Abstract
Suppressor of Hairy-wing [Su(Hw)] is a DNA-binding architectural protein that participates in the organization of insulators and repression of promoters in Drosophila. This protein contains acidic regions at both ends and a central cluster of 12 zinc finger domains, some of which are involved in the specific recognition of the binding site. One of the well-described in vivo function of Su(Hw) is the repression of transcription of neuronal genes in oocytes. Here, we have found that the same Su(Hw) C-terminal region (aa 720–892) is required for insulation as well as for promoter repression. The best characterized partners of Su(Hw), CP190 and Mod(mdg4)-67.2, are not involved in the repression of neuronal genes. Taken together, these results suggest that an unknown protein or protein complex binds to the C-terminal region of Su(Hw) and is responsible for the direct repression activity of Su(Hw).
Similar content being viewed by others
Introduction
High-resolution chromosome conformation capture techniques have provided evidence that regulatory elements form loops that are essential for gene regulation in higher eukaryotes1,2,3,4,5,6. In particular enhancers can activate target promoters at large distances (up to hundreds of kb in some cases), which raises the question of the mechanisms regulating such long-distance enhancer–promoter interactions. More than 25 years ago, a special class of regulatory elements, named insulators, was suggested to delimit the activity of enhancers7,8,9,10,11,12. Insulators are defined as regulatory elements that disrupt the communication between an enhancer and a promoter when inserted between them. Some insulator complexes contribute to higher-order organization of chromatin in topologically associated domains that are fundamental elements of the eukaryotic genomic structure13,14.
One of the first insulators was identified in the gypsy retrotransposon, whose integration into genes often resulted in inactivation of enhancers that were separated from promoters by the gypsy insertion15,16,17,18,19. The phenotypes of the gypsy-induced mutations were suppressed by inactivation of the gene encoding the Suppressor of hairy wing protein [Su(Hw)]20. The gypsy insulator consists of 12 reiterated binding sites for Su(Hw)21,22. Today, Su(Hw) is one of the best characterized insulator proteins. It has been shown that artificial reiterated binding sites for Su(Hw) or gypsy insulator can block various enhancers at all stages of Drosophila development21,23,24,25,26,27.
The Su(Hw) protein contains the N-terminal region involved in the interaction with CP190, an array of 12 C2H2-type zinc finger domains, and the C-terminal region (aa 716–892) responsible for enhancer blocking activity28,29,30,31,32. Several Su(Hw) partners were identified, including Mod(mdg4), CP190, ENY2, Shep, Rump, and HIPP128,33,34,35,36,37,38. Mod(mdg4)-67.2 is one of the isoforms encoded by the mod(mdg4) locus39,40. The Mod(mdg4)-67.2 protein contains the N-terminal BTB/POZ domain and glutamine-rich (Q-rich) region which is common to all isoforms and the unique C-terminal region that is required for interaction with the 716–892 aa region of Su(Hw)28,31,34. The BTB and Q-rich domains of the Mod(mdg4)-67.2 interact with the M domain of CP190 and the N-terminal region of Su(Hw) (aa 1–238), respectively31. The functions of Mod(mdg4)-67.2 are as yet unknown, except for its participation in recruiting the Su(Hw) complex to specific chromatin sites and masking the repression activity of Su(Hw)31,41,42.
The CP190 protein contains the N-terminal BTB domain that forms stable homodimers43,44,45,46. The BTB domain interacts with two adjacent regions at the Su(Hw) N-terminus, which is essential for CP190 recruitment to Su(Hw) sites32. CP190 is involved in the recruitment of several transcriptional complexes to chromatin, including the NURF, dREAM, and SAGA complexes47,48. CP190 has major effects on chromatin, such as the depletion of nucleosomes and the prevention of heterochromatin expansion49. Similar to CP190, ENY2 is recruited to chromatin by interacting with Su(Hw) and participates in the boundary function of Su(Hw), which protects active genes from silencing36. Shep and Rump are the RNA binding proteins that function as negative regulators of the Su(Hw) enhancer blocking activity35,37. HIPP1 (CG3680) was identified as possible partner of Su(Hw) in a recent screen of proteins interacting with HP1, one of the main heterochromatin proteins33. Strong co-localization of HIPP1 and Su(Hw) sites suggests that these proteins are in the same complex33.
In previous genome-wide studies, three classes of the Su(Hw) binding regions have been identified, which are characterized by the binding of Su(Hw) alone (SBS-O), both Su(Hw) and CP190 (SBS-C), or three proteins Su(Hw)/CP190/Mod(mdg4)-67.2 (SBS-CM)50,51,52,53. It has been shown that many SBS-O sites are involved in transcriptional repression52,54. The function of Su(Hw) as transcriptional repressor is required for female germline development54 and for sustained male fertility55.
Here, we made an attempt to identify Su(Hw) domains involved in the repression of several neuronal genes in the ovary. We found that the same C-terminal region (aa 720–892) in Su(Hw) is required for enhancer blocking and for promoter repression.
