Abstract
Seeds of soybean (Glycine max L.) are a major source of plant-derived oils. In the past, improvements have been made in the quantity and quality of seed oil. Triacylglycerols (TAGs) are the principal components of soybean seed oil, and understanding the metabolic regulation of TAGs in soybean seeds is essential. Here, we identified four soybean genes encoding TAG lipases, designated as SUGAR DEPENDENT1-1 (GmSDP1-1), GmSDP1-2, GmSDP1-3 and GmSDP1-4; these are homologous to Arabidopsis thaliana SDP1 (AtSDP1). To characterize the function of these genes during grain filling, transgenic lines of soybean were generated via RNA interference to knockdown the expression of all four GmSDP1 genes. The seed oil content of the transgenic soybean lines was significantly increased compared with the wild type (WT). Additionally, fatty acid profiles of the WT and transgenic soybean lines were altered; the content of linoleic acid, a major fatty acid in soybean seeds, was significantly reduced, whereas that of oleic acid was increased in transgenic soybean seeds compared with the WT. Substrate specificity experiments showed that TAG lipase preferentially cleaved oleic acid than linoleic acid in the oil body membrane in WT soybean. This study demonstrates that the GmSDP1 proteins regulate both the TAG content and fatty acid composition of soybean seeds during grain filling. These results provide a novel strategy for improving both the quantity and quality of soybean seed oil.
Similar content being viewed by others
Introduction
Plant oils have diverse uses; they have been utilised as dietary oils, soaps, and industrial materials. In addition, recent explosion of population and extensive interest in biofuels has accelerated the demand for plant oils1,2. Because the total agricultural area of the world remains unchanged (FAOSTAT: http://faostat3.fao.org/home/), improvement in plant oil yield per unit area is necessary to address the increase in demand. Engineering crops with high oil content is required worldwide. Oilseed crops, such as soybean (Glycine max L.), rape (Brassica napus), peanut (Arachis hypogaea) and sesame (Sesamum indicum), are cultivated for the production of plant oils. Because plant oils, in most cases, are extracted from seeds, it is therefore important to enhance the oil content of seeds.
Triacylglycerols (TAGs) are one of the primary components of seed oils, and their biosynthetic pathway has been extensively studied in seeds3,4,5,6,7. In plants, fatty acids are synthesised in plastids, and subsequently TAGs are synthesised from fatty acids and glycerol in the endoplasmic reticulum (ER). The synthesised TAGs are then stored in oil bodies, which are storage organelles for oil. Recent advances in biochemical and molecular biological studies have revealed the TAG biosynthetic pathway in seeds8,9. Several reports show that modifications of the TAG biosynthetic pathway significantly increase oil production in seeds10,11,12,13, indicating that the modification of this pathway is key to improving oil production in plants. The TAG degradation pathway has also been shown to affect the oil content of seeds. TAG degradation is essential for the post-germinative growth of oilseed crop plants14,15,16. The first step in TAG degradation is the hydrolysis of TAGs, which is catalysed by SUGAR DEPENDENT1 (SDP1), a TAG lipase present in oil body membranes17. TAG hydrolysis produces fatty acids, which are imported into the peroxisome by a peroxisomal ABC transporter PED3/CTS/PXA118,19,20 and converted into energy for post-germinative growth14,16,21,22. The sdp1 mutants have been shown to exhibit defective TAG degradation post-germination17,23. The TAG degradation pathway is activated not only post-germination but also during seed development. Defective SDP1 has been shown to enhance the seed oil content in Arabidopsis (Arabidopsis thaliana)24, rapeseed25, and Jatropha (Jatropha curcas)26. These reports suggest that TAG turnover is highly active in developing seeds, and the suppression of TAG degradation enhances the oil content of seeds.
In addition to the quantity of TAGs, the quality of TAGs is important. TAG quality is highly dependent on its fatty acid composition, which differs widely in plant species2. Olive oil and rapeseed oil, which are used as dietary oils, contain a large portion of oleic acid (18:1), whereas cottonseed oil and soybean oil contain a large amount of linoleic acid (18:2). By contrast, the primary fatty acid in palm kernel oil, which is used in soaps, is lauric acid (12:0). The fatty acid composition of seed oil determines its usage and value; therefore, conducting research aimed at modifying the fatty acid composition of seed oil is encouraged2,4,7. In fact, several crops with altered fatty acid composition such as canola, a rapeseed variant with low erucic acid (22:1) content, are wildly cultivated. This indicates that the modification of fatty acid composition is necessary for the establishment of valuable oilseed crops.
