Abstract
Molecular diagnostic methods are becoming increasingly available for assessment of acute lower respiratory illnesses (ALRI). However, nasopharyngeal/oropharyngeal (NP/OP) swabs may not accurately reflect etiologic agents from the lower respiratory tract where sputum specimens are considered as a more representative sample. The pathogen yields from NP/OP against sputum specimens have not been extensively explored, especially in tropical countries. We compared pathogen yields from NP/OP swabs and sputum specimens from patients ≥18 years hospitalized with ALRI in rural Western Kenya. Specimens were tested for 30 pathogens using TaqMan Array Cards (TAC) and results compared using McNemar’s test. The agreement for pathogen detection between NP/OP and sputum specimens ranged between 85–100%. More viruses were detected from NP/OP specimens whereas Klebsiella pneumoniae and Mycobacterium tuberculosis were more common in sputum specimens. There was no clear advantage in using sputum over NP/OP specimens to detect pathogens of ALRI in adults using TAC in the context of this tropical setting.
Similar content being viewed by others
Introduction
Acute lower respiratory illnesses (ALRI) are associated with substantial morbidity and mortality in sub-Saharan Africa1. Understanding etiology of respiratory illnesses in resource-limited countries may support clinicians with patient management and provide a framework for decision makers when prioritizing public health interventions. Recently, with availability of multi-pathogen molecular diagnostic methods, and increasing recognition of viruses in cases of ALRI, the use of nasopharyngeal (NP) and oropharyngeal (OP) swab specimens to identify pathogens associated with ALRI has increased2,3,4. However, NP/OP swabs may not accurately reflect etiologic agents from the lower respiratory tract5. Sputum specimens, on the other hand, have been considered a more representative sample of the lower respiratory tract5, but have not been extensively explored, especially in tropical countries.
A multisite study in the United States compared pathogen yield between NP/OP and sputum specimens and argued for the use of high-quality sputum specimens for greater pathogen yield6. In contrast, another multisite study in low-middle income countries, observed that the pathogen yield from NP/OP and sputum specimens were similar2. Given the paucity of data and the difference in observations in these two recent studies, and that most data from tropical countries were derived from paediatric populations, we aimed to investigate adults living in a tropical, low-income setting. Here we describe the performance of NP/OP swabs against sputum specimens for detecting respiratory pathogens among adult patients hospitalized with ALRI using the TaqMan Array Card (TAC).
Methods
Study population and design
Since August 2009, the Kenya Medical Research Institute (KEMRI), in partnership with the U.S Centers for Disease Control and Prevention (CDC), has carried out surveillance for ALRI (defined as cough or difficulty breathing with an onset within the last 14 days from hospital admission date) among patients aged ≥ 18 years, at Siaya County Referral Hospital, in Western Kenya. The study population is culturally homogeneous and the area is known for high malaria, tuberculosis and HIV prevalence7,8,9. A structured questionnaire was programmed using Microsoft Visual Studio.net (Microsoft, Redmond, USA) and installed into netbooks (Mercer classmate) for collection of demographic and clinical examination data from patients by surveillance officers i.e. trained clinical officers and nurses. The data were stored in password protected Microsoft SQL databases (Microsoft, Redmond, USA). All patients were managed and discharged according to the Kenya Ministry of Health standards and guidelines.
Specimen collection and processing
NP and OP specimens were collected from consenting patients meeting the ALRI case definition using flexible mini-tip flocked swabs, combined into a single vial containing viral transport media, and transported to the laboratory for testing as previously described7. Sputum specimens were collected through expectoration (quality sputum defined as <10 squamous epithelial cells per low-power field) in clean 3 ml tubes and transported at 2–8 °C to KEMRI/CDC laboratories in Kisumu where they were stored at −80 °C until testing. Nucleic acids were extracted from the sputum specimens, using a modified MagMaxTM (Applied Biosystems, Foster City, CA) viral RNA extraction procedure to include dithriothreital (DTT) treatment stage to break down sputum. Briefly, 100 μl of 500 mM DTT solution was added to 100 μl of sputum, vortexed and incubated at room temperature for 30 minutes. The MagMaxTM viral RNA extraction procedure was performed on 200 μl material following the manufacturer’s instructions. The extracted total nucleic acids from NP/OP and sputum were tested using TAC as described by Kodani et al. and Weinberg et al.4,10. Each target of the 30 pathogens was considered positive if it passed the internal positive control and had exponential fluorescence curves crossing the assigned threshold at a threshold cycle (CT) value of <35. For targets with two wells, the target was considered positive when either of the two wells was positive. Results with CT > 30 for Legionella spp. were disregarded due to probe degradation of the specific chemistry used in preparation of this TAC.
