Abstract
Ovulation is triggered by gonadotropin surge-induced signaling cascades. To study the role of extracellular signal-regulated kinase 1/2 (ERK1/2) in bovine ovulation, we administered the pharmacological inhibitor, PD0325901, into the preovulatory dominant follicle by intrafollicular injection. Four of five cows treated with 50 µM PD0325901 failed to ovulate. To uncover the molecular basis of anovulation in ERK1/2-inhibited cows, we collected granulosa and theca cells from Vehicle and PD0325901 treated follicles. Next-generation sequencing of granulosa cell RNA revealed 285 differentially expressed genes between Vehicle and PD0325901-treated granulosa cells at 6 h post-GnRH. Multiple inflammation-related pathways were enriched among the differentially expressed genes. The ERK1/2 dependent LH-induced genes in granulosa cells included EGR1, ADAMTS1, STAT3 and TNFAIP6. Surprisingly, PD0325901 treatment did not affect STAR expression in granulosa cells at 6 h post-GnRH. Granulosa cells had higher STAR protein and theca cells had higher levels of STAR mRNA in ERK1/2-inhibited follicles. Further, both granulosa and theca cells of ERK1/2-inhibited follicles had higher expression of SLC16A1, a monocarboxylate transporter, transporting substances including β-hydroxybutyrate across the plasma membrane. Taken together, ERK1/2 plays a significant role in mediating LH surge-induced gene expression in granulosa and theca cells of the ovulating follicle in cattle.
Similar content being viewed by others
Introduction
The process of ovulation is dependent upon the luteinizing hormone (LH) surge to elicit a plethora of signaling cascades required for the release of a fertilizable oocyte and formation of a corpus luteum (CL) needed for pregnancy1,2. Granulosa and theca cells of the ovulating follicle respond to the hormonal triggers, which in turn lead to their morphological and functional differentiation. The LH-dependent changes in the gene expression program is required for follicle rupture, cumulus cell expansion, oocyte maturation, and luteinization3.
The LH-surge has been shown to activate, in granulosa cells, signaling pathways including: phosphatidylinositide 3-kinase (PI3K/AKT), cAMP/Protein kinase A (PKA) and the extracellular signal-regulated kinase 1 and 2 (ERK1/2), also referred to as mitogen-activated protein kinase 3/14,5,6,7,8,9. The ERK1/2 pathway is well-researched in the mouse model, whereby both pharmacological inhibition in vivo10 and in vitro11, and a granulosa cell conditional knockout mouse model12 have demonstrated the pivotal role of the ERK1/2 pathway in ovulatory processes. A number of pharmacological inhibitors (U0126, PD98059, and PD0325901) have been developed to impede ERK1/2 signaling, where PD0325901 was shown to have the least off-target effects and thus, making it the ideal candidate13. Nonetheless, all three inhibitors have been used to evaluate the role of ERK1/2 signaling in theca and/or granulosa cell cultures across multiple species including mouse14, bovine7, rat15,16, and human16,17.
Mouse studies have revealed that the absence of ERK1/2 signalling reduces the expression of LH-regulated genes including: the early growth response 1 (Egr1) transcription factor, as well as genes required for follicular rupture (a disintegrin and metalloproteinase with thrombospondin motifs 1 (Adamts1) and prostaglandin-endoperoxide synthase 2 (Ptgs2)), cumulus expansion (pentraxin 3(Ptx3) and TNF alpha induced protein 6 (Tnfaip6)), oocyte maturation (amphiregulin (Areg)) and luteinization markers (steroidogenic acute regulatory protein (Star) and cytochrome P450 family 11 subfamily a member 1 (Cyp11a1))10,11,12.
Evaluation of ERK1/2 signaling in ovulatory-sized follicles in livestock species has been predominantly performed in cows, where granulosa and theca cells were pharmacologically treated in vitro by one of the aforementioned inhibitors. Furthermore, the experiments involved the collection of abattoir ovaries where theca and granulosa cells were isolated and treated with gonadotropins in the presence or absence of an ERK1/2 inhibitor for 15 minutes to 24 h4,5,7,9,18,19,20. For example, pharmacological inhibition of ERK1/2 signaling with U0126 was performed in cultured bovine granulosa cells from abattoir ovaries (follicles between 8 and 12 mm in diameter were selected). The following ovulatory genes were revealed to be down-regulated when cultured in the presence of forskolin (to induce the LH-surge) and U0126: ADAMTS1, CXCR4, ADAM71, VNN2, and RGS29,18,19,20,21. While treatment of bovine theca cells with PD98059 in vitro resulted in increased STAR expression and thus, enhanced progesterone production5. Although these bovine studies have demonstrated a key role for ERK1/2 in regulation of select LH-regulated genes including ADAMTS1 and STAR in granulosa and theca cells, the global impact of ERK1/2 signalling in bovine ovulation remains to be investigated.
Based on the aforementioned studies, we hypothesized that in the absence of ERK1/2 signaling LH-regulated genes downstream ERK1/2 would be differentially expressed leading to aberrant ovulation in cows. Therefore, our objective was to determine the role of ERK1/2 in bovine ovulation by means of developing a dynamic in vivo model, where follicular wave synchronized cows were subjected to intrafollicular injection of PD0325901 to abolish ERK1/2 signaling specifically in the ovulatory follicle. Moreover, by use of a novel approach of next generation sequencing, we performed RNA-sequencing to identify global changes in gene expression of granulosa cells of the ovulatory follicle exposed to PD0325901 and thus, gain a greater understanding of fertility in the bovine species.
