Abstract
Dryopteris fragrans (L.) Schott is a fern growing on the surface of hot rocks and lava. It is exposed to sunlight directly and bears local hot environment. We sequenced the complete nucleotide sequence of its chloroplast (cp) genome. The cp genome was 151,978 bp in length, consisting of a large single-copy region (85,332 bp), a small single-copy region (31,947 bp) and a pair of inverted repeats (17,314 bp). The cp genome contained 112 genes and 345 RNA editing sites in protein-coding genes. Simple sequence repeats (SSRs) and long repeat structure pairs (30–55 bp) were identified. The number and percent of repeat structures are extremely high in ferns. Thermal denaturation experiments showed its cp genome to have numerous, dispersed and high GC percent repeat structures, which conferred the strongest thermal stability. This repeat-heavy genome may provide the molecular basis of how D. fragrans cp survives its hot environment.
Similar content being viewed by others
Introduction
The chloroplast (cp) is a plant-specific and vital organelle that serves as the site of photosynthesis by converting light energy into chemical energy. This organelle is also involved in other biochemical processes, including sugar, amino acid, lipid, vitamin, starch and pigment syntheses, sulfate reduction and nitrogen metabolism1,2,3. The cp genome is typically circular and 120 to 160 kbp in size. Most cp genomes possess a similar structure. They typically contain a large single-copy region (LSC) and a short single-copy region (SSC) that are separated by two inverted repeats (IRs). The gene order and content of cp genomes are generally highly conserved, and the substitution rate in the cp DNA is less than that in nuclear DNA4,5. On the other hand, given their highly conserved sequences, similar structures and stable maternal heredity, cp genomes are a valuable and ideal resource for plant phylogenetics, population genetics, species identification and genetic engineering6. Gain and loss of function genes in the cp genome cause extensions or contractions, respectively, thus explaining genome size variations, which also reflect species differentiation events1.
Since the complete cp genome sequences of tobacco and liverwort were published7,8, an increasing number of seed plant cp genomes have been sequenced and reported8. With the development of next-generation sequencing (NGS), the cost of sequencing has been reduced, and the technique has become faster. NGS has the advantage of providing extremely high yield and accurate data on complete cp genomes. The number of sequenced cp genomes from various plants is increasing quickly. However, most complete current cp genome studies have focused on seed plants. Although ferns are a major group of plants, there are only 60 ferns for which complete cp genomes have been reported9,10. Other studies about ferns have been based on partial sequences or fragments of cp genomes11.
Dryopteris fragrans (L.) Schott is a perennial fern that grows on the surfaces of rocks and lava. It belongs to the Dryopteridaceae family and is found in the Far East, Europe and North America in small communities12. The Wudalianchi (N48°30′–48°51′, E126°00′–126o25′, Altitude 295–315 m) in Heilongjiang Province marks the centre of its distribution in China13. Its biotope is significantly different from that of other ferns. Most ferns prefer to grow in shady, warm and moist places. If those ferns are placed in dry and hot environments or exposed to ultraviolet (UV) ray for a short time, they quickly become crinkly and wilt or die. However, D. fragrans is exposed to sunlight and the dry surfaces of black rocks and lava directly. At the same time, it must endure strong UV rays and high surface temperatures of approximately 70 °C in summer. Therefore, this is a special species with superior stress resistance in ferns. This fern possesses special mechanisms to help it live in its severe environment. Previous studies of this species have mostly focused on its secondary metabolites and related genes14,15,16,17,18,19,20,21,22,23,24,25,26. Although lots of research has been performed on this species, its ability to survive in severe environments has largely been ignored. In our previous study, we partly sequenced its genome and obtained some contigs of its cp genome, which attracted our attention. Because the cp is the energy transducer for plants, it can induce changes in the external environment directly and react quickly. Studying the complete cp genome of D. fragrans may provide useful information about its superior heat resistance.
Here, we sequenced and annotated the complete cp genome of D. fragrans, and its cp genome features and structures were compared with related species. RNA editing sites were predicted by PREP in protein-coding genes and validated by transcripts. Simple sequence repeats (SSR) and long repeat structures were identified in this cp genome. The long repeat structures were extremely abundant compared with other species, and most were located in the intergenic spacer (IGS) region, which exhibited high GC content repeat structures and which may enhance cp genome stability. The thermal denaturation experiment showed that the D. fragrans cp genome exhibited strong thermal stability. These data would provide useful information and contribute to a better understanding of how this special fern lives in harsh environments. Furthermore, it will also be helpful in the study of secondary metabolism, genetic engineering, physiology and evolution within ferns and other species in the future.
Results
Chloroplast genome assembly and validation
Quantitative real-time polymerase chain reaction (qRT-PCR) result showed that the rbcL was detected in isolated cpDNA samples, while actin 6, the nuclear specific gene, was not detected (Supplemental Fig. 1). It showed that the cpDNA samples were pure. The sequencing run generated 2,740,440 raw reads, totaling 822,132,000 bases, with an average read length of 300 bp from the D. fragrans cp genome. A total of 2,262,910 clean reads with an average read length of 184.56 bp were de novo assembled into 31 contigs. The average sequence coverage depth of each nucleotide on the cp genome was 105X . A maximum scaffold size of 143,707 bp that spanned most of the small and large single copy region (SSC and LSC) and the entire inverted repeat (IR) region was generated. Because the IR region had double the coverage compared with the remaining scaffold, it was used twice in the complete cpDNA sequence. We submitted the annotated cp genome sequence of D. fragrans under GenBank accession number KX418656.2.
