation datasets (Ps < 0.0001) (Fig. 4). The rates of OS at 5 years in the low-risk and high-risk groups for all datasets were approximately similar to ones in final prognostic signature (Fig. 4). Furthermore, the hazard ratios were significantly higher in the high-risk group than in the low-risk group (Ps ≤ 0.001) (Table 4). The hazard ratio of the revised prognostic signature was slightly higher than that of the final prognostic signature (Tables 2 and 4).
Similar to the final prognostic gene signature, the revised prognostic signature was also independent of the IPI factors (Ps ≤ 0.001). This revised signature was by far the most powerful independent prognostic factor only in GSE31312 (HR = 0.47 [0.33–0.67]), but not in GSE10846 (HR = 0.61 [0.42–0.89]) (Table 5). In GSE10846, the hazard ratio of the revised signature was higher than that of the final signature in multivatiate analysis (0.61 vs. 0.39) (Tables 3 and 5).
Evaluation of the prognostic genes of 3 previously published signatures in GSE10846 and GSE31312
As shown in Supplementary Table 2, except for PLAU and ITPKB, the majority of the other genes in the prognostic signatures proposed by Lossos et al.2, Rosenwald et al.4, and Wright et al.6 had no consistent associations with the survival time in the multivariate Cox analysis, where these genes were associated with long survival in one dataset and with short survival in another. Additionally, in case of consistent associations, the association was principally not significant in both datasets (Ps > 0.05) or it was significant in only 1 dataset (mainly GSE31312) (Supplementary Table 2).
Discussion
In the present study, we sought to develop a gene-based prognostic predictor which could accurately predict the survival time in patients with DLBCL. Finally, we succeeded in constructing a 7-gene prognostic signature which robustly and reliably predicted the clinical outcome in our training and validation groups. As presented above, although the previously published prognostic signatures for patients with DLBCL can predict survival in their corresponding studied patients, they fail to predict the outcome in external groups of patients. Hence, we presumed that reconstruction of a prognostic signature from the genes commonly associated with the survival time in different groups of patients might resolve this problem. When mining the literature, we found that in studies with workflow similar to that in our investigation, an FDR below 10% or 15% was reasonable for the selection of significant genes. We mostly selected the common genes among genes significantly associated with survival with an approximate FDR of 0 (Supplementary Table 1), which means that the probability of a false positive was approximately 0.
We used the gene signatures of APOC1, CALD1, CD84, GPNMB, ITPKB, PLAU, and RTN1 to reconstruct the final outcome predictor. Among them, APOC1, GPNMB, and PLAU were previously defined as members of the stromal-1 signature in a 108-gene model comprising 3 gene-expression signatures termed “germinal-center B-cell”, “stromal-1”, and “stromal-2” developed by Lenz et al.8. In addition, ITPKB and PLAU appeared in the outcome gene signatures of DLBCL proposed by Wright et al.6 and Rosenwald et al.4, respectively. Chiming in with our findings, these genes were associated with a long survival time in all these studies.
APOC1 as an inflammation-related gene was found to be positively associated with the survival time in patients with DLBCL7. In addition, in breast cancer cells, this gene was regarded as an important tumor suppressor and cell proliferation inhibitor8. In contrast, it was reported that this gene was highly expressed in late-stage lung cancer9. Several studies have confirmed the potential role of ITPKB as an ideal tumor aggressiveness biomarker or favorable prognosis factor in DLBCL6,10,11. ITPKB (inositol-trisphosphate [IP3] 3-kinase B) was recently characterized as a critical tumor suppressor gene whose deficiency prompted DLBCL. Furthermore, ITPKB-activating agents can have curative potential10. This gene was among the gene cocktail used for the accurate categorization of DLBCL samples into ABC-like and GCB-like subtypes via a nuclease protection assay11. GPNMB (glycoprotein non-metastatic melanoma protein B) is highly expressed in different tumor cell types including glioma cells12, bone metastatic breast cancer cells13,14, low-metastatic melanoma cell lines15, and melanoma cells16. GPNMB was considered an important tumor suppressor in DLBCL17 and was reported to be differentially expressed in mantle cell lymphoma (MCL)18. PLAU (Plasminogen Activator, Urokinase) and CALD1 (Caldesmon 1)—accompanied by DCN, SPARC, FN1, MMPs, and PDGFRs—are members of genes related to extracellular matrix remodeling19. Concurrent overexpression of MMPs and PLAU was associated with favorable prognosis in patients with DLBCL4,7. Additionally, overexpression of PLAU and CALD1 was demonstrated in classical Hodgkin lymphoma tissues19. In contrast, high levels of MMPs and PLAU were associated with tumor invasion in some human solid tumors20,21. In our study, we found that RTN1 was a favorable prognostic gene in both final and revised signatures. A previous study confirmed upregulation of RTN1 in CXCR4− DLBCL versus CXCR4+ DLBCL and reported that CXCR4− and CXCR4+ subgroups were associated with a better and poorer survival time, respectively22.