Results
Inhibition of ERK1/2 signaling abolishes ovulation in cattle
First, we tested the impact of inhibition of ERK1/2 signaling on ovulation in cows. The dominant follicle of the synchronized follicular wave was treated by intrafollicular injection with either a Vehicle or ERK1/2 signaling inhibitor, PD0325901 thirty minutes before GnRH treatment. Transrectal ultrasonography five days after the GnRH treatment revealed that all cows treated with Vehicle, 1 µM and 10 µM doses of PD0325901 successfully ovulated, while only one of five cows treated with 50 µM PD0325901 ovulated, however this cow had low levels of circulating progesterone, suggesting her CL was not functional (Fig. 1). Additionally, we measured plasma levels of progesterone on day 5 after GnRH treatment. Cows treated with 10 µM or 50 µM had significantly lower levels of progesterone compared to Vehicle treated counterparts (P < 0.05; Fig. 1). Therefore, we used 50 µM PD0325901 for all further experiments to investigate the molecular basis of anovulation in ERK1/2 inhibited ovulatory follicles in cattle.
Effect of intrafollicular administration of the MEK inhibitor, PD0325901 on ovulation in cattle. All cows were subjected to follicular-wave synchronization and were treated with an ultrasound-guided intrafollicular administration of a Vehicle or different doses of PD0325901 30 minutes prior to intramuscular administration of GnRH. The number of cows ovulating in response to PD0325901 treatment are given in the table. Ultrasonography was used to identify the presence of a corpus luteum (CL). Progesterone levels in plasma samples collected five days after ovulation are presented in the graph. Bars with different letters are significantly different P < 0.05.
Inhibition of ERK1/2 signaling in bovine granulosa cells by 50 µM PD0325901 was confirmed by protein analysis. At 6 h post-GnRH, there was lower abundance of phospho-ERK1/2 in granulosa cells of the ovulatory follicle treated with PD0325901 compared to those of follicles treated with Vehicle (P < 0.01; Fig. 2).
Inhibition of ERK1/2 activity in granulosa cells of ovulating follicles by an intrafollicular administration of PD0325901. Protein abundance of ERK1/2 phosphorylation in bovine granulosa cells collected from the dominant follicles of GnRH stimulated cows, which were challenged with a Vehicle control or 50 µM PD0325901. Quantification by densitometry are presented in the graph. The blot was cropped and it was first used to quantify the presence of Phospho-ERK1/2 and then stripped to quantify for the presence of Total-ERK1/2. ** denotes significant difference of Phospho-ERK1/2 normalized against Total-ERK1/2 between GnRH stimulated cows, which were challenged with a Vehicle control or 50 µM PD0325901, where P < 0.01.
Differentially expressed genes (DEGs) in granulosa cells
Having established that ERK1/2 activity is indispensable for ovulation, we next performed the second experiment to determine the ERK1/2 dependent gene expression in the bovine ovulatory follicle. We performed RNA-seq analysis on granulosa cells collected from the dominant follicles of the cows in three treatment groups: Vehicle, Vehicle + GnRH, and PD0325901 + GnRH. The reads aligned at over 86% to the bovine genome and the principle component analysis (PCA) showed that the transcriptomes of the three treatment groups separated out into three unique clusters (Supplementary Fig. 1 and Supplementary Table 1). We then performed pairwise differential genes expression analyses - first by comparing Vehicle + GnRH vs Vehicle to determine LH-induced genes and then by comparing PD0325901 + GnRH vs Vehicle + GnRH to determine ERK1/2 dependent genes. The pairwise differential analyses using the DESeq 2 R package showed that 2121 genes were differentially expressed between Vehicle + GnRH and Vehicle groups and 285 genes were differentially expressed between PD0325901 + GnRH and Vehicle + GnRH (>1 or <−1 log2fold change, q-value < 0.01; Fig. 3A,B). Of the 2121 LH-regulated DEGs from the first comparison, 1426 genes were upregulated and 695 genes were downregulated in Vehicle + GnRH granulosa cells relative to those of Vehicle group (a full list is available in Supplementary Table 2). The top ten upregulated genes were IL1B, TGM3, CD69, CXCL8, SDS, IL1RN, CAPN6, CCL4, S100A12, and CXCR1 and the top ten downregulated genes included B3GALT2, GPR142, SLC30A2, HAND2, PLPPR1, ANKRD34B, GPR88, ANK3, HCN1, and ACSBG1 (Fig. 4). Of the 285 ERK1/2-dependent DEGs from the second comparison, 33 genes were upregulated and 252 genes were downregulated in PD0325901 + GnRH granulosa cells relative to those of Vehicle + GnRH group (a full list is available in Supplementary Table 3). The top ten upregulated genes were KCNJ5, CDH8, NDST3, AOX1, AGTR2, SSTR2, RLN3, NOS2, CYCS, and MMP15 CXCR1, and the top ten downregulated genes included CASS4, IL1B, GPR84, CXCL8, TNF, CSF1, MEFV, IL1RN, NLRP3, and CSF3R (Fig. 4). In addition, there were 210 DEGs that were common between the two pairwise comparisons (Supplementary Table 4). Interestingly, the majority (196) of these 210 genes were upregulated in Vehicle + GnRH (LH-induced genes) and downregulated in PD0325901 + GnRH (ERK1/2-induced genes) granulosa cells. On the other hand, six genes were downregulated in Vehicle + GnRH (LH-inhibited genes) and upregulated in PD0325901 + GnRH (ERK1/2-inhibited genes) granulosa cells. Finally, only seven genes were upregulated in Vehicle + GnRH (LH-induced genes) followed by another upregulation in PD0325901 + GnRH (ERK1/2-inhibited genes) granulosa cells and only one gene was downregulated in Vehicle + GnRH (LH-inhibited gene) followed by a downregulation in PD0325901 + GnRH (ERK1/2-induced genes) granulosa cells.