Chloroplast genome features and comparison
The cp genome of D. fragrans was 151,978 bp in length, with a typical quadripartite structure (Fig. 1). It included a pair of IRA and IRB of 17,314 bp separated by one SSC and one LSC of 31,947 bp and 85,332 bp, respectively. The D. fragrans cp genome contained 112 genes (Table 1), including 4 ribosomal RNA genes; 26 transfer RNA genes, and 22 genes encoding ribosomal subunits, of which 12 encoded the small subunit and 10 for the large subunit. It also included 3 genes encoding DNA-directed RNA polymerases, and 44 genes dedicated to photosynthesis, of which 11 encoded subunits of the NAD(P)H-quinone oxidoreductase, 4 encoded the photosystem I complex, 13 encoded the photosystem II complex, 6 encoded the cytochrome b/f complex, 6 encoded different subunits of the ATP synthase and 1 encoded the large chain of the ribulose bisphosphate carboxylase (rbcL). Three genes encoded the dark-operative protochlorophyllide oxidoreductase subunits; 5 genes (ycf1, 2, 3, 4, 12) were dedicated to open reading frames; 2 were detected to protease; 1 encoded a translational initiation factor and 5 for other proteins. Among them, 14 genes contained introns, including psaA, atpF, ndhA, ndhB, ndhE, ndhF, ndhG, rpl2, rpl20, rpoB, rpoC1, cemA, clpP and ycf3 (Table 1). Compared with Adiantum capillus-veneris, Pteridium subsp. aquilinum and Cyrtomium devexiscapulae, the D. fragrans cp genome gained orf42, trnR-ACG, rrn5, rrn4.5, rrn23, trnA-UGC and trnN-GUU in SSC. In addition, trnR-ACG, rrn5, rrn4.5, rps12 and trnI-GUG were lost in IRs, and ndhB was truncated in IRA. The psbK and trnG-UCC were lost in IRB-LSC (Fig. 2). The GC content of the cp genome was 43.15%. The IR region was the highest (44.18%), followed by the LSC (42.70%) and the SSC (43.26%). The GC contents in rRNA genes (55.75%) and tRNA genes (55.46%) were higher than those in protein coding regions (44.06%). The comparison of cp genome size, GC content, gene number and order is listed in Table 2. The D. fragrans cp genome did not contain tRNA for the amino acid codons Lys. The codon usage frequency was listed in Table 3. Among these codons, 987 (3.57%) encoding for glutamate and 166 (0.60%) for cysteine, were the most and the least amino acids codons, respectively.
The mapped D. fragrans (L.) Schott circular chloroplast genome. Genes presented outside of the outer circle are transcribed in a clockwise manner, and those inside are transcribed in a counter-clockwise manner. Functional categories of genes are colour-coded. The dashed area in the inner circle indicates the GC content of the chloroplast genomes.
Comparison of gene order and content in the LSC, IR, and SSC regions among four cp genomes. Compared with other species, the IR length of D. fragrans is shorter and its SSC is the longest. The D. fragrans cp genome lost ndhF and gained orf42, trnR-ACG, rrn5, rrn4.5, rrn23, trnA-UGC and trnN-GUU in SSC. The trnR-ACG, rrn5, rrn4.5, rps12 and trnI-GUG were lost in the IR regions, while ndhB was truncated in the IRA. The psbK and trnG-UCC were lost in IRB-LSC.
SSRs and repeat structures in the D. fragrans chloroplast genome
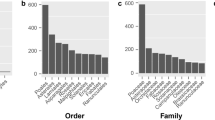
The MISA detected 44 SSRs in the D. fragrans cp genome (Supplemental Table 2), including 41 homopolymers and 3 dipolymers. Tetrapolymers, pentapolymers, and hexapolymers were not found. Sixteen SSRs were exclusively composed of A or T bases, 27 SSRs were G or C bases, and 1 was an AG base. Most of the bases were G or C bases, except for the AG dipolymer. All of these SSRs were located in the IGS, and none were located in protein-coding genes. Repeat analysis by REPuter, with the criterion of a copy size of ≥30 bp or longer and a sequence identity ≥90%, identified 80 forward, 1 reverse and 23 palindromic repeat structure pairs from 30 to 55 bp. Repeat lengths of 30 to 32 bp were most common (27.40%). A total of 53 repeat pairs were found in the coding regions, of which 6 were located in the transfer RNA genes. The remaining 151 repeat pairs were located in the IGSs (Supplemental Table 3). In addition, one of the longest repeat structure (55 bp) overlapped with the longest SSR sequence (18 bp G mononucleotide sequence) (Supplemental Table 3). The average GC content of repeat structures was 43.04%, with a maximum of 63.64% and minimum of 30%. To compare the number of repeat structures with that of other fern species, we extracted correlative sequences from 29 ferns to determine the number of repeat structures of different lengths. Repeat structures were abundantly distributed in the D. fragrans cp genome, and this species contained the most repeat structures among ferns (Fig. 3A). Compared with the other 29 genomes, the D. fragrans cp genome had the highest percentage of repeat structures (5.351%). It was higher than the other species by a factor of 1.50 to 21.28 (Fig. 3B). Thus, D. fragrans was rich in repeat structures.
