Abstract
Wildlife that exploit human-made habitats hosts and spreads bacterial pathogens. This shapes the epidemiology of infectious diseases and facilitates pathogen spill-over between wildlife and humans. This is a global problem, yet little is known about the dissemination potential of pathogen-infected animals. By combining molecular pathogen diagnosis with GPS tracking of pathogen-infected gulls, we show how this knowledge gap could be filled at regional scales. Specifically, we generated pathogen risk maps of Salmonella, Campylobacter and Chlamydia based on the spatial movements of pathogen-infected yellow-legged gulls (Larus michahellis) equipped with GPS recorders. Also, crossing this spatial information with habitat information, we identified critical habitats for the potential transmission of these bacteria in southern Europe. The use of human-made habitats by infected-gulls could potentially increase the potential risk of direct and indirect bidirectional transmission of pathogens between humans and wildlife. Our findings show that pathogen-infected wildlife equipped with GPS recorders can provide accurate information on the spatial spread risk for zoonotic bacteria. Integration of GPS-tracking with classical epidemiological approaches may help to improve zoonosis surveillance and control programs.
Similar content being viewed by others
Introduction
Wild animals host and spread pathogens, thereby shaping the epidemiology of infectious diseases1,2,3. This is particularly relevant in human-transformed landscapes, where opportunistic species reach high densities associated with the exploitation of anthropogenic food sources that could carry pathogenic bacteria4,5,6,7. This facilitates pathogen spill-over between wildlife and humans, both ways, and there are concerns that this may facilitate the evolution of new zoonotic pathogens6,8,9,10. Notably, urban gulls threaten public health because they shed bacterial pathogens, antibiotic-resistant bacteria, and viruses5,11,12,13. This has become a public health problem, yet little is known about how gulls spread zoonoses in space and time5,13,14. The lack of information on the dissemination process of zoonotic pathogens weakens risk assessments and management plans15. Specifically, spatially-explicit wildlife epidemiology is missing from existing zoonosis surveillance and control actions, such as the Zoonosis Directive of the European Union16 and the Foodborne Diseases Active Surveillance Network in the USA17.
We determined how this gap could be filled at a regional scale, by coupling conventional pathogen diagnosis in gulls with GPS-tracking of bird movements using miniature electronic tags attached to infected individuals. This allows the compilation of pathogen risk maps and the identification of critical habitats, as we show for yellow-legged gulls (Larus michahellis) in southern Spain. Due to its scavenger habits, this gull has been reported as a source and reservoir of zoonotic pathogens18,19. We GPS-tracked 14 birds that tested positive for one of three major zoonotic bacteria (five Salmonella-infected, five Campylobacter-infected and four Chlamydia-infected gulls). These bacteria are leading causes of zoonotic diseases in developed and developing countries17,20, and their incidence is increasing, even in countries with adequate public health systems. For example, Salmonella and Campylobacter cause the most common enteric zoonoses in the European Union, with 94,530 and 246,307 human clinical infections in 2016, respectively20. In the case of Chlamydia, this bacterium could affect the respiratory system of humans, wildlife and domestic animals21.
Results and Discussion
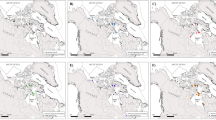
Cloacal swabs revealed that within the 19 GPS-tracked individuals, 37% (n = 5), 31% (n = 5) and 25% (n = 4) were positive for Salmonella, Campylobacter and Chlamydia, respectively, with no co-infections recorded. Previous studies found similar infection rates18,22. All movements of the infected-gulls were recorded throughout their estimated infection period [30 days23,24,25]. Pathogen risk maps and critical habitats were modeled by overlapping gull resting and foraging positions with accurate high-resolution land cover information26,27. The 27,798 recorded GPS locations revealed the greatest bacterial spread risk within 5 km of the breeding colony (Figs 1 and S1 in Supplementary Material), without significant differences in the type of habitat used between Salmonella-infected, Campylobacter-infected and Chlamydia-infected individuals (Pseudo-F = 0.78, p = 0.67). Risk spatial extent varied between infected-gulls (Fig. S1 in Supplementary Material), from areas close to the breeding colony to some infected-gull crossing-over from Spain to Portugal, stressing the importance of international health regulations and cooperation in disease control28.
(a) Study area showing the terrestrial GPS positions (red circles) of 14 GPS-tracked yellow-legged gulls during the 2015 breeding season. (b) Potential risk maps for Salmonella, Campylobacter, and Chlamydia together, based on the spatial distribution of pathogen-infected yellow-legged gulls. The white star indicates the position of the breeding colony.