Heatmaps displaying differentially expressed genes (DEGs) in bovine granulosa cells for pairwise comparisons. (A) Heatmap of 2121 DEGs in granulosa cells collected from cows prior to (Vehicle; n = 2) or 6 h after GnRH stimulation (Vehicle + GnRH; n = 3). (B) Heatmap of 285 DEGs in granulosa cells collected at 6 h post-GnRH from cows that were pre-challenged, at 30 minutes prior to GnRH, with vehicle (Vehicle + GnRH; n = 3) or MEK inhibition (PD0325901 + GnRH; n = 3). The data from rlog transformed counts with high values and low values shown in red tones and blue tones, respectively. Significantly different genes were determined based on the statistical parameters: log2fold change < −1 and >1 with a q-value < 0.01.
Venn diagram of differentially expressed genes (DEGs) for two pairwise comparisons: Vehicle + GnRH vs. Vehicle and PD0325901 + GnRH vs. Vehicle + GnRH. Top 10 up-regulated (red) and down-regulated (blue) DEGs are listed below for each specific comparison based on Log2FoldChange. The 210 DEGs that were common in both comparisons are represented with their respective direction of regulation in each comparison.
Gene ontology (GO) and pathway analysis of the DEGs
The DEGs were associated with 14 and 13 PANTHER classes (biological processes (BP)) for the comparison of Vehicle + GnRH vs Vehicle and PD0325901 + GnRH vs Vehicle + GnRH, respectively. Many of these BPs were overlapping between comparisons and include immune system processes and metabolic processes (Fig. 5A,B). To determine the pathways that were affected in the pairwise comparisons we used the DAVID tool to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. A table containing the list of top regulated pathways (P < 0.05) can be found in the Supplementary Table 5. A number of inflammatory pathways were altered between the two comparisons including amongst others the tumor necrosis factor (TNF) signaling pathways.
Inhibition of ERK1/2 signaling does not inhibit global transcription in granulosa cells
Although intrafollicular administration of PD0325901 led to abrogation of ovulation, not all genes were differentially expressed in granulosa cells of the treated follicles. Using RT-qPCR, we confirmed that the relative levels of LHR transcript were similar in granulosa cells of all three treatment groups (P > 0.05; Fig. 6). Relative mRNA abundance of FSHR was lower (P < 0.05; Fig. 6) in granulosa cells from the dominant follicles of both Vehicle and PD0325901 in GnRH-stimulated cows compared to those in cows that were not stimulated with GnRH. GnRH-induced increase in mRNA levels of PAPPA and SCARB1 occurred in granulosa cells of both Vehicle and PD0325901 treated follicles (P < 0.05; Fig. 6). Similar differential expression trend for these genes was also observed in RNA-seq data (Table 1).
The MEK inhibitor PD0325901 treatment does not alter the global gene transcription system in bovine granulosa cells. Relative mRNA abundance of LHR, FSHR, SCARB1, and PAPPA in granulosa cells of cows in Vehicle, Vehicle + GnRH and PD0325901 + GnRH groups (N = 3–6/time-point). Cows were subject to follicular-wave synchronization and were challenged with Vehicle or PD0325901 30 minutes prior to intramuscular administration of GnRH. Granulosa cell were collected from groups of individual cows at 0 h and 6 h relative to GnRH stimulus. Transcript abundance of each gene was normalized to reference genes ACTB, L19, and CYCLOPHILIN. All data are expressed as a mean ± S.E.M, where different letters represent differences at p < 0.05 after a one-way ANOVA.
Expression of LH-induced ovulatory genes in granulosa cells is reduced in the absence of ERK1/2 signaling
A multitude of genes are involved in follicle rupture, cumulus cell expansion, oocyte maturation, and luteinization, all of which are required for successful ovulation3. As previously mentioned, our bioinformatics analysis revealed that 196 genes were upregulated in Vehicle + GnRH (relative to Vehicle group) and downregulated in PD0325901 + GnRH (relative to Vehicle + GnRH group) granulosa cells (Fig. 4). From the list of 196 genes, we used RT-qPCR analysis to confirm the results of five genes (EGR1, ADAMTS1, TNFAIP6, STAT3, and TIMP1) known to be LH-induced based on studies performed in rodent10,12,22, primate23, and cattle24,25.
Granulosa cells in Vehicle + GnRH cows had higher levels of EGR1 mRNA relative to Vehicle cows, whereas granulosa cells of PD0325901 + GnRH cows had lower EGR1 mRNA abundance relative to Vehicle + GnRH cows (P < 0.05; Fig. 7). Similar mRNA abundance trend among cows of the three treatment groups was observed for ADAMTS1, TNFAIP6, STAT3, and TIMP1 genes, wherein GnRH-induced increase in their mRNA levels was abolished by PD0325901 treatment (P < 0.05 & P < 0.1; Fig. 7). Similar differential expression trend for these genes was also observed in RNA-seq data (Table 1).