Comparisons of repeat structure number and percent within 30 ferns. The sizes of the repeats are set at a repeat minimal length of ≥33 bp and maximal length of ≤55 bp with a Hamming distance of 3. The number and percentage of the repeat structures from 29 ferns were compared with those of D. fragrans. (A) The number of repeat structure in the D. fragrans cp genome was compared to that of 29 ferns. D. fragrans possesses the most repeat structures; (B) The percent of repeat structures in the D. fragrans cp genome was compared to that of 29 ferns. D. fragrans possesses the highest repeat structure percent (5.351%).
RNA editing
The transcripts obtained by PCR were used to verify the RNA editing sites predicted by PREP. The PREP prediction results showed that there were 438 RNA editing sites in protein-encoding genes, corresponding to 338 codon changes. All editing events were of the C-to-U variety. Among them, 96 non-synonymous mutations were found at the first position of the codon, while the remaining mutations were found at the second position, and none were found at the third position. However, in the transcript validation results, there were 345 RNA editing sites in the D. fragrans cp genome (Supplemental Table 4). In all, 88 mutations occurred in the first position of the codon, 208 at the second position, and 49 at the third position. The C-U mutations were the most common, reaching 305 (88.41%) mutations, followed by U-C 13 (3.77%), G-A 8 (2.32%), A-G 7 (2.03%), A-C 3 (0.87%), C-G 2 (0.58%), G-U 2 (0.58%), U-A 2 (0.58%), U-G 2 (0.58%) and G-C 1 (0.29%) (Supplemental Fig. 2). There were 318 codon changes, including 33 synonymous mutations and 285 nonsynonymous mutations. The majority of editing sites were predicted in the ndhF gene (130629-123964, 41 editing sites), followed by the atpB gene (72943-71462, 21 editing sites). The conversions of amino acids included 119 hydrophilic to hydrophobic changes (H to Y, S to L, S to F, and T to M) and 105 hydrophobic to hydrophobic changes (L to F, A to V, P to S, R to W and P to L). The codons that turned into leucine (Leu) were the most common, accounting for 125 changes (39.31%). The number of RNA editing sites (TCA (S)-TTA (L)) were the most frequent (37 editing sites), followed TCG(S)-TTG(L) (28 editing sites) and CCA(P)-CTA(L) (21 editing sites).
Comparison of cpDNA thermal stability
To confirm that repeat structures with high GC content contributed enhanced the cp genome stability of D. fragrans, the thermal denaturation for all species cp genomes were completed. In the denaturation experiment, the absorbance of all samples increased with elevated temperatures (Fig. 4). The percentage increase of all ferns was under or approximately 20% at 35 °C. Some rose quickly, such as Arabidopsis, wheat, and T. palustris. Most samples began to go up quickly at 35 °C and sharply rose at 55 °C. Almost all cp genomes could not bear 75 °C and their absorbance increased greatly. However, only D. fragrans maintained it from beginning to 55 °C and changed slightly from 65 °C to 75 °C. Even at 85 °C, D. fragrans still kept the lowest value compared to the others. It showed that its cp genome could cope with heating.
Discussion
DOGMA is the most popular software for cp genome annotation and is used widely27,28,29,30. This software can detect protein-coding genes, rRNA, and tRNA quickly. However, it also has drawbacks in detecting genes, because its ability to detect introns is not very sensitive. In our work, DOGMA annotated 110 genes but did not detect genes with intron(s). We performed another software analysis using MAKER-P. It detected 9 genes (ndhF, rpl21, rps6, cemA, ccsA, lhbA, matK, ycf1 and ycf12) that were not annotated by DOGMA. It also identified 14 genes containing intron(s), and their positions were also corrected (Table 1). The genes in the fern cp genomes are different from those of seed plants, although both of them are vascular plants. This made DOGMA perform not very well in ferns and produced incorrect result. The MAKER-P showed an advantage in the detection of protein-coding genes and introns. Thus, annotation for fern cp genomes requires the use of different software programs.
The typical size of fern cp genome is 131 to 168 kb31,32, and the D. fragrans cp genome is within this range. The gene number and order are largely similar to the cp genomes of ferns, but there are some differences among species. The genome size variation is mostly due to length variation in the IR and the SSC regions28. Some IR expansions/contractions are observed within species. Compared to other ferns, the IR regions of the D. fragrans cp genome lost a 4033 bp sequence, including trnR-ACG, rrn5, rrn4.5, rrn23, trnA-UGC and trnN-GUU. This sequence was located in IRs of other fern cp genomes. However, this sequence in the D. fragrans cp genome was moved into SSC. Thus, these genes did not exist as two copies in the D. fragrans cp genome. It is possible that the fern reduced the expression of these genes. The phenomenon causes the D. fragrans cp genome to contain the longest SSC and shorter IRs (Table 2). Alhough synteny and inversions are important, the structure of the D. fragrans cp genome does not show obvious changes. It is consistent with results of Xiang et al.9. Furthermore, overlapped genes are not notable (Fig. 1), which would reduce the cp genome usage. Thus, the D. fragrans cp genome has more intergenic sequences, leading to a more dispersed gene distribution and increasing the sequence length. These findings suggest that the fern cp genome chooses reduces the overlapping genes but extends the intergenic sequences. Thus, genes are more independent and sequence utilization is more specific.