Spread-risk areas overlapped with human-related habitats such as water ponds, fishing port or touristic beaches (Figs 2; S2 in Supplementary Material), increasing the risk of direct and indirect disease transmission to and from humans10,14 Notably, the use of water reservoirs (built for human use) by infected gulls is likely to lead to the contamination of drinking, recreational and irrigation water sources29. For this reason, it is important to ensure correct water treatment in this sensible habitats to reduce any potential risk to public health. Similarly, the extensive use of fishing ports and fish farms as feeding areas by yellow-legged gulls could point to serious infection risk for seafood30. Moreover, the use of beaches by infected-gulls (Fig. 2) exposes to pathogen spillover tens of thousands of tourists using these recreational habitats14. Moreover, the utilization of wetlands or estuaries by infected-gulls enhances the probability for pathogen transmission to other wildlife species31. Garbage dumps are also assumed to facilitate the infection of gulls by pathogens present in the human organic garbage, as well as cross-species and cross-individual transmission13,18. Yet, this habitat was seldom used by gulls in our study, due to its low availability in the area used by tracked-gulls (there are only two dumps in the area surrounding the breeding colony27). If garbage dumps are not the main pathogen source, bacterial infection of GPS-tracked gulls may be associated with the use of other food sources in decomposition, such as stranded marine animals (notably mammals) that could present pathogenic-bacteria, human organic refuse food found in recreational beaches or urban parks, or urban prey such as pigeons and rats32,33. Our results strongly indicate the need for integrated waste and pest control at a landscape scale.
Average habitat use of Salmonella-infected, Campylobacter-infected and Chlamydia-infected yellow-legged gulls GPS-tracked during the 2015 breeding season. Each pathogen is represented by a vertical bar, subdivided by the proportion of locations in each habitat (human-related or natural) in relation to all GPS positions. The picture shows a group of adult and juvenile yellow-legged gulls feeding on fish refuse at a fishing port close to the breeding colony. Photograph taken by Joan Navarro in a fishing port close to the breeding colony.
Overall, our study reveals that pathogen-infected gulls equipped with GPS recorders could provide accurate maps of zoonotic spread risk, from the local to regional and international scales. In some circumstances, this approach could be scaled up to build an international network, using gulls and other potential vectors of animal pathogens34, to achieve large scale zoonotic surveillance and to identify and implement prevention measures across potential sensitive habitats. Because this may trigger public concern, we recommend that these measures be coupled with environmental mediation work, to ensure that wildlife is not perceived as generally harmful to humans35.
Material and Methods
Fieldwork and tracking procedures
Fieldwork was carried out at the natural Biosphere reserve of Marismas de Odiel (37°13′N, 6°59′W; southwestern Iberian Peninsula; Fig. 1) in a colony of 250–300 breeding pairs of yellow-legged gulls. We deployed high-resolution GPS-trackers recording the positions of individuals at 5 minute intervals [Uva-Bits loggers36] on 19 breeding gulls more than 4-years of age during their breeding period (May 2015). Uva-BiTS loggers can recharge themselves using solar energy, allowing to track the movements of birds continuously during several years36. The age of each individual was determined from plumage characteristics. Incubating birds were caught at the nest using a walk-in wire mesh trap and GPS-trackers were attached using a wing harness fixed with a reef knot in the tracheal pit, an attachment method recommended for large gulls37. The GPS-tracker and harness weighed less than 1.8% of the body mass of the birds [16 g for the GPS and harness, mean ± standard deviation = 1072 ± 110 g for the tracked gulls]26. GPS data were automatically downloaded remotely from devices to a field-based laptop when the birds were present at the breeding colony, where a network of 3 antennas provided complete coverage of the breeding area36. GPS data was parsed into the central database and immediately available in the UvA-BiTS Virtual Lab (www.UvA-BiTS.nl) for visualization and data exploration, therefore providing tracking information in real time36. To avoid potential biases associated with differences in the number of GPS data between individuals, tracking data were analyzed only when all individuals were equipped. We focused our analyses on the 30 days following deployment (from 14-May to 15-June 2015) to cover the potential infection period of each tracked pathogen23,24,25.
All fieldwork was approved by the Ethics Committee of CSIC (Ref: 28-04-15-237), in accordance with the Spanish and EU legislation on the protection of animals used for scientific purposes.