Results
Model system for studying Su(Hw)-dependent repression
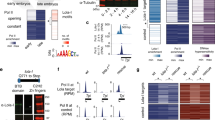
As shown previously, Su(Hw) is involved in the repression of many neural genes in the ovary54,56. These genes contain binding sites for Su(Hw) in promoter regions, suggesting the involvement of this protein in direct repression of transcription. To further characterize Su(Hw) domains involved in repression, we selected five representative neural genes whose promoters are bound by Su(Hw) alone (Rph, cg32017, and Hs3st-A), by Su(Hw) and CP190 (Syn2), or by Su(Hw), CP190, and Mod(mdg4)-67.2 (mAcR-60C) (Fig. 1a). Using ChIP, we confirmed the binding of the insulator proteins to the promoters in the ovaries and heads of females (Fig. 1b). Interestingly, ChIP with a common antibody against all Mod(mdg4) isoforms showed an enrichment of Mod(mdg4) at the promoters. However, Mod(mdg4)-67.2 binds only to the Su(Hw) site at the mAcR-60C promoter, suggesting that another Mod(mdg4) isoform was recruited to other promoters by an unknown DNA-bound protein. In general, the observed patterns of Su(Hw), CP190, and Mod(mdg4)-67.2 recruitment to the tested promoters in the ovaries and heads were similar, which is in agreement with the previous observation that these proteins stably bind to their sites in different Drosophila tissues and cell lines51,52,53,54.
Model system for studying Su(Hw)-dependent repression. (a) Genome browser view of insulator protein binding. The regions used in qPCR are indicated by vertical black arrows and numbered according to their chromosome position (FlyBase, 2006). The names of identified genomic regions are given next to the schemes of model genes. (b) ChIP-qPCR data on the binding of insulator proteins to the promoters in the ovaries and heads of females. ChIP was performed with antibodies against Su(Hw), Mod(mdg4)-67.2 (Mod-67.2, the C-terminal region corresponding to the specific isoform), CP190, Mod-com (the region common to all Mod(mdg4) isoforms), and nonspecific immunoglobulins (IgG, control). The ras64B coding region (ras) was used as a control devoid of Su(Hw) binding sites. The percent recovery of immunoprecipitated DNA (Y axis) was calculated relative to the amount of input DNA. Error bars indicate standard deviation of three independent biological replicates. Statistical analysis (Student’s t-test) was performed relative to protein binding to the ras64B coding region. Asterisks indicate significance levels of *p < 0.05, **p < 0.01, or ***p < 0.001 (here and in Figs 2–5).
We then compared the expression of the model genes in the heads and ovaries of females (Fig. 2a). As expected, the model genes were repressed in the ovaries and actively transcribed in the heads. To confirm the role of Su(Hw) in gene repression, we examined the expression of the model genes in the su(Hw)− background. In the absence of the Su(Hw) protein, transcription from the tested genes proved to be increased 10- to 20-fold in the ovaries, while its level in the heads remained unchanged (Fig. 2b). Taken together, these results confirm that the selected genes provide a good model system for the study of Su(Hw)-mediated repression in the ovaries.
Expression of model genes in the heads and ovaries of females. (a) Wild-type (wt) expression of genes, with mRNA levels in the heads shown as a fold change relative to those in the ovaries. (b) Expression of genes in the su(Hw)v/su(Hw)e04061 background (su(Hw)−), with mRNA levels shown as a fold change relative to the wild-type levels. Error bars indicate standard deviation of three independent biological replicates. Here and in Figs 3–5 and 7, mRNA levels are normalized to that of the ras64B gene, which remains unchanged in the su(Hw)− background.
Tissue-specific repression of model genes depends on the presence of Su(Hw) binding sites in promoters
There are two possible mechanisms explaining how the Su(Hw) protein can repress the promoters: (1) Su(Hw) competes for the binding site with a transcription factor that activates transcription; (2) Su(Hw) recruits a tissue-specific complex that is responsible for repression. To test these alternative models, we mutated the Su(Hw) binding sites in the promoters of the mAcR-60C and Rph genes (Fig. 3a). The lacZ gene was used as a reporter to test the activity of the promoters, and the mini-white gene, which determines eye pigmentation, was used as a marker to select positive transgenic lines. The tested genes were cloned with the attB site to allow site-specific integration with the phiC31 integrase system57. The constructs were inserted in the same genomic region 38D. The RT-qPCR analysis in the ovaries showed that mutational inactivation of Su(Hw) binding sites led to an increase in transcription, compared to the wild-type level, by a factor of 4 for mAcR-60C and by a factor of 6 for Rph (Fig. 3b). At the same time, the effect of mutations in the promoters on transcription of the reporter in the heads lacked statistical significance. These results argue against the model that Su(Hw) masks binding site for a transcriptional activator in the promoters.
Su(Hw) binding sites are required for repression of transcription. (a) Scheme of the constructs (drawn not to scale) containing the mini white gene (white), LacZ-GFP coding region (LacZ-GFP), transcription terminator from SV40 virus (SV40), and promoters of two model genes, mAcR-60C and Rph (promoter). Arrowheads show the direction of transcription. The sizes of promoter fragments cloned into the constructs are indicated. WT, wild-type sequences including Su(Hw) binding site (bold type); mut, sequences with mutation in Su(Hw) binding site. (b) Expression of two model genes in the wild-type (wt) and mutant (mut) backgrounds. (c) Expression of model genes in the heads in the Su(Hw) overexpression background. Expression levels are shown as a fold change relative to wild type. Error bars indicate standard deviation of two independent biological replicates.