RNA editing is an important post-transcriptional process in cps and is thought to be functionally significant33. In our work, the phenomenon was obvious in protein-coding genes. There were great disparities between the results of PREP and transcript validation. The number of RNA editing sites and codon transformations in the PREP results were far more than those of the validation. This result indicates that there may be some deficiencies in PREP, though the prediction results conformed to the number and quantity of predicted variations in general. This may be because the PREP database is not abundant enough, especially for ferns. Moreover, the result shows that transcript verification is necessary for RNA editing site prediction. In seed plant cps, a conversion from C-to-U is the most predominant form34, and reverse U-to-C editing is the opposite in seed plants35. Most editing events in the D. fragrans cp genome were C-to-U (88.4%) events. At the same time, its C-to-U transition is the most frequent type of base change. It has been reported that the excess of C-to-U RNA editing developed in early stages of vascular plant evolution36. Our results support this view. On the other hand, the number and percent of codons transitioning to Leu were the highest in the D. fragrans cp genome. It is similar with those of Adiantum capillus-veneris, though the genetic distance between A. capillus-veneris and D. fragrans is long in the fern clade. Leu biosynthesis occurs in cp and plays an important role in photosynthesis-related metabolism37,38. Both species account for a heavily used Leu codon, suggesting they have a great need for Leu. Their level of RNA editing is more than ten times that of any other vascular plant examined across an entire cp genome39. This reflects the fact that RNA editing occurs in different fern species and may play a major role in fern cp and cp genome processing.
Simple sequence repeats (SSRs) ranging in length from 1–6 or or more base pairs, also known as microsatellites and short tandem repeats (STRs), are important genetic molecular markers for population genetics40,41 and are widely used for plant genotyping42,43. In this work, there were 44 SSRs in the D. fragrans cp genome. The number of GC SSRs was more than the number of AT SSRs. This finding contrasts with the view that cp SSRs are generally composed of short polyadenine (poly A) or polythymine (poly T) repeats and rarely contain tandem guanine (G) or cytosine (C) repeats44. On the other hand, the number and percent of repeat structure (30–55 bp) in the D. fragrans cp genome were far more than other species (Fig. 3). This is the first time that a fern species has been shown to contain a considerable number of repeat structures. It shows that its cp genome is rich in repeat structures. At the same time, most repeat structures were located in every IGS dispersedly, and the GC percentages of most repeat structures were higher than the average value (43.04%). This indicates that the dispersed repeat structures probably play a key role in maintaining cp genome stability. D. fragrans may increase the IGS number and length of inserted repeat structures with a high active GC content. Previous has work suggested that repeat structures are very important for sequence rearrangement and variation in cp genomes by preventing illegitimate combinations and slipped-strand mispairing1,45,46. Our results could support this point of view. Furthermore, these repeat structures have also became a part of the intergenic sequences between genes, resulting in the independence of each gene. This feature allows for the selective expression of genes.
Wudalianchi was formed in great volcanic eruption. Its physiognomy is mainly consist of alkaline basalt47. This is a kind of black volcanic rock with low specific heat capacity (0.84 kJ/(kg·K)) and small thermal conductivity. The basalt would absorb lots of heat quickly under direct and long sunshine in summer. It could result in high surface temperature easily and form a local hot environment in the range of basalt geomorphology. The temperature of basalt surface in summer often reaches 70 °C. On the other hand, the basalt topography cools quickly at night. D. fragrans grows on the exposed basalt surface and is exposed to large temperature fluctuations between day and night. Most ferns cannot endure such high temperature and dramatic temperature changes, but D. fragrans is a rare fern and is highly resistant to heat. In the results mentioned above, our study revealed this fern possesses the greatest number of repeat structures, with a high GC percentage, among all ferns studied. The three hydrogen bonds between GC are stronger than the two between AT, such that the GC percentage determines the strength of the DNA double chain (i.e., loose or tight). Some researchers have noted that the higher the level of GC content, the more stable the structure of the genome DNA48. We speculate that these repeat structures with high GC content may allow the fern to cope with heat and large temperature differences. Thus, we performed a heat denaturation experiment to compare the cpDNA thermal stability of ferns species and closely related species from different habitats and families, including Nephrolepidaceae, Thelypteridaceae, Pteridaceae, Davalliaceae, Aspleniaceae, Polypodiaceae, Dryopteridaceae, Dennstaedtiaceae, Parkeriaceae and Isoetaceae. Arabidopsis and wheat showed the most obvious changes in the denaturation experiment. This indicates that their cpDNA is very sensitive to heat. S. sessilifolia, T. palustris, C. fortunei, I. sinensis and C. thalictroides changed earlier and largely under 45 °C. The habitats of these five species are swamps or humid underforest environments. Their heat resistance was weak. D. chingia, N. biserrata, P. scolopendrium, M. strigosa and P. amoena showed smaller variations and similar heat stability between 35–45 °C. However, most of them could not survive at temperatures over 45 °C and began to rise significantly with the increase of temperature (Fig. 4). D. chingia, P. amoena and M. strigosa are saxicolous ferns in forest. Their cpDNA showed heat resistance and their thermal stability was limited. In addition, there were great differences between D. fragrans and C. fortunei in terms of their thermal stability, although both of them belong to the Dryopteridaceae family. This suggests that great differences exist within species of the same family, which is caused by different environments. These results support the speculation that a considerable number of dispersed repeat structures with a high GC content (43.04%) enhance D. fragrans cpDNA thermal stability and maintain its structure in the face of thermal changes. It is one of molecular basis of D. fragrans in response to severe environments. It also provided a new scope for understanding the environmental adaption mechanisms of plants.