Intrafollicular injection of PD0325901 reduces the relative mRNA abundance of LH-induced ovulatory genes in granulosa cells. Relative mRNA abundance of EGR1, ADAMTS1, TNFAIP6, STAT3, and TIMP1 in granulosa cells of cows in Vehicle, Vehicle + GnRH and PD0325901 + GnRH groups (N = 3–6/time-point). Cows were subject to follicular-wave synchronization and were challenged with Vehicle or PD0325901 30 minutes prior intramuscular administration to GnRH. Granulosa cell were collected from groups of individual cows at 0 h and 6 h relative to GnRH stimulus. Transcript abundance of each gene was normalized to reference genes ACTB, L19, and CYCLOPHILIN. All data are expressed as a mean ± S.E.M, where different letters (A&B) represent differences at p < 0.05 and C represents tendency for differences (P < 0.1) after a one-way ANOVA.
Expression of LH-induced ovulatory genes in theca cells is altered in the absence of ERK1/2 signaling
Successful ovulation and formation of a corpus luteum are dependent on functional and morphological changes in both granulosa and theca cells in response to the LH-surge24,26. Therefore, we sought to further analyze the impact of PD0325901 on the gene expression profile in bovine theca cells by comparing GnRH–stimulated theca cells in the absence or presence of PD0325901. Similar to bovine granulosa cells, relative mRNA abundance of TIMP1 was significantly decreased in theca cells of the ovulatory follicles from the PD0325901 + GnRH group compared to the Vehicle + GnRH group (P < 0.05; Fig. 8A). Unlike in bovine granulosa cells, relative abundance of ADAMTS1 and STAT3 transcripts was higher in theca cells from the PD0325901 + GnRH follicles compared to the Vehicle + GnRH follicles (P < 0.05; Fig. 8B). Lastly, relative transcript levels of EGR1 and TNFAIP6 were unaltered between the two groups (P > 0.05; Fig. 8C).
The impact of ERK1/2 pathway inhibition by PD0325901 on gene transcription in bovine theca cells. In the presence of PD0325901 following GnRH stimulation TIMP1 is reduced in theca cells (A), while ADAMTS1 and STAT3 are induced (B). In bovine theca cells, EGR1 and TNFAIP6 remain unchanged between Vehicle + GnRH and PD0325901 + GnRH. Data were normalized to reference genes ACTB, L19, and CYCLOPHILIN (C). All data are expressed as a mean ± S.E.M, where * represents p < 0.05.
Inhibition of ERK1/2 signaling increases STAR abundance in granulosa and theca cells
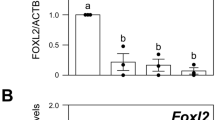
STAR is a luteinization marker expressed in bovine granulosa and theca cells27,28. Mouse studies have demonstrated that LH-induced Star expression in granulosa cells is dependent on ERK1/2 signaling11,12. As expected, our RNA-seq data showed an increase in STAR transcript abundance in Vehicle + GnRH vs Vehicle granulosa cells (Table 1). Surprisingly, there was no difference in the levels of STAR between PD0325901 + GnRH and Vehicle + GnRH granulosa cells (Table 1). Likewise, RT-qPCR analysis showed that there was no reduction in STAR mRNA levels in PD0325901 + GnRH granulosa cells when compared to Vehicle + GnRH granulosa cells (Fig. 9A). To confirm these transcript data, we used immunoblot analysis, which revealed PD0325901 + GnRH granulosa cells had higher levels of STAR protein than Vehicle + GnRH granulosa cells (P < 0.01; Fig. 9B,C). Using the same cell lysates, we observed lower abundance of EGR1 in PD0325901 + GnRH granulosa cells (P < 0.05; Fig. 9B,C). Similar to granulosa cells, relative mRNA abundance of STAR was higher in PD0325901 + GnRH theca cells (P < 0.05; Fig. 9D). Nonetheless, levels of progesterone in the follicular fluid collected at 6 h post-GnRH were similar between the two groups: 696.38 ± 193.82 ng/mL in Vehicle + GnRH and 827.80 ± 173.36 ng/mL in PD0325901 + GnRH (P > 0.05).
The impact of ERK1/2 pathway inhibition by PD0325901 on the transcription and translation of the steroidogenic enzyme STAR. (A) Relative mRNA abundance of STAR comparing Vehicle + GnRH and PD0325901 + GnRH. (B) STAR protein abundance by immunoblot in bovine granulosa cells subject to PD032590. EGR1 was used as a time-point control and ACTINB was used as a loading control. The blot was cut and cropped to analyze all three proteins by antibodies on the same blot. (C) Densitometry reveals quantitative abundance of STAR protein abundance in GnRH stimulated bovine granulosa cells in the presence of PD0325901. (D) RT-qPCR was performed on theca cells collected from cattle subject to GnRH treatment in the presence or absence of PD0325901 to establish the expression pattern of STAR. Transcript data were normalized to reference genes ACTB, L19, and CYCLOPHILIN. All data are expressed as a mean ± S.E.M, where letters (A&B) and * represents p < 0.05 and ** represents p < 0.01.
ERK1/2 appears to regulate uptake of beta-hydroxoybutyric acid in granulosa and theca cells
We found that in RNA-seq data solute carrier family 16 member A1 (SLC16A1; also known as monocarboxylate transporter-1) was one of the 33 upregulated genes in PD0325901 + GnRH granulosa cells (Table 1). Multiple studies have shown that cellular transport of β-hydroxybutyric acid (BHBA) occurs through SLC16A129,30. We explored further the expression of SLC16A1 in pre-ovulatory follicles as BHBA has been shown to increase proliferation and decrease steroidogenesis in cultured granulosa cells31. We confirmed upregulation of SLC16A1 in PD0325901 + GnRH granulosa cells by RT-qPCR analysis (Fig. 10). Likewise, mRNA abundance of SLC16A1 was significantly higher in theca cells collected from the dominant follicles of PD0325901 + GnRH compared to Vehicle + GnRH (P < 0.01; Fig. 10). No difference in the concentration levels of BHBA (0.45 ± 0.08 mmol/L Vehicle + GnRH and 0.34 ± 0.07 mmol/L PD0325901 + GnRH) and glucose (73.75 ± 17.4 mg/dL Vehicle + GnRH and 62.50 ± 17.4mgl/dL PD0325901 + GnRH) in follicular fluid was observed between the two groups of cows (P > 0.05).