Although we did not include c5orf30, LPP, CSF2RA, PDLIM4, and RGS3 in our final gene signature, they can be considered single prognostic genes. Two of these genes—namely CSF2RA and PDLIM4—were previously determined as members of the stroma-1 signature, developed to predict the outcome of patients with DLBCL7.
In the current study, we developed a potential reproducible prognostic gene signature which was able to robustly discriminate low-risk patients with DLBCL from high-risk ones. In addition, we reconstructed a 3-gene signature from the final prognostic signature. Although the revised signature was not as powerful as the final signature, it was able to efficiently predict the outcome in both training and validation groups and was considered an independent prognostic parameter. Not only can these signatures be drawn upon in clinical approaches in tandem with other routine prognostic factors, but also they can be deemed molecular targets with a critical role in the biology of cancer.
Methods
A schematic diagram depicting the analysis pipeline in our study is presented in Fig. 5.
Training and validation datasets
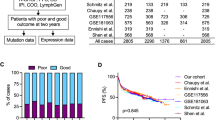
The Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database was searched to find the gene expression profiling datasets of patients with DLBCL. Only datasets containing clinical metadata (especially the survival time) (11 datasets) were retained, and the rest was excluded. Additionally, every effort was made to select expression datasets from all types of microarray chips such as Affymetrix and Illumina, if possible. The datasets were downloaded in the SOFT file format and were subsequently transformed logarithmically using tools provided in the geWorkbench 2.5.1 package23, if necessary. We employed various strategies to integrate different datasets used in our study. First, most of our datasets were generated using Affymetrix chip and GPL570 platform (Table 6). Hence, gene expression data were generated using similar approaches in these datasets. Furthermore, we only analyzed genes, which are existed in all chips and platforms. As another step, we normalize expression data using MAS 5 algorithm in all datasets. Hence, if a dataset were originally normalized using another method, we downloaded that dataset in raw format and then normalized it using MAS 5 method. More details on the clinical characteristics of the studied datasets are provided in Table 6. Some datasets with clinical metadata such as GSE57611, GSE23501, GSE93984, and GSE21846 were deleted for a specific reason (Table 6). The datasets were randomly divided into training (n = 1219) and validation (n = 417) datasets. In brief, GSE10846 (n = 420), GSE31312 (n = 470), GSE32918 (n = 172), and GSE69051 (n = 157) were used as training datasets, while GSE4475 (n = 123), GSE11318 (n = 203), and GSE34171 (n = 91) were utilized as validation datasets. Since GSE32918 and GSE69051 have originated from a similar research study24 and had some common samples, they were merged as a single dataset and named as GSE32918&69051. Number of samples for these datasets was determined after correction based on the common samples (172 samples for GSE32918 and 157 samples for GSE69051).
Identification of the common genes associated with survival in the training datasets
The association between gene expression and OS was examined using the univariate Cox proportional hazards analysis. In this analysis, the association between a group of covariates (genes) and the response variable (the survival time) was evaluated. The univariate Cox analysis was performed using the BRB-Array tools developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. In this analysis, the findings were strengthened by employing a strict pipeline and retaining only genes with a P value less than 0.001 and a false discovery rate (FDR) less than 5%. Then, the common genes which were significantly associated with OS between the training datasets were extracted. For this purpose, only common genes with consistent associations were selected, while genes with inconsistent associations (negatively associated with OS in a dataset and positively associated with OS in another) were excluded. We also considered therapeutic regimens in the datasets used in our survival analysis. Hence, in each dataset, only common genes associated with the survival between patients with distinct treatments were selected for subsequent analysis. Additionally, for the confirmation of whether these genes were commonly associated with OS in all the training datasets, a class prediction analysis was also performed using 2 algorithms—namely support vector machine (SVM) and diagonal linear discriminant analysis (DLDA). In this analysis, 2 classes (ie, long survival [≥5 y] and short survival [<5 y]) were defined and thereafter classifiers, which could predict the 2 classes, were identified using 2 class prediction algorithms (ie, SVM and DLDA). The class prediction analysis was performed using the methods incorporated in BRB-Array tools.