Methods
Plants, cp DNA extraction, sequencing and assembly
Fresh leaves of D. fragrans from Wudalianchi, Heilongjiang Province were collected and frozen in liquid nitrogen after cleaning. Professor Baodong Liu of Harbin Normal University provided the leaves of Nephrolepis biserrata, Polypodiodes amoena, Isoetes sinensis, Cyrtomium fortunei, Phyllitis scolopendrium, Davallodes chingiae, Scutellaria sessilifolia, Microlepia strigosa, and Thelypteris palustris. The Arabidopsis thaliana, wheat (Triticum aestivum L.) and Ceratopteris thalictroides were collected in our lab. The cp isolation methods were modified based on previous methods49,50,51. Five grams of complete leaves from all species were picked and rinsed. The leaves were then crushed in liquid nitrogen and added to a separation solution (0.33 M D-Sorbitol, 50 mM Tris-HCl pH 7.6, 5 mM MgCl2, 10 mM NaCl, 2 mM EDTA, 2 mM D-sodium erythorbat and 0.2% beta-mercaptol) for grinding. The cell suspensions were filtered, and the filtrate was centrifuged at 1000 rpm for 10 min to eliminate large-sized cell fragments. The supernatant was collected and centrifuged at 4000 rpm for 10 min to separate the cp. We obtained cps and extracted pure cpDNA using the CTAB-based method52. Every DNA sample was treated with RNase. To assess the contamination of the nuclear genome, nuclear special gene, actin6, and cp special gene, rbcL, were selected to performed qRT-PCR. The specific primer pairs or degenerate primer pairs were designed based on special sequence or homologous sequences (Supplemental Table 1). LineGene 9620 instrument (HANGZHOU BIOER TECHNOLOGY Co., LTD. China) and TransStart Green qPCR SuperMix (TRANSGEN BIOTECH, China) were used for detection. The qRT-PCR program was set as follows, 5 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at 60 °C. The cpDNA samples of D. fragrans cp genome were sequenced using Illumina technology on HiSeq. 2000 at Genewiz (China). To improve the Illumina sequence read quality and accuracy of the sequences, we performed Trimmomatic (version 0.30)53 to optimize the processing for filtering the adaptor sequence. The software SSPACE (version 3.0)54, GapFiller (version 1.10)55 and Velvet (version 1.2.10)56 were used to examine the raw reads and assemble them into contigs and scaffolds with default parameters. There were some gaps left after assembly. To finish the assembly of the whole genome, gaps were filled by PCR. PCR reactions were performed in a total volume of 20 μL containing 6 μL of deionized sterile water, 10 μL of EasyTaq Mix buffer, 1 μL of each primer at 10 pmol/μL (TransGen Biotech, Beijing, China) and 2 μL of cp DNA. PCR products were purified and sequenced by Bio-Serve (Harbin, China). All primers used for gap filling are listed in Supplemental Table 1.
Chloroplast genome annotation and comparative analyses
Gene location and annotations of the D. fragrans cp were performed using the Dual Organellar GenoMe Annotator (DOGMA) (http://dogma.ccbb.utexas.edu)57 and MAKER-P58,59, including protein-coding and rRNA and tRNA genes. All genes, rRNAs, and tRNAs were identified using the plastid/bacterial genetic code. The predicted annotations were verified using Chloroplast Genome DB (http://chloroplast.cbio.psu.edu/)60 and Blast61. tRNAscan-SE was used to identify the tRNAs62. Codon usage and relative synonymous codon usage (RSCU) were calculated by CodonW 1.4.2 (http://codonw.sourceforge.net)63. The annotated sequence was submitted to GenBank. The circular gene map of the D. fragrans cp was generated using OGDRAW64.
The GC%, LSC, SSC, IR regions, gene number and length of complete genome of the D. fragrans cp genome were compared to the cp genomes from Adiantum capillus-veneris (NC_004766), Osmundastrum cinnamomeum (NC_024157), Cyrtomium devexiscapulae (KT599100), Woodwardia unigemmata (KT599101), Alsophila spinulosa (NC_012818), Psilotum nudum (NC_003386), Pteridium aquilinum subsp. aquilinum (NC_104348), Angiopteris evecta (NC_008829), Isoetes flaccida (NC_014675), Huperzia lucidula (NC_006861), Athyrium anisopterum (NC_035738.1), Athyrium opacum (NC_035841.1), Austroblechnum melanocaulon (NC_035840.1), Deparia lancea (NC_035844.1), Diplazium dushanense (NC_035851.1), Homalosorus pycnocarpos (NC_035855.1), Macrothelypteris torresiana (NC_035858.1), Matteuccia struthiopteris (NC_035859.1), Onoclea sensibilis (NC_035861.1), Pseudophegopteris aurita (NC_035861.1), Ginkgo biloba (AB684440), Arabidopsis thaliana (NC_000932) and Oryza sativa (NC_001320). Furthermore, we compared the borders, gene content and order of the LSC, SSC and IRs regions with those of A. capillus-veneris, P. aquilinum subsp. aquilinum and C. devexiscapulae.