Intrafollicular administration of PD0325901 impact on SLC16A1. PD0325901 induces the relative mRNA abundance of solute carrier family 16 member 1 (SLC16A1), a suggested transporter of beta-hydroxybutyric acid, in both granulosa and theca cells. Transcript data were normalized to reference genes ACTB, L19, and CYCLOPHILIN. All data are expressed as a mean ± S.E.M, where different letters represents p < 0.05 and ** represents p < 0.01.
Discussion
In dairy cattle, anovulation on average persists in nearly 20% of dairy cows beyond the voluntary waiting period of breeding at 60 days in milk32. Detailed understanding of ovarian physiology is crucial to develop therapeutic and management strategies to enhance reproductive performance of lactating cows and to ensure sustainable dairy farming. Successful ovulation is dependent on the trigger elicited through the LH surge which in turn activates a cascade of signalling pathways, among others the ERK1/2 pathway10. This study is novel in its approach in using an in vivo model to investigate the role of ERK1/2 in bovine ovulation. We have shown for the first time in a monoovulator species, that the intrafollicular treatment with an inhibitor of ERK1/2 pathway (50 µM PD0325901) abrogates ovulation in cattle. Using next generation sequencing we were able to generate large data sets of transcript changes in bovine granulosa cells, which allowed us to confirm that anovulation is caused by downregulation of genes required for ovulation. To our knowledge, we are among few groups to analyze the transcriptome of bovine granulosa cells through RNA-sequencing33,34.
Our pairwise analysis of Vehicle + GnRH vs Vehicle cows revealed 2121 LH-regulated genes in granulosa cells of ovulating follicles. The immune system processes was one of the biological processes that was found to be regulated by the LH surge. Furthermore, the KEGG pathway analysis suggested that the JAK-STAT, TNF, and MAPK signaling pathways were among the pathways enriched among LH-regulated genes. All these biological processes and the pathways have been linked to inflammatory processes in multiple cell types35,36. Ovulation is an inflammation-like process and in line with this the genes involved in inflammation, such as STAT3 and TNFAIP68,37,38,39 were upregulated in response to the LH surge. Pairwise comparison of PD0325901 + GnRH vs Vehicle + GnRH cows showed that similar pathways were enriched among ERK1/2-dependent LH-regulated genes. These overall pairwise comparisons demonstrate that the ERK1/2 pathway regulates ovulation, similar to the mouse, by altering LH-regulated gene expression in granulosa cells of the ovulating follicle in cattle.
Some of the LH-induced gene were SCARB1, PAPPA, EGR1, ADAMTS1, TNFAIP6, STAT3, and TIMP1, which were also confirmed by RT-qPCR analyses. It has been previously shown that the expression of SCARB1, which is responsible for cellular uptake of high-density lipoproteins in steroidogenic tissues, increases as the bovine follicle size increases, using abattoir-collected ovaries40. Moreover, Scarb1-null mice are infertile despite normal ovarian morphology, estrus cycles, progesterone levels, and number of ovulated follicles41. However, the ovulated oocytes are dysfunctional and preimplantation embryos die shortly after mating, which may be caused due to abnormal lipoprotein metabolism41,42. Elsewhere, PAPPA expression has been shown in granulosa cells of the dominant and also in the ovulating follicle, suggesting that pathways downstream of LH may mediate PAPPA expression in cattle43. Additionally, Pappa knockout mice exhibit compromised fertility due to aberrant expression of steroidogenic genes, including STAR44. Our data along with these observations show that SCARB1 and PAPPA may play critical roles in bovine ovulation. However, neither SCARB1 nor PAPPA were downregulated in ERK1/2 inhibited granulosa cells. Similar lack of effect of ERK1/2 inhibition on Scarb1 and Pappa expression was shown in a mouse study10. Therefore, these studies demonstrated that LH induction of SCARB1 and PAPPA expression does not require ERK1/2 signaling and alternative signaling such as by the PKA pathway could be involved in their regulation43.