Data analysis
We described demographic characteristics and laboratory outcomes of patients using proportions, means, and medians. The paired specimen results were compared between NP/OP and sputum specimens using the McNemar’s test and further stratified by HIV status, malaria infection, clinical severity (≤ vs. >7 days hospitalized) and duration of illness (< vs. ≥5 days from onset to admission) which are provided as supplementary tables. Percent agreement between NP/OP and sputum specimens were calculated as the total number of times the NP/OP and sputum specimens agreed for each TAC target divided by the total number of patients tested. Tests with p-values <0.05 were considered statistically significant. The CT values for each target were summarized using box-and-whiskers plots. Analysis was performed using SAS (version 9.2, Cary, NC).
Ethical considerations
This study was reviewed and approved by the Kenya Medical Research Institute’s Scientific Ethics Review Unit (SSC no. 2558) and US Centers for Disease Control and Prevention’s Institutional Review Board (CDC IRB no. 6543). Written informed consents were obtained from all participants and all research was performed in accordance with the relevant guidelines/regulations.
Disclaimer
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention (CDC) or the Kenya Medical Research Institute (KEMRI).
Results
From March 2014 through July 2015, we tested 294 pairs of NP/OP swabs and sputum specimens using TAC. On average, NP/OP swabs and sputum specimens were collected 29 (Standard Deviation [SD] = 14) and 38 (SD = 31) hours after admission, respectively.
Most patients (198/294; 67%) were between ages 18–49 years, and 81/294 (28%) were male. Fever was either self-reported or measured (≥38 °C) in 40% of the patients, and malaria was diagnosed in 19% of the patients. More than half the patients (179/294; 61%) had underlying chronic illness; 39% and 22% for HIV and other chronic conditions respectively. Of the 294 patients, 116 (39%) sought care in <5 days from respiratory illness onset, 99 (34%) sought care 5–10 days and 79 (27%) sought care after 10 days. Of all cases, 88 (30%) were hospitalized for >7 days and 24 (8%) died in hospital (Table 1). Of the 24 who died, 9 (38%) were HIV-positive, 3 (13%) had malaria and 3 (13%) had other chronic illnesses.
Of the 294 pairs of specimens tested, 248 bacterial and 136 viral pathogens were detected from 141 and 115 NP/OP specimens respectively whereas 213 bacterial and 47 viral pathogens were detected from 141 and 45 sputum specimens respectively. NP/OP specimens were more likely to test positive for any viral pathogen compared to sputum specimens, p < 0.01. There was no difference in bacterial pathogen detection in NP/OP vs. sputum specimens, and the pathogen specific agreements ranged between 85–100% (Table 2).
Among the bacterial pathogens detected, Streptococcus pneumoniae and Moraxella catarrhalis were more significantly present in NP/OP specimens compared to sputum specimens. However, Klebsiella pneumoniae and Mycobacterium tuberculosis were more significantly present in the sputum specimen compared to the NP/OP specimen. Among the viral pathogens, Respiratory Syncytial Virus (RSV), Rhinovirus, and Human Coronavirus 2 (NL63) were commonly detected in NP/OP than sputum specimens (Table 2). When stratified by HIV status, malaria infection, prolonged hospitalization and duration from onset to admission, there was not much difference in pathogen yield. Although, Mycobacterium tuberculosis (detected in sputum) was associated with HIV positive, malaria negative, and severe cases, and with patients admitted ≥5 days from illness onset. Also, patients with malaria were more likely to test positive for Pseudomonas aeruginosa from sputum compared to NP/OP specimens, p = 0.02 (Supplementary Tables S1–S4). Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis were the most co-detected bacterial pathogens and rhinovirus was the most co-detected viral pathogen (Supplementary Tables S5 and S6).