We also tested whether the overexpression of Su(Hw) can affect transcription of the model genes in the heads. To induce Su(Hw) overexpression, we used the hsp70 promoter and two-time heat shock treatment for 2 hours at the embryonic and larval stages. The results were confirmed by Western blot analysis (Fig. S1A) and RT-qPCR assay, which revealed approximately 10-fold increase in su(Hw) gene transcription, compared to the wild-type level (Fig. S1B). Despite the high level of Su(Hw), we observed a slight repression of only one model gene, Syn2 (Fig. 3c). This result suggests that, in most cases, the amount of Su(Hw) is not critical for repression. Thus, it is likely that derepression of model genes in the heads is not associated with Su(Hw) displacement from the target promoters.
The CP190 and Mod(mdg4)-67.2 proteins are not involved in Su(Hw)-mediated repression
It has been shown53,54 that the CP190 and Mod(mdg4)-67.2 partners of Su(Hw) are not usually essential for repression, since Su(Hw) alone is present at many repressed promoters. As noted above, the mAcR-60C promoter is bound by Su(Hw), CP190, and Mod(mdg4)-67.2, while the Syn2 promoter, only by Su(Hw) and CP190. Hence, the question has arisen whether CP190 and Mod(mdg4)-67.2 are involved in the regulation of these promoters. According to our previous data32, the BTB domain of CP190 binds to two regions located between aa 88 and 202 at the N-terminus of Su(Hw) (Fig. 4a). The deletion of these regions in the Su(Hw)Δ114 mutants prevents CP190 recruitment to the Su(Hw) complex. We compared the recruitment of Su(Hw) and CP190 proteins to the promoters of model genes in pupae of transgenic lines expressing full-length Su(Hw)+ and Su(Hw)Δ114 in the absence or presence of the mod(mdg4)u1 mutation inactivating the Mod(mdg4)-67.2 isoform (Fig. 4b). Unexpectedly, no significant effect of the 114-bp deletion on Su(Hw) recruitment was observed even in the absence of Mod(mdg4)-67.2. Thus, Mod(mdg4)-67.2 and CP190 are not required for the Su(Hw) binding to the mAcR-60C (Su(Hw)/CP190/Mod(mdg4)-67.2) and Syn2 (Su(Hw)/CP190) promoters. In the Su(Hw)Δ114 mutants, CP190 did not bind to the mAcR-60C promoter, confirming the role of Su(Hw) in the recruitment of this protein. In contrast, CP190 still bound to the Syn2 promoter in these mutants, suggesting that CP190 is recruited to this promoter independently of Su(Hw).
Mod(mdg4)-67.2 and CP190 proteins are not involved in repression. (a) Structural scheme of the Su(Hw) protein showing its domains and the corresponding numbers of amino acid residues: CID, CP190 interaction domain; Zf, zinc-finger domain; LZ, leucine zipper motif; CTAD, C-terminal acidic domain. Areas interacting with CP190 and Mod(mdg4)-67.2 proteins are indicated by arrowheads. The horizontal line below shows the Su(Hw) regions contained in the Su(Hw)Δ114 derivative, with the dotted fragment indicating the deletion. Numbers in brackets indicate the amino acid residues flanking the protein regions included in the derivative. (b) Binding of Su(Hw) and CP190 proteins to the promoters of model genes in pupae of transgenic lines expressing full-length Su(Hw)+ and Su(Hw)Δ114 in the absence (wt) or presence (mod−) of the mod(mdg4)u1/mod(mdg4)u1 mutation. Su(Hw)− refers to the y2 w1118scD1ct6; su(Hw)v/su(Hw)e04061 background. The Su(Hw)Δ114 derivative is designated Δ114. Different variants of the Su(Hw) protein were expressed in the lines y2 w1118scD1ct6; P{Su(Hw)}-38D/P{Su(Hw)}-38D; su(Hw)v/su(Hw)e04061, where P{Su(Hw)} refers to Su(Hw)+ – P{w+;UbqW-Su(Hw)1–945-FLAG}; or Su(Hw)Δ114 – P{w+;UbqW-Su(Hw)1–88/202–945-FLAG}. The ras64B coding region (ras) was used as a control devoid of Su(Hw) binding sites. The percent recovery of immunoprecipitated DNA (Y axis) was calculated relative to the amount of input DNA. Error bars indicate standard deviation of three independent biological replicates. (c) RT-qPCR analysis of model gene expression in the ovaries and heads in the absence or presence (mod−) of the mod(mdg4)u1/mod(mdg4)u1 mutation. Expression levels are shown as fold change relative to the Su(Hw)+ variant. Error bars indicate standard deviation of three independent biological replicates. Significance leves are shown for treatment comparisons indicated by horizontal brackets.
The RT-qPCR analysis in the ovaries and heads showed that the Su(Hw)Δ114 mutation did not lead to derepression of model genes (Fig. 4c). In contrast, we observed that in the case of the Rph promoter (Su(Hw) alone) and the mAcR-60C promoter (Su(Hw)/CP190/Mod(mdg4)-67.2) this mutation resulted in a noticeable decrease of transcription, suggesting the positive role of the corresponding Su(Hw) domain in stimulation of transcription from the promoters. Taken together, these results confirm that the Mod(mdg4)-67.2 and CP190 proteins are not involved in repression.