Comparing GnRH + PD0325901 to GnRH + Vehicle granulosa cells showed that many of the LH-induced genes, such as EGR1, ADAMTS1, TNFAIP6, STAT3, and TIMP1, were downregulated in ERK1/2 inhibited granulosa cells. Importantly, these genes were also downregulated in the absence of ERK1/2 signaling in granulosa cells of mice10,12. First, EGR1 plays an important role related to proliferation, differentiation, apoptosis, and gene regulation. Mice with Egr1 deletion are sterile due to both ovarian and pituitary abnormalities45. In bovine ovulating follicles, EGR1 is induced in granulosa cells at 6 h post-human chorionic gonadotropin (hCG) and suggested to play an additional role in prostaglandin biosynthesis pathway46. Our group previously reported that in mice, Egr1 regulates Ptgs2 expression by binding to its promoter10. Second, Adamts1-null mice experience subfertility47, which was attributed to aberrant extracellular matrix remodelling of the follicle wall required for ovulation48. In bovine granulosa cells in vitro, treatment with forskolin and ERK1/2 inhibitor (U0126) decreased ADAMTS1 expression21. Third, TNFAIP6 expression is induced between 4 h and 8 h post-hCG in rat granulosa cells49 and Tnfaip6-deficient mice are sterile due to abnormal remodelling of the cumulus extracellular matrix50. In cattle, TNFAIP6 expression is up-regulated in the preovulatory follicle starting at 6 h post-hCG8. Moreover, this same group revealed by in vitro bovine granulosa cell culture that the PKA pathway was responsible for TNFAIP6 promoter activity and transcription. However, they also confirmed that the activator protein-1 (AP-1) transcription factor was indispensable for TNFAIP6 expression8. Previous work has suggested that ERK1/2 may phosphorylate the transcription factor AP-151. In Supplementary Table 3, we show that FOS like 1, AP-1 transcription factor subunit (ENSBTAG00000006194) is downregulated in GnRH + PD0325901 vs GnRH + Vehicle granulosa cells, providing a justification for the decrease in TNFAIP6 expression. Fourth, TIMP1 is a key regulator of matrix metalloproteinases activity and production, and Timp1-null mice have shortened estrus period and altered serum levels of progesterone and estradiol52, yet surprisingly, these mice are fertile suggesting that there are redundant mechanisms that compensate for Timp153. Lastly, STAT3 is a transcription factor involved in cytokine mediation and Stat3-null mice are embryonic lethal confirming the wide-spread expression of the gene54. Mice that are deficient of the progesterone receptor, which are infertile, experience a significant decrease in Stat3 expression in ovaries55. Thus, STAT3, a regulator of inflammation, may play an important role in ovulation. On the contrary, there was an increase in STAT3 expression in ERK1/2 inhibited theca cells. This difference in ERK1/2 regulation of STAT3 in response to ERK1/2 inhibition between granulosa and theca cells is not surprising. In fact, ERK1/2 appears to have different regulatory effect on STAT3 expression and function in different cell types. Inhibition of ERK1/2 has been shown to increase STAT3 expression in human hepatoma cell lines56 and cardiomyocytes57. In oral squamous cell carcinoma cells, it was shown that in the absence of ERK1/2 signaling, there was a decrease in phosphorylated -serine STAT3, but an increase in phosphorylated -tyrosine STAT358.
The cow is an excellent model to study the impact of signaling pathways in theca cells, which is difficult in the murine model59,60. Christenson et al.24 have put together a list of differentially expressed genes comparing 0 h and 21 h GnRH-induced LH surge in both bovine granulosa and theca cells. This group identified TIMP1 as being upregulated by LH in theca cells. Our data show that TIMP1 expression in theca cells was downregulated as in granulosa cells of cows from the PD0325901 + GnRH vs Vehicle + GnRH. Similar to STAT3, ADAMTS1 expression increased in ERK1/2 inhibited theca cells. Previous studies have shown that ADAMTS1 has been detected in bovine theca cells and is increased by 24 h post-GnRH61, while STAT3 has only yet been localized in theca cells of antral follicles in swine62. Also, expression of EGR1 and TNFAIP6 was not affected by the inhibitor treatment in theca cells. EGR1 expression has been previously detected in bovine theca cells and induced by 6 h post-hCG46,63, while TNFAIP6 mRNA abundance has been detected in both bovine and equine theca cells64,65. Theca cell transcriptome in bovine remains poorly studied, however these results contribute to the field of knowledge addressing signaling between follicular cell compartments.
The proper function of steroidogenic genes is crucial for ovulation and the formation of the corpus luteum. Of the multiple steroidogenic enzymes, STAR is the rate limiting enzyme and it facilitates the transport of intracellular cholesterol to the inner mitochondrial membrane for the onset of steroidogenesis and thus, progesterone synthesis. Mice deficient of Star have been shown to experience impaired ovulation66,67. Murine studies have shown that in the absence of ERK1/2 signaling there is a decrease in Star abundance and circulating progesterone concentrations11,12. On the contrary, immortalized steroidogenic granulosa cells and primary rat and human granulosa cells show an increase in STAR expression and progesterone production when cells were subjected to ERK1/2 inhibition15,16. Nonetheless, another study has shown that ERK1/2 inhibition attenuated STAR expression and progesterone synthesis in primary cell cultures of human granulosa cells68. In our in vivo bovine model, the LH-induced increase in STAR mRNA granulosa cells was not abolished by ERK1/2 inhibition. In fact, protein abundance was higher in both granulosa and theca cells of GnRH + PD0325901. In line with our data, treatment of bovine theca cells with PD98059 (ERK1/2 inhibitor) for 24 h increased STAR mRNA and protein abundance and progesterone production in vitro5,7.
RNA-sequencing of granulosa cells provided a diverse list of differentially expressed genes between different comparisons. One of the up-regulated genes in GnRH + PD0325901 granulosa cells was SLC16A1, which was further confirmed by RT-qPCR in both granulosa and theca cells. To our knowledge, this is the first study to report the presence of SLC16A1 in both bovine granulosa and theca cells. Although limited data exists on the role of SLC16A1, it has been demonstrated that in the anterior pituitary cells of dairy cows cellular uptake of BHBA occurred through SLC16A129. Moreover, elevated levels of BHBA reduced PKA activity, which subsequently prevented growth hormone and prolactin transcription and secretion from the pituitary cells29. Dairy cows during early lactation have elevated levels of circulating BHBA69. This ketone has been described as a source of energy used under conditions of starvation70. Previous research has shown that high levels of BHBA increased proliferation, but decreased estradiol and progesterone production in bovine granulosa cells31. As the preovulatory LH surge terminates proliferation and induces progesterone synthesis in granulosa cells, it is plausible that SLC16A1 may play a role in the process through regulation of BHBA uptake. Therefore, our data indicate that ERK1/2 mediates the LH-driven reduction in BHBA uptake to bring about luteinisation during ovulation.