Pathogen determination
Cloacal swabs from each GPS-tracked gull were collected and placed in PBS medium (Deltalab, Barcelona, Spain), and stored frozen at −80 °C. The detection of each pathogen was performed in the Ecophysiology Laboratory of the Estación Biológica de Doñana CSIC (http://ebd.csic.es/lef/web/) using real-time PCR assays for each bacterial genus (Salmonella, Campylobacter and Chlamydia) following established protocols38,39,40. Before each PCR assay, DNA was extracted from each cloacal swab using a commercial DNA purification kit (Promega Maxwell®). CT values of 40 were used as cut-off points. As we used non-specific PCR primers, we only detected the genus of the pathogen. We selected these three bacteria because they are leading causes of zoonotic enteric diseases (Salmonella and Campylobacter) and respiratory diseases (Chlamydia) in developed and developing countries, affecting humans, wildlife and domestic animals20,21. The primers for Salmonella were able to detect 99.4% of 630 strains belonging to over 100 serovars40. The primers for Campylobacter successfully amplify C. jejuni and C. coli, but not other Campylobacter species. The primers for Chlamydia and Chlamydophila successfully detect the nine known species for these genus. However, as we only evaluate the presence of these bacteria at genus level, we unknown if all individuals infected with Salmonella, Campylobacter or Chlamydia are really infected with pathogens that can also infect humans.
Potential pathogen risk maps and habitat use
We only considered locations recorded outside the gull breeding colony (using a radius of 500 m around each nest. see26). Further, we assumed that gulls mainly shed pathogens to the environment through their feces. Consequently, a high risk of infection was assumed to occur within feeding and resting areas. Therefore, we removed all locations associated with gull travelling behavior [speed >4 km·h−1]26 and those location on the sea. Habitat use was assigned to each gull location by overlapping locations with land cover information. High resolution information on land cover was obtained from the program SIOSE (Soil Information System of Spain, Junta de Andalucía, last update 2013) and geographical references of waste dumps from the Spatial Reference Databases of Andalucía (DERA, last update 21/02/2014). This habitat classification was subsequently visually reviewed using the most recent satellite images offered by Google Earth V 7.1.2.2041 at a 0.5 m spatial resolution. All GPS foraging locations were finally classified into eleven categories: Estuary, wetland, touristic beach, natural beach, fishing port, salt mine, fish farm, water pond, agricultural area, urban area and garbage dump. Pathogen risk maps were constructed on the basis of the current distribution of GPS-tracked gulls infected by each pathogen. The transmission risk was estimated from the number of locations of infected gulls collected on a spatial grid of 750 × 750 m over the entire study area. Differences in habitat use (%) between Salmonella-infected, Campylobacter-infected and Chlamydia-infected yellow-legged gulls were tested using one way semiparametric permutation multivariate analyses of variance tests (PERMANOVA tests) on the Euclidean distance matrix41. PERMANOVA allows for the analysis of statistical designs without the constraints of multivariate normality, homoscedasticity and greater number of variables than sampling units. The method calculates a pseudo-F-statistic directly analogous to the traditional F-statistic for ANOVA tests, using permutation procedures to obtain P-values for each term in the model41.
Data Availability
All data are available in a central PostgreSQL database at UvA-BiTS (http://www.uva-bits.nl/virtual-lab).
References
Daszak, P., Cunningham, A. A. & Hyatt, A. D. Emerging Infectious Diseases of Wildlife- Threats to Biodiversity and Human Health. Science 287, 443–449 (2000).
Boulinier, T. et al. Migration, Prospecting, Dispersal? What Host Movement Matters for Infectious Agent Circulation? Integr. Comp. Biol. 56, 330–342 (2016).
Becker, D. J., Hall, R. J., Forbes, K. M., Plowright, R. K. & Altizer, S. Anthropogenic resource subsidies and host–parasite dynamics in wildlife. Philos. Trans. R. Soc. B Biol. Sci. 373, 20170086 (2018).
Bradley, C. A. & Altizer, S. Urbanization and the ecology of wildlife diseases. Trends Ecol. Evol. 22, 95–102 (2007).
Fuirst, M., Veit, R. R., Hahn, M., Dheilly, N. & Thorne, L. H. Effects of urbanization on the foraging ecology and microbiota of the generalist seabird Larus argentatus. PLoS ONE 13, e0209200 (2018).