The C-terminal domain (aa 720–892) of Su(Hw) is responsible for repression of the promoter
Next, we focused on the C-terminal domain of the Su(Hw) protein that contains the acidic and enhancer-blocking regions (aa 892–945 and 716–892, respectively)28,29,30. To map the domains involved in transcriptional repression, we generated transgenes expressing different variants of Su(Hw) under control of the ubiqitin63-E promoter (Fig. 5a). The transgenes were inserted in the same 38D genomic region using the phiC31 integrase system57. The transgenes were crossed into the su(Hw)− (su(Hw)v/su(Hw)e04061 trans-heterozygous) background. Western blot analysis showed that all Su(Hw) variants were expressed at similar levels (Fig. S2). The Su(Hw)ΔC line expressed the protein with deletion of the terminal 53 aa, named acidic domain, which was previously shown to be involved in transcriptional repression32. The Su(Hw)Δ283 line expressed the protein lacking 16 aa (from 760 to 778) that are critical for the enhancer-blocking activity of Su(Hw)29,30. In addition, we generated two transgenes mimicking the su(Hw)J and su(Hw)e7 mutations described previously29. The Su(Hw)e7 and Su(Hw)J transgenes produced truncated proteins lacking 223 and 150 aa, respectively. Both deletions affect the domain that is essential for interaction with the C-terminal domain of Mod(mdg4)-67.228,31,34. ChIP with pupae showed that all Su(Hw) variants bound to the tested promoters at a level comparable to that of wild-type Su(Hw) (Fig. 5b).
The C-terminal region (720–892 aa) of Su(Hw) is responsible for repression. (a) Structural scheme of the Su(Hw) protein, with the lines below representing its derivatives included in analysis. All designations are as in Fig. 4. (b) ChIP-qPCR data on Su(Hw) binding to the promoters of model genes in the transgenic lines Su(Hw)+ (+), Su(Hw)ΔC (ΔC), Su(Hw)ΔN (ΔN), Su(Hw)Δ283 (Δ283), Su(Hw)e7 (e7), Su(Hw)J (J), and Su(Hw)− – y2 w1118scD1ct6; su(Hw)v/su(Hw)e04061 background (−). Different variants of the Su(Hw) protein were expressed in the lines y2 w1118scD1ct6; P{Su(Hw)}-38D/P{Su(Hw)}-38D; su(Hw)v/su(Hw)e04061, where P{Su(Hw)} refers to Su(Hw)+ – P{w+;UbqW-Su(Hw)1–945-FLAG}; Su(Hw)ΔC – P{w+;UbqW-Su(Hw)1–892-FLAG}; Su(Hw)ΔN – P{w+;UbqW-Su(Hw)238–945-FLAG}; Su(Hw)Δ283 – P{w+;UbqW-Su(Hw)1–760/778–945-FLAG}; Su(Hw)e7 – P{w+;UbqW-Su(Hw)1–720-FLAG}; or Su(Hw)J – P{w+;UbqW-Su(Hw)1–801-FLAG}. The ras64B coding region (ras) was used as a control devoid of Su(Hw) binding sites. The percent recovery of immunoprecipitated DNA (Y axis) was calculated relative to the amount of input DNA. Error bars indicate standard deviation of three independent biological replicates. Statistical analysis was performed relative to Su(Hw) binding in the Su(Hw)+ line. Significance leves are shown for treatment comparisons indicated by horizontal brackets. (c) RT-qPCR data on the expression of model genes in the ovaries and heads in the Su(Hw) mutant backgrounds. Expression levels are shown as fold change relative to those in the Su(Hw)+ transgenic line. Error bars indicate standard deviation of three independent biological replicates.
We then compared the effect of Su(Hw) mutants on the expression of model genes in the ovaries and heads (Fig. 5c). In Su(Hw)J, Su(Hw)e7, or Su(Hw)Δ283 females, their expression level in the ovaries proved to be increased up to 5-fold, as in su(Hw)− females, while that in the heads expectedly remained unchanged. In contrast, deletion of the C-terminal acidic domain had no effect on the repression activity of the mutant Su(Hw) protein. The expression of different Su(Hw) variants directly correlated with female fertility: Su(Hw)J, Su(Hw)e7, and Su(Hw)Δ283 females were sterile, while Su(Hw)ΔC females were fertile. These results show that the enhancer-blocking domain mediates the Su(Hw) dependent repression of the model promoters.
Discussion
We have mapped the 720–892 aa region of the Su(Hw) protein, which is responsible for tissue-specific repression of neural genes in the ovaries. This region overlaps with the Su(Hw) enhancer-blocking domain (aa 716–892) characterized previously. Our results show that Su(Hw) overexpression does not lead to increased repression of promoters in the head, suggesting that Su(Hw) itself is not involved in this process. The same deletions in Su(Hw) affect insulation, transcriptional repression and lead to female sterility. As found previously, one copy of the gypsy insulator fails to insulate the eye enhancer in the transgenic lines25. The Su(Hw) protein in the head fails to function either as an insulator or as a repressor. This provides a basis for the hypothesis that the same unknown complex bound to the 720–892 aa region of Su(Hw) is responsible for the promoter repression and enhancer blocking. It seems likely that this complex is bound to Su(Hw) in many tissues, with the exception of cells attributed to the nervous system. This hypothesis is confirmed by the observation that the gypsy insulator cannot block communication between the iab cis-regulatory domains and Abd-B promoter in the CNS58. In addition, the RNA-binding protein Shep has been shown to antagonize Su(Hw) activity in the CNS but not in other tissues35. It is possible that Shep competes with the hypothetical enhancer-blocking/repression complex for binding to the Su(Hw) protein.