Examination of the repeat sequences and RNA editing
MISA, a microsatellite identification tool (http://pgrc.ipk-gatersleben.de/misa/misa.html), was used to detect SSRs65, with thresholds of mononucleotide repeats ≥10 bases, dinucleotide repeats ≥6 bases, tri- and tetranucleotide repeats ≥5 bases, and hexanucleotide or greater repeats ≥5 bases. The max distance between two SSRs was 100 base pairs. Based on these analyses, we identified the location of the SSRs. The REPuter program66 was used to assess long repeat sequences on the forward, reverse and palindrome sequences within the cp genomes. The sizes of the repeats were set at a repeat minimal length of ≥30 bp and a maximal length of ≤55 bp with a Hamming distance of 3. Furthermore, we selected 29 ferns, including Adiantum capillus-veneris, Alsophila spinulosa, Angiopteris evecta, Athyrium opacum, A. sinense, A. sheareri, Austroblechnum melanocaulon, Cyrtomium falcatum, C. devexiscapulae, Deparia pycnosora, Diplaziopsis cavaleriana, Diplazium dilatatum, D. dushanense, D. lancea, D. striatum, Dryopteris decipiens, Homalosorus pycnocarpos, Lygodium japonicum, Macrothelypteris torresiana, Matteuccia struthiopteris, Marsilea crenata, Osmundastrum cinnamomeum, Onoclea sensibilis, Psilotum nudum, Pteridium aquilinum subsp. aquilinum, Schizaea elegans, S. pectinat, Thelypteris aurita and W. unigemmata to calculate the long repeat sequences using the same parameters. We compared differences in the repeat numbers of different lengths from those ferns.
Prediction and Transcript validation of RNA editing sites
The predictive RNA Editor for Plants (PREP) was used to predict potential RNA editing sites in protein-coding genes with a cutoff value of 0.867. The protein-coding genes were accD, atpA, atpB, atpI, ccsA, clpP, matK, ndhB, ndhD, ndhF, ndhG, petB, petD, petD, petL, psaI, psbB, psbE, psbF, psbL, rpoA, rpoB, rpoC1, rps14, rps2, rps2 and ycf3. Total RNA was isolated from leaves using Tiangen™ Plant Total RNA Kit (China). The quality and concentration of RNA samples were examined by agarose gel electrophoresis and spectrophotometer analysis, respectively. The first-strand cDNA was prepared with 3 μg of total RNA using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (Transgen Biotech, China). Primer pairs for each gene were designed based on extracted gene sequences and are listed in Supplemental Table 5. The PCR was carried out as follows: 5 min at 95 °C, 30 cycles of 30 s at 95 °C, 30 s at 56–63 °C, 60 s at 72 °C and 10 min at 72 °C. The PCR products were sequenced at HaiGene (Harbin, China). The sequences were aligned with extracted gene sequences. The RNA editing sites validated by PCR were collected and compared to the PREP results.
Thermal denaturation and renaturation of cp genomes
DNA denaturation produces hyperchromic effect. The isolated cpDNA from all species was used. For denaturation, the absorbance increase could reflect the thermal stability of cpDNA by gradient thermal treatment. All cpDNA from species were dissolved in Tris-EDTA (TE) buffer. The concentration was adjusted to 50 ng/mL. Absorbance at 260 nm was used to monitor the denaturation processes, and the TE buffer was used as a blank control. Each sample was treated in different temperatures water bath (25 °C, 35 °C, 45 °C, 55 °C, 65 °C, 75 °C, 85 °C, 95 °C), and each temperature treatment went for 10 min. The absorbance of the initial temperature treatment was set as A0 (15 °C), and the value of highest temperature treatment was set at Amax (95 °C). The cp DNA was heated to 95 °C in order to melt the DNA completely and determine limit values for cp genomes, such that the standards for each cp genome can be provided. The formula to determine the increase percentages of the absorbance increase (AI) of the hyperchromic effects as: AI = (Atemp - A0)/(Amax - A0) × 100%. The recorded data were repeated 3 times and collected to calculate.
References
Bausher, M. G., Singh, N. D., Lee, S. B., Jansen, R. K. & Daniell, H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 6, 21 (2006).
Jarvis, P. & Soll, J. Toc, Tic, and chloroplast protein import. Biochim. Biophys. Acta. 1541, 64–79 (2001).
Leister, D. Chloroplast research in the genomic age. Trends Genet 19, 47–56 (2003).
Korpelainen, H. The evolutionary processes of mitochondrial and chloroplast genomes differ from those of nuclear genomes. Naturwissenschaften 91, 505–518 (2004).
Wolfe, K. H., Li, W. H. & Sharp, P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84, 9054–9058 (1987).
Catherine, J. N., Abdul, B. & Graham, J. K. Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae. BMC Genomics 15(Suppl 9), S13 (2014).
Shinozaki, K. et al. The complete nucleotide sequence of tobacco chloroplast genome: its gene organization and expression. EMBO. J. 5, 2043–2049 (1986).
Ohyama, K. et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574 (1986).
Lu, J. M., Zhang, N., Du, X. Y., Wen, J. & Li, D. Z. Chloroplast phylogenomics resolves key relationships in ferns. Journal of Systematics and Evolution 53(5), 448–457 (2015).
Wei, R. Plastid Phylogenomics Resolve Deep Relationships among Eupolypod II Ferns with Rapid Radiation and Rate Heterogeneity. Genome Biol. Evol. 9 ( 6 ),1646–1657. https://doi.org/10.1093/gbe/evx107.