Soybeans (Glycine max) are one of the most important crops for vegetable protein supplies, and their breeding for increased seed protein has been studied. At the same time, soybeans are also a major oilseed crop, and soybean oil is the world’s second mostly highly produced oil after palm oil (FAOSTAT: http://faostat3.fao.org/home/). However, the oil content of soybean seed is relatively low compared with that of other oilseed crops. Therefore, studies focussed on identifying ways to increase the seed oil content in soybean are required27. Polyunsaturated fatty acids (PUFAs), including linoleic and linolenic acids, are susceptible to oxidation. Large amounts of linoleic and linolenic acids reduce the oxidative stability of soybean oil; thus, it is important to reduce the amount of these fatty acids27. Further investigation of the TAG metabolic pathway is needed to develop soybean cultivars with high oil and low PUFA content.
In this study, we identified SDP1 genes in soybean and generated transgenic soybean lines with suppressed SDP1 expression, specifically during seed development. Our data show that GmSDP1 regulates both the oil content and fatty acid composition of soybean seeds.
Results
Identification and characterisation of SDP1 genes in soybean
This study aimed to understand TAG degradation during seed development in soybean. The reference genome of soybean cv. Williams 82 is 1.1 Gbp, and approximately 75% of the genes exist as multiple copies28. To identify soybean genes orthologous to Arabidopsis SDP1 (AtSDP1) gene, amino acid sequences containing patatin-like phospholipase domain (PF01734), were acquired from the soybean genome using Phytozome v12.1.629. Phylogenetic analysis of 29 sequences containing the domain of PF01734 from soybean genome was performed, which revealed that four lipase sequences (Glyma.02g190000.1, Glyma10g105200.1, Glyma19g132900.1 and Glyma03g130900.1) belonged to the same clade as AtSDP1 (Fig. 1); we designated these four sequences as GmSDP1-1, GmSDP1-2, GmSDP1-3 and GmSDP1-4, respectively. All four GmSDP1 amino acid sequences showed over 70% homology with the AtSDP1 sequence (Supplementary Fig. S1).
Phylogenetic analysis of the triacylglycerol (TAG) lipase in Arabidopsis thaliana (AtSDP1) and 29 lipases containing patatin-like phospholipase domain in soybean. Amino acid sequences of all proteins were obtained from Phytozome v12.1.6. Phylogenetic analysis was performed using MEGA7. Bootstrap values were determined based on 1,000 repetitions.
The expression of GmSDP1 genes was analysed during seed germination. After 1 day of seed imbibition, expression of GmSDP1-4, GmSDP1-1 and GmSDP1-3 increased by 4-, 3- and 2-fold, respectively, compared with those at 0 day of seed imbibition (Fig. 2A). After 3 days of seed imbibition, expression of GmSDP1-3 and GmSDP1-4 was similar to those at day 1, whereas the expression of GmSDP1-1 was significantly lower than that at day one (Fig. 2A). After 5 days of seed imbibition, expression of GmSDP1-3 and GmSDP1-4 was higher than those at day 3 (Fig. 2A), whereas the expression of GmSDP1-1 was similar to that at day 3 (Fig. 2A). No increase in GmSDP1-2 expression was detected during germination (Fig. 2A). Expressions of the GmSDP1 genes were also measured during seed development. In developing seeds, expressions of all GmSDP1 genes increased until 35 days after flowering (DAF) but decreased at 45 DAF (Fig. 2B). These results show that multiple copies of GmSDP1 are transcribed in soybean seeds.
Quantitative analysis of GmSDP1-1, GmSDP1-2, GmSDP1-3 and GmSDP1-4 expression levels during seed germination and development. Expression levels of GmSDP1-1, 1-2, 1-3 and 1-4 were measured via quantitative real-time PCR (qRT-PCR) analysis during germination (A) and seed development (B). The GmEF1b gene was used as an internal control41. Standard curves were generated with plasmids containing GmSDP1-1, 1-2, 1-3, 1-4 or GmEF1b cDNA. Values represent mean ± standard deviation (SD) of three independent experiments.