The Mod(mdg4)-67.2 protein is not required for female fertility and is not involved in the Su(Hw)-mediated repression of the promoters56. At the same time, Mod(mdg4)-67.2 affects the enhancer-blocking activity of Su(Hw)-dependent insulators25,34,38,41,59,60. Mod(mdg4)-67.2 can interact with proteins responsible for long-distance communication between enhancers and promoters. For example, Mod(mdg4)-67.2 interacts with Zeste, which is essential for long-range interactions between the eye enhancer and white promoter25,61.
Interestingly, Mod(mdg4)-67.2 protein interacts with the 716–892 aa region of Su(Hw) that includes a leucine zipper motif and completely overlaps with the enhancer blocking/repression domain28,34. In the y2 model system62,63, the Su(Hw) complex acts as an insulator, while in the absence of Mod(mdg4)-67.2 it becomes a repressor41,64, suggesting that Mod(mdg4)-67.2 competes with the hypothetical enhancer-blocking/repression complex. This hypothesis is consistent with the observation that the genomic sites that bind the Su(Hw) protein in the absence of Mod (mdg4)-67.2 act as negative regulators of transcription52,54.
As shown previously, CP190 mediates protein–protein interactions necessary for the establishment of long-term contacts between insulator complexes46. Moreover, CP190 protein is critical for the functioning of the Fub insulator element that topologically separates the Ubx gene from the posterior abd-A gene60. CP190 is also involved in the recruitment of transcriptional complexes that open chromatin and stimulate transcription49,65. We have demonstrated that deletion of the Su(Hw) N-terminal domain responsible for the interaction with CP190 leads to a significant decrease in the expression of model genes, suggesting that CP190 is involved in transcription activation rather than in Su(Hw)-dependent repression.
The CP190 and Mod(mdg4)-67.2 proteins facilitate the binding of Su(Hw) to many SBS-CM sites and, in particular, to the gypsy insulator31,42. Su(Hw) binds to the mAcR-60C promoter in complex with CP190 and Mod(mdg4)-67.2. However, deletion of the Su(Hw) N-terminal domain even in combination with the mod(mdg4)u1 mutation has no effect either on Su(Hw) binding or on promoter repression. Thus, Su(Hw) recruitment to the mAcR-60C promoter does not depend on the presence of Mod(mdg4)-67.2 or CP190. It is noteworthy in this context that CP190 is recruited to the Syn promoter independently of Su(Hw). Our previous data show that multiple interactions between CP190, Mod(mdg4)-67.2, and Su(Hw) are involved in the formation of a stable DNA-bound chromatin complex31,32. Based on this fact, we hypothesize that only the Su(Hw)/Mod(mdg4)-67.2/CP190 complex or Su(Hw) in combination with as yet unknown proteins are bound to chromatin, while SBS-C sites are formed by independent binding of the Su(Hw) protein and CP190 in complex with another architectural protein.
It was previously speculated that Su(Hw) repression might depend on the recruitment of the LINT complex54. This transcriptional repressor complex of Drosophila contains three subunits: CoREST, lethal (3) malignant brain tumor protein (L(3)mbt), and L(3)mbt interacting protein 1 (dLint-1)66,67. As shown previously, over a third of CNS-enriched repressed target genes contain SBSs that colocalize with L(3)mbt and half of them contain SBSs that colocalize with dLint-154. Based on this finding, we examined whether L(3)mbt colocalized with the promoters of our model genes (Fig. S3). We analyzed L(3)mbt occupancy in the larval CNS and in Kc cells (female embryonic cell line) in comparison with Su(Hw) and CP190 localization53,67,68. We did not observe precise tissue specific occupancy of L(3)mbt on the tested promoters: this proteint was either enriched in the larval CNS and Kc cells (Rph) or absent in both cases (Hs3st-A, Syn2, and mAcR-60). It was only on the cg32017 promoter that L(3)mbt was enriched in Kc cells and not detected in the CNS. Apparently, L(3)mbt is not directly recruited to SBSs by Su(Hw). This assumption is confirmed by results of yeast two hybrid assay (Suppl. Table 1), which show that L(3)mbt directly interacted with CP190 but not with Su(Hw). Since CP190 is not directly involved in transcriptional repression, L(3)mbt is unlikely to play a general role in repression of tested genes. A recent screen of proteins interacting with HP1 has identified HIPP1 as a possible partner of Su(Hw)33. We have shown that HIPP1 dirrectly interacts with Su(Hw) region responsible for repression (unpublished data). As demonstrated redcently, however, HIPP1 is dispensable for the repression activity of Su(Hw)69. Taken together, these results suggest that additional, as yet unidentified complex is responsible for tissue-specific repression of neural genes in the ovaries. Further studies are needed to identify components of this complex and mechanism of its formation on SBSs.
Materials and Methods
The transgenic constructs are described in the Supplementary Materials.
Germ-line transformation, genetic crosses, and phenotypic analysis
All flies were maintained at 25 °C on the standard yeast medium. To obtain transgenic flies with insertion in 38D, the DNA of reporter constructs was injected into preblastoderm embryos of y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP′}ZH-38D genotype57. The generation of transgenic lines and construct introduction into the Su(Hw)v/Su(Hw)e04061 background were performed as described41.