In conclusion, ovulation in cattle, similar to the mouse, is dependent on proper ERK1/2 signaling, as a number of genes in granulosa cells required for different cascades in the ovulatory process are dysregulated in its absence. In contrast to mice, ERK1/2 inhibition increased STAR protein abundance in granulosa and theca cells. Similarly, genes such a SLC16A1 were also elevated in granulosa cells lacking ERK1/2 signaling in both cell types. Moreover, some genes that were downregulated in response to ERK1/2 inhibition in granulosa cells were upregulated in theca cells. Taken together, ERK1/2 plays a significant role in transducing downstream signaling of LH surge during ovulation in cattle.
Materials and Methods
Cows and follicular synchronization
All beef cows in this study were used in accordance with procedures approved by the Ethics and Animal Welfare Committee of the Federal University of Santa Maria, Brazil. Follicular wave synchronization was performed by first (Day - 9) inserting an intravaginal device with 1 g progesterone (DIB; Intervet/Schering-Plough, Brazil), and intramuscular injection of 2 mg estradiol benzoate and 500 µg Sodium Cloprostenol. On day 0, the intravaginal device was removed and the ovaries were examined by transrectal ultrasonography using an 8 MHz linear-array transducer (AquilaVet® scanner, Pie Medical, the Netherlands), where cows having a follicle with a diameter >12 mm were subject to intrafollicular treatment described below.
Experimental design
We conducted two experiments to establish the role of ERK1/2 activity on ovulation in cattle.
Experiment 1
We first tested whether or not inhibition of ERK1/2 activity in an ovulatory follicle would inhibit ovulation and formation of corpus luteum. As demonstrated by a study comparing three ERK1/2 pathway inhibitors13, unlike other inhibitors (U0126 and PD98059), PD0325901 does not appear to have off-target effects, thus making it an ideal ERK1/2 signaling inhibitor. Our group has previously shown that intraperitoneal administration of a single dose of PD0325901 in mice using a concentration of 25 µg/g body weight led to abrogated ovulation in mice without cytotoxicity10. Since intraperitoneal administration of the inhibitor in cows is not feasible, we sought to inhibit ERK1/2 activity by intrafollicular administration of PD0325901 (Selleckchem, Houston, Texas, USA).
Experiment 1 was performed to determine the effective dose for inhibition of ovulation through dose trial. The dominant follicle of the synchronized follicular wave in separate groups of cows was treated on the morning of Day 0 with either a Vehicle control (Phosphate-buffered saline with DMSO; N = 5), or three different doses of PD0325901 (1 µM (N = 5), 10 µM (N = 5), or 50 µM (N = 5)). The diameter of the dominant follicle of each cow was measured by ultrasonography prior to injection. The accurate injection volume to obtain a final intrafollicular concentration of 1 µM, 10 µM, or 50 µM was determined based on the linear regression model equation for the follicular volume as previously described71: V = −685.1 + 120.7D, where V corresponds to the estimated follicular volume and D to the diameter of the follicle to be injected. The inhibitor was initially dissolved in dimethyl sulfoxide (DMSO) (Fisher Scientific, Saint-Laurent, QC, Canada). Thirty minutes following Vehicle or PD0325901 treatment (Day 0), cows were stimulated with an intramuscular dose of gonadotropin releasing hormone (GnRH) (100 µg gonadorelin acetate from Profertil®, Tortuga, Brazil) to induce the endogenous LH surge and thus, ovulation. It is well established that the peak of the endogenous LH surge occurs at 1 h after GnRH treatment43. Five days after GnRH treatment, a blood sample was collected from the coccygeal vein for plasma progesterone analysis and ultrasonography was performed to assess the presence of a corpus luteum. Progesterone concentrations were determined with an electrochemiluminescence immunoassay (Roche, Brazil) in a commercial lab (Laboratorio Pasin, Santa Maria, RS, Brazil). See Fig. 11 for experimental design.
Experiment 2
A second study was conducted to determine the molecular basis of ERK1/2 regulation of ovulation. We used the effective dose of 50 µM PD032901, which abrogated ovulation, to achieve ERK1/2 inhibition. Follicular wave synchronization and intrafollicular treatments were performed as described above. We collected ovaries from each cow at a specific developmental stage of the dominant follicle by colpotomy under caudal epidural anesthesia using 80 mg lidocaine chlorhydrate (4 ml 2% lidocaine) as previously described72. Ovary collection times included 0 h and 6 h relative to GnRH treatment representing before and after LH surge. The three groups of cattle involved in ovary collection included: 1) 0 h relative to GnRH treatment (Vehicle) (N = 3) – these cows were not treated with GnRH, but ovaries were collected at the expected time of GnRH treatment during the follicular wave synchronization protocol; 2) 6 h post-GnRH treatment with intrafollicular Vehicle (Vehicle + GnRH) (N = 6) and 3) 6 h post-GnRH treatment with intrafollicular PD0325901 (PD0325901 + GnRH) (N = 5). See Fig. 11 for experimental design. Upon collection, all ovaries were washed with a 0.9% saline solution followed by follicular puncture of the dominant follicle to collect samples for downstream analysis. Granulosa cells were collected through flushing of the follicle with phosphate buffered saline (PBS) for protein and gene expression analysis, followed by theca cell collection for gene expression analysis via dissection of the follicular wall. Follicular fluid was also collected. All samples, were stored in liquid nitrogen until return to the laboratory, where they were stored at −80 °C until further processing.