Reconstruction of the prognostic gene signature
The prognostic signature was developed as described previously2,25,26. In brief, the prognostic signature was reconstructed as a linear combination of the expression levels of the common genes and the z-score in the multivariate Cox regression analysis. Hence, at the first step, a multivariate Cox proportional-hazards regression analysis was performed for each gene, where all the individual components of the IPI (ie, age, stage, lactate dehydrogenase level, Eastern Cooperative Oncology Group [ECOG] performance status, and number of extranodal sites)27 and gene expression were entered as covariate variables. Additionally, sex and molecular subtype (ie, ABC-like, GCB-like, and type 3) were entered as another 2 variables into the multivariate analysis. The multivariate analysis was solely performed on the datasets with the clinical IPI data (ie, the GSE10846 and GSE31312). Afterward, the log-transformed normalized expression value of each gene was multiplied by the z-score. Finally, the prediction score was calculated for each patient as described in the following equation:
Subsequently, the patients were first ranked based on their prediction scores before they were classified into 2 groups (>median value and <median value) and the survival times were compared between the groups using the Kaplan–Meier analysis and log-rank test at a P value less than 0.01. The survival analyses were performed using Survival (http://cran.r-project.org/package=survival) and SPSS 16.0 (Chicago, USA) packages.
Evaluation of the prognostic gene signature in the validation datasets
The prognostic efficacy of the final developed gene signature was assessed externally in 417 patients as 3 validation datasets (GSE4475, GSE11318, and GSE34171). A workflow similar to the training datasets was performed. Similarly, the predictor score was calculated in the validation samples based on the details provided above. Subsequently, 2 groups were constituted after ranking patients based on their predictor score and then the survival time was compared between the groups using the Kaplan–Meier analysis and log-rank test at a P value less than 0.01.
Prognostic signature and subtype of diffuse large-B-cell lymphoma
Whether the survival time was significantly different between the groups constituted based on our signature in each molecular subtype of DLBCL (ie, ABC-like, GCB-like, and type 3) was also investigated using the Kaplan–Meier analysis. Additionally, the expressions of the members of the outcome predictor were compared between these subgroups using the one-way ANOVA test at a P value less than 0.05.
Extraction of a revised prognostic signature from the final prognostic signature
The goal of this step was to minimize the number of the genes to obtain a more practical signature which could be technically simple and applicable for routine clinical practice. Efforts were made to find a signature with a minimal number of genes, which could predict the patients’ clinical outcome with a statistical power similar to that of the final prognostic signature. To that end, in each round, 1 gene was deleted from the final signature and then the prediction ability of the remaining genes was tested using the Kaplan–Meier analysis and the log-rank test. A gene was considered a critical (hub) gene when its absence significantly reduced the prediction ability of the outcome predictor. Finally, critical (hub) genes were used to reconstruct a revised prognostic signature with the method applied for the final signature.
Association between the prognostic genes in the signatures of Lossos et al. (2004), Rosenwald et al. (2002), and Wright et al. (2003) and overall survival in the GSE31312 and GSE10846 datasets
The ability of the prognostic genes in the previously published outcome predictors in the estimation of survival as well as the consistency of their associations with survival in the 2 big training datasets was evaluated by determining the association between the prognostic genes in the signatures of Lossos et al.2 (n = 6), Rosenwald et al.4 (n = 17), and Wright et al.6 (n = 14) and the OS time using the multivariate Cox proportional-hazards regression analysis in GSE31312 and GSE10846, as described above. Again, the IPI components and each gene were used as predictor variables and OS as the response variable.
Ethical standards
Our study was performed using datasets deposited in GEO database. Hence, no ethical approval was required.
Data Availability
The datasets in the manuscript were deposited in GEO database (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE10856, GSE31312, GSE69051, GSE32918, GSE4475, GSE11318, and GSE34171. Other supporting data are included as Supplementary Files.
References
Lenz, G. & Staudt, L. M. Aggressive lymphomas. The New England Journal of Medicine 362, 1417–1429, https://doi.org/10.1056/NEJMra0807082 (2010).
Lossos, I. S. et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. The New England Journal of Medicine 350, 1828–1837, https://doi.org/10.1056/NEJMoa032520 (2004).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511, https://doi.org/10.1038/35000501 (2000).
Rosenwald, A. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England Journal of Medicine 346, 1937–1947, https://doi.org/10.1056/NEJMoa012914 (2002).
Shipp, M. A. et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nature Medicine 8, 68–74, https://doi.org/10.1038/nm0102-68 (2002).
Wright, G. et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America 100, 9991–9996, https://doi.org/10.1073/pnas.1732008100 (2003).
Lenz, G. et al. Stromal gene signatures in large-B-cell lymphomas. The New England Journal of Medicine 359, 2313–2323, https://doi.org/10.1056/NEJMoa0802885 (2008).
Sun, Y. et al. Identification of Apolipoprotein C-I Peptides as a Potential Biomarker and its Biological Roles in Breast Cancer. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research 22, 1152–1160, https://doi.org/10.12659/msm.896531 (2016).
Ko, H.-L. et al. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for lung cancer: A marker phase I trial. Thoracic Cancer 5, 500–508, https://doi.org/10.1111/1759-7714.12117 (2014).
Sauer, K. et al. IP3 3-Kinase B Suppresses B Cell Lymphoma by Antagonizing PI3K/mTOR in B cells. The Journal of Immunology 196(142), 2–142.2 (2016).