Dissection of flies
For each replicate, approximately 100 ovaries from 4- to 6-hour-old virgin females were dissected as described previously51,52. After dissection, the flies were collected into a 50-mL Falcon tube, frozen in liquid nitrogen, and vigorously vortexes for 1 min. The contents of the tube were poured onto a sheet of white paper, and the heads of flies were separated from the bodies with a bird feather.
RNA isolation and RT-qPCR analysis
For RT-qPCR experiments, total RNA from the heads and ovaries of females was isolated with TRIzol reagent (Invitrogen). Genomic DNA was removed by treatment with DNase I (Fermentas, 1 U per 10 μg) followed by purification with a QIAGEN RNeasy kit. RNA was reverse transcribed into cDNA with a RevertAid H Minus RT Revert Transcriptase (Fermentas) following the manufacturer’s instructions. The resulting cDNA was analyzed by quantitative PCR (Bio-Rad CFX 96 Cycler) using SYBR Green. Relative steady-state mRNA levels were determined from the threshold cycle for amplification using the ΔΔCT method. Each experiment was performed in two to three independent biological replicates, and the results were averaged. The expression level of each gene was determined using ras64B as an internal control. Primer sequences used in real-time PCR analysis are listed in Suppl. Table 2.
Chromatin preparation and ChIP analysis
Chromatin was prepared either from the heads and ovaries of female flies or from middle pupae. The material (150–200 mg) was homogenized in 5 mL of buffer A (15 mM HEPES, pH 7.6 with 60 mM KCl, 15 mM NaCl, 4 mM MgCl2, 0.5% Triton X100, 0.5 mM DTT, 10 mM sodium butyrate, EDTA-free protease inhibitors cocktail, and 1.8% formaldehyde) at room temperature using first a Potter homogenizer and then a Dounce homogenizer with type A pestle (three strokes). The protease inhibitors cocktail (Roche, Cat #1873 580) was used following the manufacturer’s instructions. After 15 minutes (total time starting from beginning of homogenization), glycine solution was added to a concentration of 225 mM, and the mixture was stirred, incubated for 5 minutes, and then centrifuged at 4000 g and 4 °C for 5 minutes. The pellet was washed three times with buffer A at 4 °C and resuspended in 0.5 mL of lysis buffer (15 mM HEPES, pH 7.6 with 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.5 mM DTT, 0.1% sodium deoxycholate, 0.1% SDS, 0.5% N-lauroylsarcosine, 10 mM sodium butyrate, and protease inhibitors cocktail). The mixture was incubated on a rotary shaker at 4 °C for 10 minutes, sonicated on ice with a Branson Sonifier 450 (power 2, duty cycle 100%, time 4630 seconds at 2-second intervals), rotated for another 10 minutes, and then centrifuged at 15,000 g for 5 minutes. The supernatant was transferred to a new tube. The pellet was resuspended in 0.5 mL of lysis buffer, incubated on a rotary shaker for 10 minutes, and centrifuged again. The first and second supernatants were pooled and centrifuged twice at 15,000 g for 10 minutes. The supernatant was diluted with ten volumes of ChiP Dilution Buffer (16.7 mM Tris-HCl, pH 8.0 with 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, and 167 mM NaCl) and, to reduce nonspecific background, pre-cleared by incubation with protein A or protein G agarose beads for 30 min at 4 °C, with constant stirring. Agarose was pelleted by brief centrifugation, and the supernatant was collected for chromatin immunoprecipitation with appropriate antibodies (see below). After overnight incubation at 4 °C on a rotary shaker, protein A or protein G agarose beads were added to collect the precipitated complexes, and incubation was continued for 2 hours under the same conditions. Agarose was pelleted by centrifugation (700–1000 rpm at 4 °C, ~1 minute), the supernatant was carefully removed, and the pellet was washed with the following buffers (1 mL each, for 3–5 minutes on a rotary shaker): Low Salt Wash Buffer (20 mM Tris-HCl, pH 8.0 with 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 150 mM NaCl), High Salt Wash Buffer (20 mM Tris-HCl, pH 8.0, with 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 500 mM NaCl), LiCl Wash Buffer (10 mM Tris-HCl, pH 8.0, with 0.25 M LiCl, 1% NP40, 1% deoxycholate, and 1 mM EDTA,), and two portions of TE Buffer. The complex was removed from the agarose by two rounds of incubation with 250 μL of elution buffer (1%SDS, 0.1 M NaHCO3) at room temperature for 15 minutes, with rotation. The eluates were pooled, supplemented with 20 μL of 5 M NaCl, and heated at 65 °C for 4 hours to reverse the complex–DNA crosslinks. Then 20 μL of 1 M Tris-HCl (pH 6.5), 10 μL of 0.5 M EDTA, and 2 μL of Proteinase K solution (10 mg/mL) were added, and the mixture was incubated at 45 °C for 1 hour. DNA was recovered by phenol/chloroform extraction and ethanol precipitation and solubilized in water for PCR. PCR products were amplified from at least two separate immunoprecipitates from three different chromatin preparations. Primer sequences used in PCR for ChIP analysis are shown in Suppl. Table 3.
Antibodies
Specific antibodies and working dilutions were as follows: rat anti-CP190 (1:500), rabbit anti-Mod(mdg4)-67.2 (1:500), mouse anti-Mod-common (1:500), and rabbit anti Su(Hw) N-terminal domain (1:200) raised in our lab42,70; rabbit antibodies against the C-terminal domain of Su(Hw) (1:200) kindly provided by M. Erokhin32.