Establishment of transgenic soybean lines with suppressed GmSDP1 gene expression
Multiple copies of GmSDP1 were expressed during seed development. To verify the function of GmSDP1 genes during seed development, we generated transgenic soybean lines via RNA interference (RNAi) to knockdown the expression of all four GmSDP1 genes. The patatin-like phospholipase domain (PF01734) is present in all 29 lipases (Fig. 1); therefore, the DNA sequence of this domain was an unsuitable candidate for RNAi-mediated knockdown of the GmSDP1 genes. We, therefore, screened highly conserved sequences specifically present in the GmSDP1 genes. Two such sequences were identified at the 5′ and 3′ termini of these genes (Supplementary Fig. S2). The sequence identified at the 5′ terminus (S1; Fig. 3A) was cloned from the coding sequence (CDS) of GmSDP1-1, and the sequence at 3′ terminus (S2; Fig. 3A) was cloned from GmSDP1-4 CDS. The cloned S1 and S2 sequences were fused in tandem, and the fused sequence was cloned into an RNAi vector under the control of soybean 11S globulin gene promoter, one of the genes encoding a major seed storage protein in soybean (Fig. 3A). The RNAi vector was introduced into WT soybean plants via infiltration using Agrobacterium tumefaciens, and three independent transgenic lines were established (SDP1i_2, _15 and _19). The germination percentage of the transgenic lines was approximately 80%, which was similar to that of the WT (data not shown). Expressions of each GmSDP1 gene were measured at 35 DAF, and significant reduction in the expression of each gene was observed in the transgenic lines during seed development (Fig. 3B). On the other hand, expressions of all four GmSDP1 genes in transgenic lines were similar to those in the WT during germination (Fig. 3C). These results show that the expression of GmSDP1 genes in transgenic lines was significantly suppressed, specifically during seed development.
Establishment and characterisation of GmSDP1 knockdown lines in soybean. (A) Schematic diagrams of the RNAi construct used for GmSDP1 knockdown. (B,C) qRT-PCR analysis of relative expression levels of GmSDP1-1, 1-2, 1-3 and 1-4 in seeds at 35 DAF (B) and 3-day-old seedlings (C) of the WT and transgenic lines. GmEF1b was used as an internal control. Standard curves were generated using plasmids containing GmSDP1-1, 1-2, 1-3, 1-4 or GmEF1b cDNA. Values represent mean ± SD of three independent experiments.
Seed yield and oil content of transgenic soybean lines
Transgenic lines of soybean were grown in the growth chamber, and seed production was investigated. Seeds of line SDP1i_15 were larger than that of WT, and seed coat rupture was also observed (arrow heads in Fig. 4A). The weight of WT seeds was 0.313 g, whereas seeds of lines SDP1i_2, _15 and _19 weighed 0.343, 0.357 and 0.326 g, respectively (Fig. 4B). By contrast, seed number showed no differences between the WT and transgenic lines (Fig. 4C). The seed yield, which represents weight of seeds harvested from a plant, of transgenic lines was significantly higher than that of the WT; seed yield of lines SDP1i_2, _15, and _19 was 15.2%, 18.6% and 5.2%, respectively, higher than that of the WT (Fig. 4D). Additionally, the seed oil content of the transgenic lines SDP1i_2, _15 and_19 was 2.2%, 3.1% and 0.4% higher than that of the WT, respectively (Fig. 4E). We also estimated the oil yield per plant, based on the seed yield and oil content. The oil yield of lines SDP1i_2, _15 and_19 was 29.2%, 30.5% and 7.8% higher, respectively, than that of the WT (Fig. 4F). Taken together, these results show that the knockdown of multiple GmSDP1 genes increases the seed oil content in soybean.