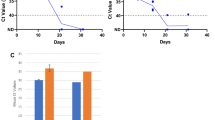
The median CT values shown in box-and-whiskers plot were comparable for NP/OP and sputum specimens for all bacterial targets (Fig. 1). This was also observed for most viral pathogens, except that a higher concentration of viruses (corresponding to lower CT values) was observed for influenza B, RSV and human metapneumovirus in the NP/OP specimens compared to sputum specimens. The median CT values were similar in the NP/OP and sputum specimens for the RNP target.
Discussion
We found good agreement (85–100%) in the detection of respiratory pathogens between NP/OP swabs and sputum specimens when using TAC. There was no overall benefit of using sputum over NP/OP swab specimens for etiologic assessment of respiratory illness, except that we found improved detection of Mycobacterium tuberculosis and Klebsiella pneumoniae from sputum specimens compared to NP/OP. When considering viral pathogens, rhinovirus, RSV, and coronavirus (HCoV NL63) were more often identified in NP/OP swabs. The use of molecular tests in NP/OP and sputum specimens warrants further investigation as a tool to identify etiology of ALRI in hospitalized adults.
Although previous pneumonia related studies have observed more bacteria yield from sputum11,12, arguing that this would be the ideal specimen for identification of lower respiratory tract infection etiology. We did not observe substantial differences in pathogen detection from NP/OP vs. sputum specimens. Mycobacterium tuberculosis, was mostly detected in sputum specimens, however, one case was detected in the NP/OP swab and not found in the sputum specimen. Though others suggested that this could happen in cases of extra-pulmonary tuberculosis13, we found that this patient was positive by GeneXpert (data not shown) which may suggest some shortfalls of testing for Mycobacterium tuberculosis in sputum using TAC.
Jeong et al. argued that as viruses play an important role in lower respiratory tract infections, sputum would be the best choice for etiologic diagnosis of virus-associated respiratory illness. The authors looked at adults who were hospitalized with severe acute respiratory illness, although the difference in yield from the two types of specimens was not impressive (15% increase in viral detection using sputum). Moreover, they only collected NP swabs which could have led to less representation of respiratory viruses with tropism to the oropharynx14. Wolff et al. also observed the detection of more viruses in sputum, where CT values were earlier for almost all pathogens compared to that found in NP/OP swabs6, leading the authors to argue for the benefit of sputum use in diagnosis of lower respiratory tract infections. However this position was not supported in the recent work done by Thea et al.2, where they looked at children <5 years hospitalized with ALRI and pneumonia, or by our study done among adults. Perhaps in tropical countries, the likelihood of multi-pathogen detection may be higher than that seen in temperate countries, for children and adults alike, which may merit further investigation.
Both Wolff et al. and Thea et al. used high quality induced-sputum specimens in their studies but acknowledged that contamination would still be possible2,6. Furthermore, Thea et al. noted that the yield of pathogens from high quality induced sputum was similar to that of low quality induced sputum suggesting that specimen collected from the upper respiratory tract (i.e., NP/OP) could be used when investigating ALRI. The biggest challenge in these studies, including ours, is the interpretation of multi-pathogen detection, particularly when trying to attribute a specific pathogen as the cause of an episode of ALRI. Many bacterial pathogens detected from respiratory specimens could reflect carriage, being part of the normal microbiome, and not necessarily associated with the disease process. Similarly, viral pathogens detected in respiratory samples could indicate recent past infections. The early CT values from the upper tract could indicate higher pathogen load and be associated with pneumonia15, which would suggest that NP/OP swabs could be used when investigating lower ALRI etiology if interpretation is combined with CT values. However, establishing causality can be challenging even when considering organism density (here we used CT values as proxy). The use of radiographic images, epidemiologic data, clinical course of illness, and the possibility to investigate lower respiratory tract specimens collected aseptically could be important tools to add to the investigation of ALRI/pneumonia.
A main limitation in our study is that direct comparison of our findings with those from the literature is complicated due to varying case definitions, severity of cases investigated, age of patients, study settings (temperate vs. tropical, less vs. more resourced countries), and laboratory tests used. Secondly, we did not use induced sputum collection as we worked with a population of adult patients, most of whom were accustomed to providing sputum specimen for tuberculosis testing. Moreover, as used by Thea et al.2, we considered <10 squamous epithelial cells per low-power field to determine specimen quality and observed that RNP CT values were similar for NP/OP and sputum specimens, giving us confidence that results from sputum specimens were credible.