Data Availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information file.
References
Bouwman, B. A. & de Laat, W. Architectural hallmarks of the pluripotent genome. FEBS Lett., https://doi.org/10.1016/j.febslet.2015.04.055 (2015).
Chetverina, D., Aoki, T., Erokhin, M., Georgiev, P. & Schedl, P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays. 36, 163–172 (2014).
Ghavi-Helm, Y. et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 512, 96–100 (2014).
Gibcus, J. H. & Dekker, J. The hierarchy of the 3D genome. Mol Cell. 49, 773–782 (2013).
Maksimenko, O. & Georgiev, P. Mechanisms and proteins involved in long-distance interactions. Front Genet. 5, 28 (2014).
Schwartz, Y. B. & Cavalli, G. Three-Dimensional Genome Organization and Function in Drosophila. Genetics. 205, 5–24 (2017).
Ali, T., Renkawitz, R. & Bartkuhn, M. Insulators and domains of gene expression. Curr Opin Genet Dev. 37, 17–26 (2016).
Chetverina, D. et al. Boundaries of loop domains (insulators): Determinants of chromosome form and function in multicellular eukaryotes. Bioessays. 39, https://doi.org/10.1002/bies.201600233 (2017).
Ghirlando, R. & Felsenfeld, G. CTCF: making the right connections. Genes Dev. 30, 881–891 (2016).
Hnisz, D., Day, D. S. & Young, R. A. Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell. 167, 1188–1200 (2016).
Kyrchanova, O. & Georgiev, P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 588, 8–14 (2014).
Matzat, L. H. & Lei, E. P. Surviving an identity crisis: a revised view of chromatin insulators in the genomics era. Biochim Biophys Acta. 1839, 203–214 (2014).
Wang, Q., Sun, Q., Czajkowsky, D. M. & Shao, Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat Commun. 9, 188 (2018).
Yang, J. & Corces, V. G. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev. 22, 86–92 (2012).
Bender, W. et al. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 221, 23–29 (1983).
Golovnin, A. et al. Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila melanogaster. Mol Cell Biol. 27, 963–974 (2007).
Hoover, K. K., Gerasimova, T. I., Chien, A. J. & Corces, V. G. Dominant effects of suppressor of Hairy-wing mutations on gypsy-induced alleles of forked and cut in Drosophila melanogaster. Genetics. 132, 691–697 (1992).
Parkhurst, S. M. et al. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2, 1205–1215 (1988).
Peifer, M. & Bender, W. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 5, 2293–2303 (1986).
Modolell, J., Bender, W. & Meselson, M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci USA 80, 1678–1682 (1983).
Geyer, P. K. & Corces, V. G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6, 1865–1873 (1992).
Jack, J., Dorsett, D., Delotto, Y. & Liu, S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 113, 735–747 (1991).
Golovnin, A. et al. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 130, 3249–3258 (2003).
Holdridge, C. & Dorsett, D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol. 11, 1894–1900 (1991).
Kyrchanova, O. et al. Effective blocking of the white enhancer requires cooperation between two main mechanisms suggested for the insulator function. PLoS Genet. 9, e1003606 (2013).
Parnell, T. J. et al. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol. 26, 5983–5993 (2006).
Scott, K. C., Taubman, A. D. & Geyer, P. K. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 153, 787–798 (1999).
Ghosh, D., Gerasimova, T. I. & Corces, V. G. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20, 2518–2527 (2001).
Harrison, D. A., Gdula, D. A., Coyne, R. S. & Corces, V. G. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7, 1966–1978 (1993).
Kim, J., Shen, B., Rosen, C. & Dorsett, D. The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol Cell Biol. 16, 3381–3392 (1996).
Melnikova, L. et al. Multiple interactions are involved in a highly specific association of the Mod(mdg4)-67.2 isoform with the Su(Hw) sites in Drosophila. Open Biol. 7, https://doi.org/10.1098/rsob.170150 (2017).
Melnikova, L. et al. Interactions between BTB domain of CP190 and two adjacent regions in Su(Hw) are required for the insulator complex formation. Chromosoma. 127, 59–71 (2018).
Alekseyenko, A. A. et al. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 28, 1445–1460 (2014).
Gause, M., Morcillo, P. & Dorsett, D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 21, 4807–4817 (2001).
King, M. R., Matzat, L. H., Dale, R. K., Lim, S. J. & Lei, E. P. The RNA-binding protein Rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner. J Cell Sci. 127, 2956–2966 (2014).
Kurshakova, M. et al. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell. 27, 332–338 (2007).
Matzat, L. H., Dale, R. K., Moshkovich, N. & Lei, E. P. Tissue-specific regulation of chromatin insulator function. PLoS Genet. 8, e1003069 (2012).
Pai, C. Y., Lei, E. P., Ghosh, D. & Corces, V. G. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 16, 737–748 (2004).
Buchner, K. et al. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 155, 141–157 (2000).
Gerasimova, T. I., Gdula, D. A., Gerasimov, D. V., Simonova, O. & Corces, V. G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 82, 587–597 (1995).
Georgiev, P. & Kozycina, M. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics. 142, 425-436 (1996).
Golovnin, A., Volkov, I. & Georgiev, P. SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J Cell Sci. 125, 2064–2074 (2012).