Rimsza, L. M. et al. Accurate classification of diffuse large B-cell lymphoma into germinal center and activated B-cell subtypes using a nuclease protection assay on formalin-fixed, paraffin-embedded tissues. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research 17, 3727–3732, https://doi.org/10.1158/1078-0432.ccr-10-2573 (2011).
Kuan, C.-T. et al. Glycoprotein nonmetastatic melanoma protein B, a potential molecular therapeutic target in patients with glioblastoma multiforme. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research 12, 1970–1982, https://doi.org/10.1158/1078-0432.ccr-05-2797 (2006).
Rose, A. A. N. et al. Osteoactivin promotes breast cancer metastasis to bone. Molecular cancer research: MCR 5, 1001–1014, https://doi.org/10.1158/1541-7786.mcr-07-0119 (2007).
Rose, A. A. N. & Siegel, P. M. Osteoactivin/HGFIN: is it a tumor suppressor or mediator of metastasis in breast cancer? Breast cancer research: BCR 9, 403, https://doi.org/10.1186/bcr1791 (2007).
Weterman, M. A. et al. nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. International Journal of Cancer 60, 73–81 (1995).
Tse, K. F. et al. CR011, a fully human monoclonal antibody-auristatin E conjugate, for the treatment of melanoma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research 12, 1373–1382, https://doi.org/10.1158/1078-0432.ccr-05-2018 (2006).
Mahadevan, D. et al. Transcript profiling in peripheral T-cell lymphoma, not otherwise specified, and diffuse large B-cell lymphoma identifies distinct tumor profile signatures. Molecular Cancer Therapeutics 4, 1867–1879, https://doi.org/10.1158/1535-7163.mct-05-0146 (2005).
Henson, S. E., Morford, T., Stein, M.-P., Wall, R. & Malone, C. S. Candidate genes contributing to the aggressive phenotype of mantle cell lymphoma. Acta Histochemica 113, 729–742, https://doi.org/10.1016/j.acthis.2010.11.001 (2011).
Chetaille, B. et al. Molecular profiling of classical Hodgkin lymphoma tissues uncovers variations in the tumor microenvironment and correlations with EBV infection and outcome. Blood 113, 2765–3775, https://doi.org/10.1182/blood-2008-07-168096 (2009).
Huang, C.-Y. et al. Urokinase-type plasminogen activator resulting from endometrial carcinogenesis enhances tumor invasion and correlates with poor outcome of endometrial carcinoma patients. Scientific Reports 5, 10680, https://doi.org/10.1038/srep10680 (2015).
McKee, C. M. et al. Protease nexin 1 inhibits hedgehog signaling in prostate adenocarcinoma. The Journal of Clinical Investigation 122, 4025–4036, https://doi.org/10.1172/jci59348 (2012).
Chen, J. et al. Dysregulated CXCR4 expression promotes lymphoma cell survival and independently predicts disease progression in germinal center B-cell-like diffuse large B-cell lymphoma. Oncotarget 6, 5597–5614, https://doi.org/10.18632/oncotarget.3343 (2015).
Floratos, A., Smith, K., Ji, Z., Watkinson, J. & Califano, A. geWorkbench: an open source platform for integrative genomics. Bioinformatics 26, 1779–1780, https://doi.org/10.1093/bioinformatics/btq282 (2010).
Barrans, S. L. et al. Whole genome expression profiling based on paraffin embedded tissue can be used to classify diffuse large B-cell lymphoma and predict clinical outcome. British Journal of Haematology 159, 441–453, https://doi.org/10.1111/bjh.12045 (2012).
Zamani-Ahmadmahmudi, M., Dabiri, S. & Nadimi, N. Identification of pathway-based prognostic gene signatures in patients with multiple myeloma. Translational Research: The Journal of Laboratory and Clinical Medicine 185, 47–57, https://doi.org/10.1016/j.trsl.2017.05.001 (2017).
Sun, J., et al. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Scientific Reports 6, https://doi.org/10.1038/srep27842 (2016).
Sehn, L. H. & Gascoyne, R. D. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood 125, 22–32, https://doi.org/10.1182/blood-2014-05-577189 (2015).
Acknowledgements
Authors wish to thank Mr. Pedram Amouzadeh who assisted in the proof-reading of the manuscript.
Author information
Authors and Affiliations
Contributions
M.Z.A. and S.M.N. participated in the study design and analysis of the data. M.Z.A. and S.M.A. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zamani-Ahmadmahmudi, M., Nassiri, S.M. Development of a Reproducible Prognostic Gene Signature to Predict the Clinical Outcome in Patients with Diffuse Large B-Cell Lymphoma. Sci Rep 9, 12198 (2019). https://doi.org/10.1038/s41598-019-48721-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48721-0
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.