McMahon, B. J., Morand, S. & Gray, J. S. Ecosystem change and zoonoses in the Anthropocene. Zoonoses Public Health 65, 755–765 (2018).
Wolfe, N. D., Dunavan, C. P. & Diamond, J. Origins of major human infectious diseases. Nature 447, 279–283 (2007).
Kuiken, T. Pathogen Surveillance in Animals. Science 309, 1680–1681 (2005).
Cross, A. R. et al. Zoonoses under our noses. Microbes Infect. https://doi.org/10.1016/j.micinf.2018.06.001 (2018).
Cerdà-Cuéllar, M. et al. Do humans spread zoonotic enteric bacteria in Antarctica? Sci. Total Environ. 654, 190–196 (2019).
Bonnedahl, J. et al. Dissemination of Escherichia coli with CTX-M Type ESBL between Humans and Yellow-Legged Gulls in the South of France. PLoS ONE 4, e5958 (2009).
Vergara, A. et al. Prevalence of ESBL and/or carbapenemase-producing Escherichia coli isolated from yellow–legged gulls from Barcelona, Spain. Antimicrob. Agents Chemother. 61, AAC.02071–16 (2016).
Gamble, A. et al. Exposure of yellow-legged gulls to Toxoplasma gondii along the Western Mediterranean coasts: tales from a sentinel. Int. J. Parasitol Parasites Wildl. 8, 221–228.
Alm, E. W. et al. Potential for gulls to transport bacteria from human waste sites to beaches. Sci. Total Environ. 615, 123–130 (2018).
Taff, C. C. et al. Influence of Host Ecology and Behavior on Campylobacter jejuni Prevalence and Environmental Contamination Risk in a Synanthropic Wild Bird Species. Appl. Environ. Microbiol. 82, 4811–4820 (2016).
Directive 2003/99/EC of the European Parliament and of the Council of 17 November 2003 on the monitoring of zoonoses and zoonotic agents, amending Council Decision 90/424/EEC and repealing Council Directive 92/117/EEC. OJ L 325, 12.12.2003, p. 31–40.
Wenberg, M. F. & Porretta, M. Foodborne Diseases Active Surveillance Network (FoodNet). J. Am. Diet. Assoc. 98, A101 (1998).
Ramos, R., Cerdà-Cuéllar, M., Ramírez, F., Jover, L. & Ruiz, X. Influence of Refuse Sites on the Prevalence of Campylobacter spp. and Salmonella Serovars in Seagulls. Appl. Environ. Microbiol. 76, 3052–3056 (2010).
Migura-Garcia, L., Ramos, R. & Cerdà-Cuéllar, M. Antimicrobial Resistance of Salmonella Serovars and Campylobacter spp. Isolated from an Opportunistic Gull Species, Yellow-legged Gull (Larus michahellis). J. Wildl. Dis. 53, 148–152 (2017).
European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2016. EFSA J. 15 (2017).
Shewen, P. E. Chlamydial infection in animals: a review. Can. Vet. J. 21, 2–11 (1980).
Aaziz, R. et al. Chlamydiaceae in North Atlantic Seabirds Admitted to a Wildlife Rescue Center in Western France. Appl. Environ. Microbiol. 81, 4581–4590 (2015).
Glünder, G., Neumann, U. & Braune, S. Occurrence of Campylobacter spp. in Young Gulls, Duration of Campylobacter Infection and Reinfection by. Contact. J. Vet. Med. Ser. B 39, 119–122 (1992).
Girdwood, R. W., Fricker, C. R., Munro, D., Shedden, C. B. & Monaghan, P. The incidence and significance of salmonella carriage by gulls (Larus spp.) in Scotland. J. Hyg. 95, 229–241 (1985).
Harkinezhad, T., Geens, T. & Vanrompay, D. Chlamydophila psittaci infections in birds: A review with emphasis on zoonotic consequences. Vet. Microbiol. 135, 68–77 (2009).
Navarro, J. et al. Shifting individual habitat specialization of a successful predator living in anthropogenic landscapes. Mar. Ecol. Prog. Ser. 578, 243–251 (2017).
Navarro, J. et al. Feathered Detectives: Real-Time GPS Tracking of Scavenging Gulls Pinpoints Illegal Waste Dumping. PLoS ONE 11, e0159974 (2016).
de Jong, B. & Ekdahl, K. The comparative burden of salmonellosis in the European Union member states, associated and candidate countries. BMC Public Health 6, 4 (2006).