Productivity of GmSDP1 knockdown lines. (A) Images of WT and transgenic SDP1i-15 seeds. Scale bars = 0.5 mm. (B) Weight of transgenic seeds. Values represent mean ± SD of three independent experiments with 130–150 seeds harvested from individual plants. Significant difference between WT and transgenic lines was determined using Student’s t-test and is denoted as **(P < 0.05). (C) Seed numbers harvested from transgenic lines. Values represent mean ± SD of three individual plants. (D) Seed yield of transgenic lines. Values represent mean ± SD of three individual plants. Significant difference between WT and transgenic lines was determined using Student’s t-test and is denoted as *(P < 0.1) or **(P < 0.05). (E) Fatty acid (FA) concentrations in seeds of transgenic lines measured via gas chromatography–mass spectrometry (GC–MS). Values represent mean ± SD of three independent experiments, with 20 seeds per experiment. Significant difference between WT and transgenic plants determined using Student’s t-test is denoted as **(P < 0.05). (F) Oil yield of transgenic lines estimated from the seed yield and FA concentration. Values represent mean ± SD of three individual plants. Significant difference between WT and transgenic lines determined using Student’s t-test is denoted as *(P < 0.1) or **(P < 0.05).
Fatty acid profiles of transgenic soybean lines
In addition to the seed oil content, the fatty acid composition is also important for the breeding of superior oilseed crops. We thus analysed the fatty acid composition of seed TAGs. Results showed that linoleic acid (18:2) was a major component of TAGs, comprising 56% of the total fatty acids in WT seeds; however, it was significantly reduced by 4.9%, 7.3% and 2.1% in the transgenic lines SDP1i_2, _15, and _19, respectively, compared with the WT (Fig. 5). By contrast, the content of oleic acid (18:1) was significantly increased by 6.9%, 10.6% and 3.4% in the transgenic lines SDP1i_2, _15, and _19, respectively, compared with the WT (Fig. 5). No major differences in the amount of palmitic acid (16:0), stearic acid (18:0) and linolenic acid (18:3) were detected between the WT and transgenic lines (Fig. 5). These results indicate that GmSDP1 genes regulate the ratio of oleic acid to linoleic acid in TAGs.
Fatty acid profiles of GmSDP1 knockdown lines. Fatty acid profiles of TAGs extracted from mature seeds. TAGs were extracted from seeds using thin layer chromatography (TLC). Fatty acid profiles of TAGs were measured via GC–MS. Values represent mean ± SD of three independent experiments, with 20 seeds per experiment. Significant difference between WT and transgenic plants determined using Student’s t-test is denoted as ##(P < 0.01) or ###(P < 0.001).
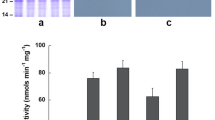
Considering that the SDP1 gene encodes TAG lipase17, we hypothesised that the substrate specificity of GmSDP1 plays a major role in determining the ratio of oleic acid to linoleic acid in TAGs. To elucidate the difference in the substrate specificity of GmSDP1 between oleic and linoleic acid, TAG lipase activity in oil body membranes was measured using an equimolar mixture of triolein and trilinolein as substrates. In the WT, the ratio of oleic acid to linoleic acid hydrolysed in this experiment was 2.45:1.00. By contrast, ratios of oleic acid to linoleic acid in the transgenic lines SDP1i_2, _15 and _19 were 1.4:1.0, 1.1:1.0, and 1.6:1.0, respectively, which were significantly lower than that of the WT (Fig. 6). In addition, the amounts of released fatty acids in the transgenic lines SDP1i_2, _15 and _19 were decreased to 31.2%, 31.1% and 62.4% of that in WT, respectively (Supplementary Fig. S4). These results indicate that GmSDP1 proteins are one of the major TAG lipases in the oil body membrane, and preferentially cleave oleic acid than linoleic acid.
Substrate specificity of TAG lipase in oil body membranes in soybean. Substrate preference of TAG lipase was determined using oil body membranes isolated from seeds at 35 DAF. The isolated membranes were incubated with a TAG mixture (triolein:trilinolein = 1:1). Fatty acids released from the TAG mixture were analysed by GC–MS. The amounts of the isolated membranes were normalized to total protein contents. Values represent mean ± SD of three independent experiments, with 5 seeds per experiment. Significant difference between WT and transgenic plants determined using Student’s t-test is denoted as *(P < 0.1) or **(P < 0.05).