In conclusion, our findings suggest that there is no clear advantage in using sputum specimens over NP/OP swabs to investigate the etiology of ALRI using molecular diagnostics. Interpretation of multi-pathogen molecular laboratory tests, however, needs further investigation, especially in tropical settings where co-detections seem very common.
Data Availability
All the data are included in this manuscript through tables, figures and supplementary tables but should additional data be required, requests can be made to the director KEMRI through the corresponding author.
References
Lagare, A., Maïnassara, H. B., Issaka, B., Sidiki, A. & Tempia, S. Viral and bacterial etiology of severe acute respiratory illness among children <5 years of age without influenza in Niger. BMC Infect. Dis. 15, 1–7 (2015).
Thea, D. M. et al. Limited utility of polymerase chain reaction in induced sputum specimens for determining the causes of childhood pneumonia in resource-poor settings: Findings from the pneumonia etiology research for child health (PERCH) study. Clin. Infect. Dis. 64, S289–S300 (2017).
Jain, S. et al. Community-Acquired Pneumonia Requiring Hospitalization among USAdults. N. Engl. J. Med. 373, 415–427 (2015).
Weinberg, G. A. et al. Field evaluation of TaqMan Array Card (TAC) for the simultaneous detection of multiple respiratory viruses in children with acute respiratory infection. J. Clin. Virol. 57, 254–260 (2013).
Falsey, A. R., Formica, M. A. & Walsha, E. E. Yield of sputum for viral detection by reverse transcriptase PCR in adults hospitalized with respiratory illness. J. Clin. Microbiol. 50, 21–24 (2012).
Wolff, B. J. et al. Improved detection of respiratory pathogens by use of high-quality sputum with TaqMan array card technology. J. Clin. Microbiol. 55, 110–121 (2017).
Nyawanda, B. O. et al. Evaluation of case definitions to detect respiratory syncytial virus infection in hospitalized children below 5 years in Rural Western Kenya, 2009–2013. BMC Infect. Dis. 16, 1–10 (2016).
Odhiambo, F. O. et al. Profile: The KEMRI/CDC health and demographic surveillance system-Western Kenya. Int. J. Epidemiol. 41, 977–987 (2012).
Hamel, M. J. et al. A reversal in reductions of child mortality in Western Kenya, 2003–2009. Am. J. Trop. Med. Hyg. 85, 597–605 (2011).
Kodani, M. et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J. Clin. Microbiol. 49, 2175–2182 (2011).
Hammitt, L. L. et al. A preliminary study of pneumonia etiology among hospitalized children in Kenya. Clin. Infect. Dis. 54 (2012).
Zar, H. J. et al. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: A nested case-control study of the Drakenstein Child Health Study. Lancet Respir. Med. 4, 463–472 (2016).
Mishra, R. K., Prasad, B. K. & Mathew, S. Nasopharyngeal tuberculosis. Med J Armed Forces India 71, S586–9 (2015).
Jeong, J. H. et al. Comparison of sputum and nasopharyngeal swabs for detection of respiratory viruses. J Med Virol 86, 2122–7 (2014).
Feikin, D. R. et al. Is higher viral load in the upper respiratory tract associated with severe pneumonia? Findings from the PERCH study. Clin. Infect. Dis. 64, S337–S346 (2017).
Acknowledgements
We thank the KEMRI/CDC field, clinic and data room staff involved for their diligence in data collection and management and Benjamin Ochieng together with the laboratory staff for performing the tests. We also thank the Kenya Ministry of Health staff and the study surveillance officers who helped collect the data and samples used for this analysis and the study participants at Siaya County Referral Hospital. This manuscript is published with the permission of the Director of KEMRI. This work was supported by funding from the U.S. Centers for Disease Control and Prevention [Grant number 5U19GH000041-04].
Author information
Authors and Affiliations
Contributions
B.O.N., J.A.M., C.M., H.N.N. and S.S.C. conceived the study. All contributed to study design and implementation, B.O.N. and S.S.C. analysed and interpreted the data. B.O.N. drafted the manuscript under supervision of S.S.C., and all authors critically reviewed the manuscript for intellectual content and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nyawanda, B.O., Njuguna, H.N., Onyango, C.O. et al. Comparison of respiratory pathogen yields from Nasopharyngeal/Oropharyngeal swabs and sputum specimens collected from hospitalized adults in rural Western Kenya. Sci Rep 9, 11237 (2019). https://doi.org/10.1038/s41598-019-47713-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47713-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.