Lévesque, B., Brousseau, P., Bernier, F., Dewailly, É. & Joly, J. Study of the bacterial content of ring-billed gull droppings in relation to recreational water quality. Water Res. 34, 1089–1096 (2000).
Berg, R. W. & Anderson, A. W. Salmonellae and Edwardsiella tarda in gull feces: a source of contamination in fish processing plants. Appl. Microbiol. 24 (1974).
Woolhouse, M. E. J., Haydon, D. T. & Antia, R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244 (2005).
Valderrama Vasquez, C. A., Macgregor, S. K., Rowcliffe, J. M. & Jepson, P. D. Occurrence of a monophasic strain of Salmonella group B isolated from cetaceans in England and Wales between 1990 and 2002. Environ. Microbiol. 10, 2462–2468 (2008).
Vázquez, B. et al. Screening for several potential pathogens in feral pigeons (Columba livia) in Madrid. Acta Vet. Scand. 52, 45 (2010).
Ostfeld, R., Glass, G. & Keesing, F. Spatial epidemiology: an emerging (or re-emerging) discipline. Trends Ecol. Evol. 20, 328–336 (2005).
Chan, K. M. A. et al. When Agendas Collide: Human Welfare and Biological Conservation. Conserv. Biol. 21, 59–68 (2007).
Bouten, W., Baaij, E. W., Shamoun-Baranes, J. & Camphuysen, K. C. J. A flexible GPS tracking system for studying bird behaviour at multiple scales. J. Ornithol. 154, 571–580 (2013).
Thaxter, C. B. et al. A trial of three harness attachment methods and their suitability for long-term use on Lesser Black-backed Gulls and Great Skuas. Ringing Migr. 29, 65–76 (2014).
Rahn, K. et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6, 271–279 (1992).
Ridley, A. M., Allen, V. M., Sharma, M., Harris, J. A. & Newell, D. G. Real-Time PCR Approach for Detection of Environmental Sources of Campylobacter Strains Colonizing Broiler Flocks. Appl. Environ. Microbiol. 74, 2492–2504 (2008).
Nordentoft, S., Kabell, S. & Pedersen, K. Real-Time Detection and Identification of Chlamydophila Species in Veterinary Specimens by Using SYBR Green-Based PCR Assays. Appl. Environ. Microbiol. 77, 6323–6330 (2011).
Anderson, M., Gorley, R. & Clarke, K. PERMANOVA+ for PRIMER: guide to software and statistical methods. PRIMER-E Ltd, (2008).
Acknowledgements
We like to thank to the staff of the Biosphere Reserve of Marismas del Odiel (Enrique Martínez, Laura Refojo and José. M. Sayago) and to all people involved in the fieldwork (Rafa Silva, Manuel de la Riva, Laura Gangoso, Juan Jesús Moreno, Joan Giménez, Alazne Díez, Antonio Palma, Carlos Rodríguez and Francisco Ramírez). Sarah Young revised the English grammar. Tracking devices were funded by ICTS-RBD through a demonstrative project for the ESFRI-LifeWatch (Science and Technology Infrastructure for Biodiversity Data and Observatories; Ref: SP34567) project supported by European Regional Funds. JN was partially funded by the Andalucía Talent Hub Program (Andalusian Knowledge Agency and European Union’s Seventh Framework Program; Ref: 291780) and by the Spanish National Program Ramón y Cajal (RYC-2015-17809).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: J.N., J.F. and W.B. Performed the experiments: J.N., I.A. and F.M. Analysed the data: J.N., I.A. and F.M. Wrote the paper: J.N., D.G., I.A., F.M., M.G.F., W.B. and J.F.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Navarro, J., Grémillet, D., Afán, I. et al. Pathogen transmission risk by opportunistic gulls moving across human landscapes. Sci Rep 9, 10659 (2019). https://doi.org/10.1038/s41598-019-46326-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-46326-1
This article is cited by
-
Rapid loss of maternal immunity and increase in environmentally mediated antibody generation in urban gulls
Scientific Reports (2024)
-
Unravelling urban nesting site selection in an opportunistic gull: an integrated analysis of micro-spatial habitat and litter quantification
European Journal of Wildlife Research (2024)
-
Importance of predictable anthropogenic food subsidies for an opportunistic gull inhabiting urban ecosystems
European Journal of Wildlife Research (2021)
-
Adapting to urban ecosystems: unravelling the foraging ecology of an opportunistic predator living in cities
Urban Ecosystems (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.