Discussion
Increasing the seed oil content has been a major focus in plant breeding. To meet this objective, TAG biosynthetic pathways have been extensively studied in plants10,12,13. TAG degradation activity is increased during seed development, although large amounts of TAGs are also produced in the same period. Therefore, the significance of TAG degradation during seed development has been discussed previously30,31. Recently, the TAG lipase SDP1 was identified in the oil body membrane17, and the suppression of SDP1 gene expression has been shown to increase the oil content of seeds25,26. These reports clearly show that the inhibition of TAG degradation during seed development increases the seed oil content. Moreover, these results indicate that the TAG degradation pathway is activated during seed development, resulting in the degradation of considerable amounts of TAGs. However, the biological significance of TAG degradation in developing seeds remains unclear. In this study, we aimed to understand the significance of TAG degradation during seed development in soybean via RNAi-mediated knockdown of GmSDP1 expression.
Transgenic soybean seeds were enlarged, and their oil content was higher than that of the WT (Fig. 4A, B, E). Coincidentally, the protein content of the transgenic seeds was smaller than that of WT (Supplementary Fig. S5) This result confirms that the inhibition of TAG degradation increases seed oil content in soybean, which is major oilseed crop in the world. In addition to the increase in seed oil content in transgenic lines, concentrations of oleic and linoleic acids were significantly altered between the transgenic lines and WT (Fig. 5). The concentration of oleic acid in line SDP1i_15 was approximately 1.6-fold higher than that of the WT, whereas that of linoleic acid was lower for the increment of oleic acid (Fig. 5). Because GmSDP1 encodes TAG lipase, this result suggests that GmSDP1 preferentially cleaves oleic acid than linoleic acid. This substrate specificity of GmSDP1 was experimentally verified in this study (Fig. 6). Thus, we propose that the cleavage of oleic acid contributes to the low oleic and high linoleic acid content of WT soybean seeds. The fatty acid composition of seeds varies widely among plant species2, suggesting that enzymes in the TAG biosynthetic pathway, particularly fatty acid desaturases and elongases, play a key role in the diversity of TAGs1,2,4,7,9. By contrast, evidence for the role of the TAG degradation pathway in the fatty acid composition of seeds is lacking. Our results indicate that the substrate specificity of GmSDP1 contributes towards a biased fatty acid composition (low oleic and high linoleic acid) of soybean seeds (Fig. 7). This demonstrates that the TAG degradation pathway, in addition to the TAG biosynthetic pathway, regulates the fatty acid composition of seeds. Thus, the control of fatty acid composition is a major biological significance of TAG degradation during seed development. Coordination between the biosynthesis and degradation of particular fatty acid species leads to a biased and species-specific composition of fatty acids in plant seeds.
Soybean is one of the major oilseed crops; soybean seed oil accounts for one-fourth of the total annual production of plant oil (FAOSTAT: http://faostat3.fao.org/home/). Thus, increasing the oil content of soybean seeds is directly linked to the enhancement of plant oil production. This study showed that the knockdown of GmSDP1 genes increased the oil yield in soybean (Fig. 4A–F). This increment is consistent with the results in oilseed rape25 and jatropha26 and confirms that the inhibition of TAG degradation during seed development is one of the effective strategies for producing crop cultivars with high oil content. Additionally, soybean oil is used as cooking oil; thus its fatty acid composition, along with its content, is of commercial importance. Soybean oil contains relatively higher amounts of PUFAs, such as linoleic and linolenic acids, which are prone to oxidation. It is therefore important to decrease the PUFA content of soybean oil to improve its oxidative stability27. Deletion of genes encoding fatty acid desaturases has been used as a breeding strategy for reducing the PUFA content of soybean seed oil27. This strategy is designed to depress oleic acid desaturation during grain filling32,33,34. In contrast, the knockdown of GmSDP1 genes is aimed to control oleic acid degradation. Therefore, these two strategies could be compatible; introduction of SDP1 knockdown may further increase oleic acid content in the conventional high-oleate soybean. Moreover, the knockdown could improve seed oil content without reducing the seed yield.
This shows that genetic modification of GmSDP1 genes is a novel strategy for generating soybean cultivars with superior oil quantity and quality.
Methods
Plant material and growth conditions
The Japanese soybean variety Kariyutaka was obtained from the Hokkaido Prefectural Tokachi Agricultural Experiment Station, Japan and is identical to the resource JP 86520 available from Genebank, National Institute of Agrobiological Science, Japan. Kariyutaka was used as the WT. For germination, seeds were placed on wet paper towel and incubated for 72 h at 22 °C in the dark. Three-day-old seedlings were transferred to 1/5,000a Wagner pots filled with cultured soil and grown under long day conditions (16 h light/8 h dark) at 25 °C in the growth chamber (NK systems, Japan). One-tenth strength of Hoagland solution (1 L) was supplied to the plants once every 3 days in the first month; thereafter, 2 L of the solution was supplied once every 2 days.