Bonchuk, A., Denisov, S., Georgiev, P. & Maksimenko, O. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J Mol Biol. 412, 423–436 (2011).
Oliver, D., Sheehan, B., South, H., Akbari, O. & Pai, C. Y. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 11, 101 (2010).
Plevock, K. M., Galletta, B. J., Slep, K. C. & Rusan, N. M. Newly Characterized Region of CP190 Associates with Microtubules and Mediates Proper Spindle Morphology in Drosophila Stem Cells. PLoS One. 10, e0144174 (2015).
Vogelmann, J. et al. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 10, e1004544 (2014).
Ali, T. et al. Chromatin binding of Gcn5 in Drosophila is largely mediated by CP190. Nucleic Acids Res., https://doi.org/10.1093/nar/gkw1178 (2016).
Vorobyeva, N. E. et al. Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome. Nucleic Acids Res. 41, 5717–5730 (2013).
Bartkuhn, M. et al. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 28, 877–888 (2009).
Negre, N. et al. A cis-regulatory map of the Drosophila genome. Nature. 471, 527–531 (2011).
Negre, N. et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6, e1000814 (2010).
Schwartz, Y. B. et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22, 2188–2198 (2012).
Soshnev, A. A. et al. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 40, 5415–5431 (2012).
Soshnev, A. A., Baxley, R. M., Manak, J. R., Tan, K. & Geyer, P. K. The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development. 140, 3613–3623 (2013).
Duan, T. & Geyer, P. K. Spermiogenesis and Male Fertility Require the Function of Suppressor of Hairy-Wing in Somatic Cyst Cells of Drosophila. Genetics. 209, 757–772 (2018).
Baxley, R. M., Soshnev, A. A., Koryakov, D. E., Zhimulev, I. F. & Geyer, P. K. The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev Biol. 356, 398–410 (2011).
Bischof, J., Maeda, R. K., Hediger, M., Karch, F. & Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA 104, 3312–3317 (2007).
Hogga, I., Mihaly, J., Barges, S. & Karch, F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol Cell. 8, 1145–1151 (2001).
Gohl, D. et al. Mechanism of chromosomal boundary action: roadblock, sink, or loop? Genetics. 187, 731–748 (2011).
Savitsky, M., Kim, M., Kravchuk, O. & Schwartz, Y. B. Distinct Roles of Chromatin Insulator Proteins in Control of the Drosophila Bithorax Complex. Genetics. 202, 601–617 (2016).
Kostyuchenko, M. et al. Zeste can facilitate long-range enhancer-promoter communication and insulator bypass in Drosophila melanogaster. Chromosoma. 118, 665–674 (2009).
Geyer, P. K. & Corces, V. G. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1, 996–1004 (1987).
Geyer, P. K., Spana, C. & Corces, V. G. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5, 2657–2662 (1986).
Gdula, D. A. & Corces, V. G. Characterization of functional domains of the su(Hw) protein that mediate the silencing effect of mod(mdg4) mutations. Genetics. 145, 153–161 (1997).
Bohla, D. et al. A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking. PLoS One. 9, e107765 (2014).
Meier, K. et al. LINT, a novel dL(3)mbt-containing complex, represses malignant brain tumour signature genes. PLoS Genet. 8, e1002676 (2012).
Richter, C., Oktaba, K., Steinmann, J., Muller, J. & Knoblich, J. A. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat Cell Biol. 13, 1029–1039 (2011).
Van Bortle, K. et al. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 22, 2176–2187 (2012).
Glenn, S. E. & Geyer, P. K. Investigation of the Developmental Requirements of Drosophila HP1 and Insulator Protein Partner, HIPP1. G3 (Bethesda) 9, 345–357 (2019).
Golovnin, A. et al. ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 9, 440–445 (2008).
Acknowledgements
The authors are grateful to N.A. Gorgolyuk for his help in preparing the manuscript. All the experiments except the genetic analysis experiments were supported by the Russian Science Foundation (http://www.rscf.ru/), project no. 18-14-00295 (to A.G.). The genetic analysis experiments were supported by the Russian Foundation for Basic Research, project no. 16-04-00580 (to L.M.).
Author information
Authors and Affiliations
Contributions
L.M. generated transgenes, performed molecular and genetic analysis of flies, developed the design of experiments and prepared figures. ChIP analysis was performed by V.M. and L.M.; RT-qPCR assay, by M.K. and A.G.; statistical analysis of the results, by A.G. Experiments with mutations of Su(Hw) binding sites in the promoters of model genes and with overexpression of the Su(Hw) protein were performed by P.E., M.E. and D.C. A.G. and P.G. conceived the study, developed the design of experiments and wrote the manuscript. All authors participated in drafting the manuscript and have read and approved its final version.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Melnikova, L., Elizar’ev, P., Erokhin, M. et al. The same domain of Su(Hw) is required for enhancer blocking and direct promoter repression. Sci Rep 9, 5314 (2019). https://doi.org/10.1038/s41598-019-41761-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41761-6
This article is cited by
-
Su(Hw) interacts with Combgap to establish long-range chromatin contacts
Epigenetics & Chromatin (2024)
-
Su(Hw) primes 66D and 7F Drosophila chorion genes loci for amplification through chromatin decondensation
Scientific Reports (2021)
-
HIPP1 stabilizes the interaction between CP190 and Su(Hw) in the Drosophila insulator complex
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.