Zhang, L. B. et al. Molecular circumscription and major evolutionary lineages of the fern genus Dryopteris (Dryopteridaceae). BMC Evolutionary Biology 12, 180 (2012).
Widen, C. J. & Britton, D. M. Chemotaxonomic investigations on Dryopteris fragrans. Can. J. Bot. 49, 989–992 (1971).
Shen, Z. B., Luo, W. Y., Yan, Y. S. & Zhu, J. F. Studies on phloroglucinol derivatives of Dryopteris fragrans L. Zhong Yao Cai 29, 560–561 (2006).
Kuang, H. et al. A new phenolic glycoside from the aerial parts of Dryopteris fragrans. Fitoterapia 79, 319–320 (2008).
Zhao, D. D. et al. Compounds from Dryopteris Fragrans (L.) Schott with Cytotoxic Activity. Molecules 19, 3345–3355 (2014).
Ying, Z. et al. Dryofragin, a phloroglucinol derivative, induces apoptosis in human breast cancer MCF-7 cells through ROS-mediated mitochondrial pathway. Chemico-Biological Interactions 199, 129–136 (2012).
Li, X. J. et al. Preparative separation of dryofragin and aspidin BB from Dryopteris fragrans extracts by macroporous resin column chromatography. Journal of Pharmaceutical and Biomedical Analysis 61, 199–206 (2012).
Yao, S. et al. Aspidin BB, a phloroglucinol derivative, induces cell cycle arrest and apoptosis in human ovarian HO-8910 cells. Chemico-Biological Interactions 204, 88–97 (2013).
Haixue, K. et al. Three drimane sesquiterpene glucoside from the aerial parts of Dryopteris fragrans (L.) Schot. Fitoterapia 80, 134–137 (2009).
Lee, K. K., Kim, J. H., Cho, J. J. & Choi, J. D. Inhibitory effects of 150 plant extracts on elastase activity, and their anti-inflammatory effects. Int. J. Cosmet. Sci. 21, 71–82 (1999).
Fan, H. Q. et al. Study on antifungal susceptibility of different extract of Dryopteris fragrans. Zhong Yao Cai 35, 1981–1985 (2012).
Huang, Y. H. et al. Characterization of a new sesquiterpene and antifungal activities of chemical constituents from Dryopteris fragrans (L.) Schott. Molecules 19, 507–513 (2013).
Li, B., Zhu, J. F., Zou, Z. J., Yin, Y. Q. & Shen, Z. B. Studies on the chemical constituents of Dryopteris fragrans. Zhong Yao Cai 32, 1232–1233 (2009).
Gao, R. Cloning and Expression the Gene of Squalene Synthase from Dryopteris fragrans (L.) Schott. Northeast Agricultural University, Heilongjiang Province, PRC (2014).
Li, S. S., Li, Y., Sun, L. L., Hu, B. Z. & Chang, Y. Identification and expression analysis of 4-Coumarate Coenzyme A ligase gene family in Dryopteris fragrans. Cell. Mol. Biol. 61(4), 25–33 (2015).
Li, Y. et al. Cloning and expression analysis of phenylalanine ammonia-lyase (PAL) gene family and cinnamate 4-hydroxylase (C4H) from Dryopteris fragrans. Biologia 70(5), 606–614 (2015).
Wu, Z., Gu, C., Tembrock, L. R., Zhang, D. & Ge, S. Characterization of the whole chloroplast genome of Chikusichloa mutica and its comparison with other rice tribe (Oryzeae) species. PLoS One 12, 5:e0177553, https://doi.org/10.1371/journal.pone.0177553 (2017).
Asaf, S. et al. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci Rep. 7, 7556, https://doi.org/10.1038/s41598-017-07891-5 (2017).
Wang, C. L. et al. Comparative Analysis of Four Buckwheat Species Based on Morphology and Complete Chloroplast Genome Sequences. Sci Rep. 7, 6514, https://doi.org/10.1038/s41598-017-06638-6 (2017).
Liu L. X. et al. The Complete Chloroplast Genome of Chinese Bayberry (Morella rubra, Myricaceae): Implications for Understanding the Evolution of Fagales. Front Plant Sci., 8, https://doi.org/10.3389/fpls.2017.00968 (2017).
Grewe, F., Guo, W., Gubbels, E. A., Hansen, A. K. & Mower, J. P. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evolutionary Biology 13, 8 (2013).
Zhong, B. J., Fong, R., Collins, L. J., McLenachan, P. A. & Penny, D. Two new fern chloroplasts and decelerated evolution linked to the long generation time in tree ferns. Genome Biology and Evolution 6, 1166–1173 (2014).
Sasaki, Y., Kozaki, A., Ohmori, A., Iguchi, H. & Nagano, Y. Chloroplast RNA Editing Required for Functional Acetyl-CoA Carboxylase in Plants. J. Biol. Chem., Feb 9 276(6), 3937–40 (2001).
Chen, H., Deng, L., Jiang, Y., Lu, P. & Yu, J. RNA editing sites exist in protein-coding genes in the chloroplast genome of Cycas taitungensis. J. Integr. Plant Biol. 53, 961–970 (2011).