Generation of transgenic plants
The S1 and S2 nucleotide sequences were amplified by PCR using sequence-specific primer sets (Supplementary Table S1) and fused using the fusion PCR method. The fused S1S2 sequence was cloned into the Gateway entry vector pDONR221 (ThermoFisher Scientific, USA) and then transferred to the binary vector pMDC123-GFP35. The cauliflower mosaic virus 35S promoter, green fluorescent protein (sGFP) gene and nopaline synthase (nos) terminator in the binary vector were replaced with the soybean 11S globulin gene promoter.
Recombinant plasmids were infiltrated into WT soybean via Agrobacterium-mediated transformation, as described by Yamada et al.35,36. The primary transformants were designated as T0. Transformed soybean lines were selected for Basta resistance and designated as T1. The T2 progeny exhibiting Basta resistance was used in this study.
Quantitative real-time PCR (qRT-PCR)
To examine gene expression, qRT-PCR analysis was performed as described previously37, with slight modifications. Total RNA was extracted from three independent samples of each transgenic line, according to the method of Kanai et al.38. Subsequently, cDNA was synthesized from 1 μg of total RNA using the PrimeScript RT Reagent Kit (Takara Bio., Japan) and used for qRT-PCR analysis with KAPA SYBR FAST Universal Kit (KAPA BIOSYSTEMS, USA) and ABI 7500 Fast Real-Time PCR system (ThermoFisher Scientific,), according to the manufacturer’s instructions. Primer sets used for qRT-PCR are listed in Supplementary Table 1.
Analysis of TAGs and free fatty acids
The analysis of seed TAGs and free fatty acids was performed as described previously37,39, with slight modifications. A total of 20 soybean seeds were crushed using a wooden hammer and ground with mortar and pestle. Approximately 10 mg of powdered soybean seeds was ground in 400 μl of chloroform:methanol (2:1) mixture using a stainless steel homogenizer. The resulting extract was centrifuged at 2,000 × g for 5 min. The supernatant was collected, and the pellet was extracted with 400 μl of chloroform:methanol (2:1) mixture by vortexing. The centrifugation step was repeated, and the supernatant was combined with the first supernatant, dried and stored at –20 °C. Dried lipids were dissolved in 25 μl hexane and spotted onto a silica gel thin-layer chromatography (TLC) plate (Merck Millipore; http://www.merckmillipore.com) to separate TAGs and free fatty acids from total lipids. Triheptadecanoin (Wako, Japan) and oleic acid (Sigma-Aldrich, USA) was used as standards. TLC separation was carried out using hexane:diethylether:acetic acid (80:20:1) mixture. TAG and free fatty acid spots were collected and extracted using 100 μl hexane. Fatty acid methyl esterification was performed using a Fatty Acid Methylation Kit (Nacalai Tesque, Japan). Fatty acid methyl esters were quantified via gas chromatography (GC-2010, Shimadzu, Japan)–mass spectrometry (JMSDX-300, JEOL, Japan) using a DB-23 column (Agilent Technology, USA).
Enzyme assay
Oil body membrane was purified from developing seeds of the WT and transgenic lines 35 DAF, as described previously by Edmond40. Protein content of the oil body fraction was measured using the Bio-Rad Protein Assay (Bio-Rad, USA). Lipase activity was determined on the basis of the amount of fatty acids produced after TAG hydrolysis. Reaction buffer (400 μl) containing 50 mM Bis-Tris propane (pH 8.1), 2 mM DTT, 1 mM CaCl2, 5 mM triolein (Sigma-Aldrich) and 5 mM trilinolein (Sigma-Aldrich, USA). Substrates were emulsified with 5% (w/v) gum arabic (Wako, Japan), followed by ultrasonic treatment for 30 s at 20 W using Sonifier (Branson, USA). Reactions were carried out on a shaking incubator for 60 min at 30 °C and 120 rpm.
References
Durrett, T. P., Benning, C. & Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J 54, 593–607, https://doi.org/10.1111/j.1365-313X.2008.03442.x (2008).
Dyer, J. M., Stymne, S., Green, A. G. & Carlsson, A. S. High-value oils from plants. Plant J: for cell and molecular biology 54, 640–655, https://doi.org/10.1111/j.1365-313X.2008.03430.x (2008).
Baud, S. & Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog Lipid Res 49, 235–249, https://doi.org/10.1016/j.plipres.2010.01.001 (2010).
Cahoon, E. B. et al. Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10, 236–244, https://doi.org/10.1016/j.pbi.2007.04.005 (2007).
Chapman, K. D. & Ohlrogge, J. B. Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287, 2288–2294, https://doi.org/10.1074/jbc.R111.290072 (2012).
Lung, S. C. & Weselake, R. J. Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41, 1073–1088 (2006).
Napier, J. A. & Graham, I. A. Tailoring plant lipid composition: designer oilseeds come of age. Curr Opin Plant Biol 13, 330–337, https://doi.org/10.1016/j.pbi.2010.01.008 (2010).
Bates, P. D., Stymne, S. & Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol 16, 358–364, https://doi.org/10.1016/j.pbi.2013.02.015 (2013).
Napier, J. A., Haslam, R. P., Beaudoin, F. & Cahoon, E. B. Understanding and manipulating plant lipid composition: Metabolic engineering leads the way. Curr Opin Plant Biol 19, 68–75, https://doi.org/10.1016/j.pbi.2014.04.001 (2014).
Hernandez, M. L. et al. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol 160, 215–225, https://doi.org/10.1104/pp.112.201541 (2012).
Maisonneuve, S., Bessoule, J. J., Lessire, R., Delseny, M. & Roscoe, T. J. Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol 152, 670–684, https://doi.org/10.1104/pp.109.148247 (2010).
Shen, B. et al. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153, 980–987, https://doi.org/10.1104/pp.110.157537 (2010).
Tan, H. et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol 156, 1577–1588, https://doi.org/10.1104/pp.111.175000 (2011).
Graham, I. A. Seed storage oil mobilization. Annu Rev Plant Biol 59, 115–142, https://doi.org/10.1146/annurev.arplant.59.032607.092938 (2008).
Mano, S. & Nishimura, M. Plant peroxisomes. Vitam Horm 72, 111–154, https://doi.org/10.1016/S0083-6729(05)72004-5 (2005).
Theodoulou, F. L. & Eastmond, P. J. Seed storage oil catabolism: a story of give and take. Curr Opin Plant Biol 15, 322–328, https://doi.org/10.1016/j.pbi.2012.03.017 (2012).
Eastmond, P. J. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675, https://doi.org/10.1105/tpc.105.040543 (2006).
Footitt, S. et al. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. Embo J 21, 2912–2922, https://doi.org/10.1093/emboj/cdf300 (2002).
Hayashi, M. et al. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiol 43, 1–11 (2002).
Zolman, B. K., Silva, I. D. & Bartel, B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol 127, 1266–1278 (2001).
Baker, A., Graham, I. A., Holdsworth, M., Smith, S. M. & Theodoulou, F. L. Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci 11, 124–132, https://doi.org/10.1016/j.tplants.2006.01.005 (2006).
Hayashi, M. & Nishimura, M. Arabidopsis thaliana–a model organism to study plant peroxisomes. Biochim Biophys Acta 1763, 1382–1391, https://doi.org/10.1016/j.bbamcr.2006.08.014 (2006).
Kelly, A. A., Quettier, A. L., Shaw, E. & Eastmond, P. J. Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157, 866–875, https://doi.org/10.1104/pp.111.181784 (2011).
van Erp, H., Kelly, A. A., Menard, G. & Eastmond, P. J. Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiol 165, 30–36, https://doi.org/10.1104/pp.114.236430 (2014).
Kelly, A. A., Shaw, E., Powers, S. J., Kurup, S. & Eastmond, P. J. Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol J 11, 355–361, https://doi.org/10.1111/pbi.12021 (2013).
Kim, M. J. et al. Gene silencing of Sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol Biofuels 7, 36, https://doi.org/10.1186/1754-6834-7-36 (2014).
Clemente, T. E. & Cahoon, E. B. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol 151, 1030–1040, https://doi.org/10.1104/pp.109.146282 (2009).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183, https://doi.org/10.1038/nature08670 (2010).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40, D1178–1186, https://doi.org/10.1093/nar/gkr944 (2012).