Knie, N., Grewe, F., Fischer, S. & Knoop, V. Reverse U-to-C editing exceeds C-to-U RNA editing in some ferns - a monilophyte-wide comparison of chloroplast and mitochondrial RNA editing suggests independent evolution of the two processes in both organelles. BMC Evol Biol. 16, 1, https://doi.org/10.1186/s12862-016-0707-z (2016).
Tsunewaki, K., Matsuoka, Y., Yamazaki, Y. & Ogihara, Y. Evolutionary dynamics of wheat mitochondrial gene structure with special remarks on the origin and effects of RNA editing in cereals. Genes Genet. Syst. 83(4), 301–20 (2008).
Knill, T., Reichelt, M., Paetz, C., Gershenzon, J. & Binder, S. Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 71, 227–239 (2009).
He, Y. et al. A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 60, 679–690 (2009).
Paul, W., Carol, R. & Mitsuyasu, H. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene, Sep 15(339), 89–97 (2004).
Doorduin, L. et al. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18, 93–105 (2011).
He, S., Wang, Y., Volis, S., Li, D. & Yi, T. Genetic diversity and population structure: implications for conservation of wild soybean (Glycinesoja Sieb.et Zucc) based on nuclear and chloroplast microsatellite variation. Int. J. Mol. Sci. 13, 12608–12628 (2012).
Xue, J., Wang, S. & Zhou, S. L. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae). Am. J. Bot 99, e240–e244 (2012).
Yang, A. H., Zhang, J. J., Yao, X. H. & Huang, H. W. Chloroplast microsatellite markers in Liriodendron tulipifera (Magnoliaceae) and cross-species amplification in L. chinense. Am. J. Bot 98, e123–e126 (2011).
Kuang, D. Y. et al. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome 54, 663–673 (2011).
Saski, C. et al. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 115, 571–590 (2007).
Huang, H., Shi, C., Liu, Y., Mao, S. Y. & Gao, L. Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol.Biol. 14, 151 (2014).
Ma Z. J. Experimental study on physical weathering of basalt in Wudalianchi, Heilongjiang province. China University of Geosciences, Beijing, PRC (2018).
Nicolas, G. & Lobry, J. Relationships Between Genomic G + C Content, RNA Secondary Structures, and Optimal Growth Temperature in Prokaryotes. Journal of Molecular Evolution 44, 632–636 (1997).
Mills, W. R. & Joy, K. W. A rapid method for isolation of purified, physiologically active chloroplasts, used to study the intracellular distribution of amino acids in pea leaves. Planta 148, 75–83 (1980).
Sandbrink, J. M. et al. A method for evolutionary studies of RFLP on chloroplast DNA, applicable to a range of plant species. Biochem. Syst. Ecol. 17, 45–49 (1989).
Triboush, S. O., Danilenko, N. G. & Davydenko, O. G. A method for isolation of chloroplast DNA and mitochondrial DNA from sunflower. Plant Mol. Biol. Rep. 16, 183–189 (1998).
Doyle, J. J. & Doyle, J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15 (1987).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D. & Pirovano, W. Scaffolding preassembled contigs using SSPACE. Bioinformatics 27(4), 578–579 (2010).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18, 821–829 (2008).
Wyman, S. K., Jansen, R. K. & Boore, J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–5 (2004).
Campbell, M. S. et al. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 2, https://doi.org/10.1104/pp.113.230144 (2014).
Campbell, M. S., Holt, C., Moore, B. & Yandell, M. Genome Annotation and Curation Using MAKER and MAKER-P. Curr Protoc Bioinformatics 12, 48, https://doi.org/10.1002/0471250953.bi0411s48 (2014).
Cui, L. et al. Chloroplast DB: the chloroplast genome database. Nucleic Acids Research 34, 692–696 (2006).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. Mol Biol. 215, 403–410 (1990).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Chao, F. et al. Codon usage patterns in Chinese bayberry (Myrica rubra) based on RNA-Seq data. BMC Genomics 14, 732 (2013).
Lohse, M., Drechsel, O. & Bock, R. Organellar Genome DRAW (OGDRAW) - a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274 (2007).
Thiel, T., Michalek, W., Varshney, R. K. & Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422 (2003).
Kurtz, S. et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642 (2001).
Mower, J. P. The PREP suite: predictiveRNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucl Acids Res. 37, 253–259 (2009).
Acknowledgements
We give special thanks to Professor Baodong Liu of Harbin Normal University for supplying materials of different fern species and Professor Xi Bai, Chunzhu Xu and Yongsheng Chen of Northeast Agricultural University for providing valuable assistance. This work was supported by the National Natural Science Foundation of China (31570189) and the sub-project of Fund for Fostering Talents in Basic Science of the National Natural Science Foundation of China (J1210069).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Ying Chang, Rui Gao, and Wenzhong Wang. Performed the experiments: Rui Gao, Wenzhong Wang, Guangming Zhao, Shuang Bian, and Hongli Ren. Analyzed the data: Rui Gao and Xu Wang. Contributed reagents/materials/analysis tools: Ying Chang, Rui Gao, Wenzhong Wang, Xu Wang and Peng Feng. Wrote the paper: Ying Chang and Rui Gao. Provided raw sequences: Ruifeng Fan and Qingyang Huang.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, R., Wang, W., Huang, Q. et al. Complete chloroplast genome sequence of Dryopteris fragrans (L.) Schott and the repeat structures against the thermal environment. Sci Rep 8, 16635 (2018). https://doi.org/10.1038/s41598-018-35061-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-35061-8
Keywords
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.