Chia, T. Y., Pike, M. J. & Rawsthorne, S. Storage oil breakdown during embryo development of Brassica napus (L.). J Exp Bot 56, 1285–1296, https://doi.org/10.1093/jxb/eri129 (2005).
Poirier, Y., Ventre, G. & Caldelari, D. Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121, 1359–1366 (1999).
Brace, R. C., Fehr, W. R. & Schnebly, S. R. Agronomic and seed traits of soybean lines with high oleate concentration. Crop Sci 51, 534–541, https://doi.org/10.2135/cropsci2010.04.0234 (2011).
Graef, G. et al. A high-oleic-acid and low-palmitic-acid soybean: agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnol J 7, 411–421, https://doi.org/10.1111/j.1467-7652.2009.00408.x (2009).
Kinney, A. J. Development of genetically engineered soybean oils for food applications. J Food Lipids 3, 273–292, https://doi.org/10.1111/j.1745-4522.1996.tb00074.x (1996).
Yamada, T. et al. Knockdown of the 7S globulin subunits shifts distribution of nitrogen sources to the residual protein fraction in transgenic soybean seeds. Plant Cell Rep 33, 1963–1976, https://doi.org/10.1007/s00299-014-1671-y (2014).
Yamada, T., Watanabe, S., Arai, M., Harada, K. & Kitamura, K. Cotyledonary node pre-wounding with a micro-brush increased frequency of Agrobacterium-mediated transformation in soybean. Plant. Biotechnol 27, 217–220, https://doi.org/10.5511/plantbiotechnology.27.217 (2010).
Kanai, M., Mano, S., Kondo, M., Hayashi, M. & Nishimura, M. Extension of oil biosynthesis during the mid-phase of seed development enhances oil content in Arabidopsis seeds. Plant Biotechnol J 14, 1241–1250, https://doi.org/10.1111/pbi.12489 (2016).
Kanai, M., Mano, S. & Nishimura, M. An efficient method for the isolation of highly purified RNA from seeds for use in quantitative transcriptome analysis. J Vis Exp, ARTN e55008, https://doi.org/10.3791/55008 (2017).
Kanai, M., Hayashi, M., Kondo, M. & Nishimura, M. The Plastidic DEAD-box RNA Helicase 22, HS3, is Essential for Plastid Functions Both in Seed Development and in Seedling Growth. Plant Cell Physiol 54, 1431–1440, https://doi.org/10.1093/Pcp/Pct091 (2013).
Eastmond, P. J. Cloning and characterization of the acid lipase from castor beans. J Biol Chem 279, 45540–45545, https://doi.org/10.1074/jbc.M408686200 (2004).
Hu, R. B., Fan, C. M., Li, H. Y., Zhang, Q. Z. & Fu, Y. F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. Bmc Mol Biol 10, Artn 93, https://doi.org/10.1186/1471-2199-10-93 (2009).
Acknowledgements
We thank the staff of Functional Genomics Facility and Spectrography and Bioimaging Facility, NIBB Core Research Facilities, and Model Plant Research Facility, NIBB Bioresource Center. This work was supported by CREST, JST and JSPS Grant-in-Aid for Scientific Research on Innovative Areas (22120007 to M.N.).
Author information
Authors and Affiliations
Contributions
M.K., T.Y., S.M., M.H. and M.N. devised and designed the experiments. M.K. performed the experiments. T.Y. produced and established the transgenic soybean lines. M.K., T.Y., S.M. and M.N. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kanai, M., Yamada, T., Hayashi, M. et al. Soybean (Glycine max L.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci Rep 9, 8924 (2019). https://doi.org/10.1038/s41598-019-45331-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45331-8
This article is cited by
-
Rice lipases: a conundrum in rice bran stabilization: a review on their impact and biotechnological interventions
Physiology and Molecular Biology of Plants (2023)
-
Regulation of seed traits in soybean
aBIOTECH (2023)
-
Advances in improvement of soybean seed composition traits using genetic, genomic and biotechnological approaches
Euphytica (2022)
-
Genome-wide association study reveals a patatin-like lipase relating to the reduction of seed oil content in Brassica napus
BMC Plant Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.