RNA extraction and library preparation for RNA-sequencing of granulosa cells
To purify total RNA from granulosa cells we used the commercially available kit NucleoSpin® RNA Plus (Machery-Nagel, D-Mark Biosciences, ON, Canada) according to the manufacturer’s protocol. All samples, were quantified by absorbance at 260 nm using the NanoDrop 20000 spectrophotometer (Thermo Fischer Scientific Inc., Waltham, MA, USA). Total RNA was sent to the Functional Genomics Platform of McGill University and Génome Québec Innovation Centre, Canada for cDNA library preparation and sequencing with Illumina HiSeq 2500 PE 125, using 250 ng as input material. Only top-quality samples were prepared for sequencing, whereby a minimum RNA integrated Number (RIN) of 8.5 was accepted and library quality and quantity was confirmed using a bioanalyzer at Genome Quebec Research Center (N = 2-3 samples/treatment group). A total of 8 samples were selected based on quality assessment: Vehicle (N = 2), Vehicle + 6hGnRH (N = 3) and PD0325901 + 6hGnRH (N = 3) and they were run together on one lane. The RNA-seq data used in this study are available from GEO data repository (GSE121588). Supplementary Table 1 and Supplementary Fig. 1 provide details on RNA-seq quality.
Bioinformatics analysis: read cleaning and genome alignment
FastQC v0.11.5 was used to visualize the quality of the sequence reads73. These reads were cleaned using the software Trimmomatic v0.36, where illumina adapters and reads with average PHRED scores below 20 and lengths below 63 base pairs (bp) were removed74. The FastQC software revealed clean per tile sequence and per base sequence quality. Quality of a representative sample can be seen in Supplementary Fig. 1A,B. The index of the bovine genome, Bos taurus genome B.tau 4.6.1, was built using Bowtie v2.2.1.0 and the cleaned reads were aligned to the genome using TopHat v2.0.11 with the following modifications to the default settings: final read alignments having more than 3 mismatches were removed, 100 base pairs (bp) as the expected inner distance between mate pairs, 50 bp as the standard deviation for the distribution on inner distances between mate pairs, 5 as an anchor length, 1 as the maximum number of mismatches in the anchor, 50 as the minimum intron length, 2 as the number of threads to align reads, 3 as the segment alignment mismatches, 20 as a minimum intron length found during split-segment search75. Finally, the number of reads mapped to each gene were counted using HTSeq v0.6.176.
Differentially expressed genes (DEGs)
R studio v3.3.0 was used for statistical analysis of granulosa cell sequencing data, where R packages were used to determine differently expressed genes (DEGs) between treatment groups. The counts were normalized using the estimateSizeFactors and variance were stabilized using varianceStabilizingTransformaton commands in the statistical package DESeq. 2 v1.12.377. A principle component analysis (PCA) was used to detect treatment clustering (Supplementary Fig. 1C). DESeq 2 was used to build a list of DEGs by a negative binomial generalized linear model with the following criteria: Log2Fold Change of >1 or <−1 and a Benjamini-Hochberg false discover rate (FDR)/q-value < 0.01. Heatmaps were constructed using the R package Pheatmap v1.0.8 and a Venn diagram for pairwise comparisons of DEGs within treatments was assembled with the Bioinformatics & Evolutionary Genomics website (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Gene ontology and pathway enrichment
The identified DEGs were analyzed in the PANTHER classification system (13.1) to determine enriched biological systems. For functional annotation clustering and enrichment score representation of Gene Ontology (GO), we used the Data-base for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 to generate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
RT-qPCR of granulosa and theca cells
Total RNA from granulosa cells was extracted as previously described. Total RNA from theca cells was purified using TRizol® (Thermo Fisher Scientific [Life Technologies, Inc.], Burlington, ON, Canada) according to the manufacturer’s instructions. All samples, were quantified by absorbance at 260 nm using the NanoDrop 20000 spectrophotometer (Thermo Fischer Scientific Inc., Waltham, MA, USA). Complementary DNA was synthesized from 250 ng of total RNA using the iScript cDNA Synthesis kit (Bio-Rad, Mississauga, Canada) using the following temperature program: 25 °C for 5 min (Priming), 46 °C for 20 min (Reverse transcription) and 95 °C for 1 min (Reverse transcription inactivation).
A larger number of granulosa cells samples was used to perform RT-qPCR analysis compared to RNA-seq: Vehicle (N = 3), Vehicle + GnRH (N = 6) and PD0325901 + GnRH (N = 4). Gene expression of CYP17A1 was used as a marker for theca cell contamination and thus, affected granulosa cell samples were removed from the analysis (one sample from PD0325901 + GnRH group was removed). The mRNA abundance of theca cells was compared between two treatment groups, Vehicle + GnRH (N = 6) and PD0325901 + GnRH (N = 5). All RT-qPCR assays were performed using previously described protocols78. Relative transcript abundance for each gene of interest was calculated by dividing their respective starting quantity (SQ) values by the mean SQ values of three reference genes (ACTB, L19, and CYCLOPHILIN). The primer sequences of transcripts measured in this study can be found in Supplementary Table 6, where primer design was performed using the NCBI Primer-BLAST. If variants of a gene are present, the primers were designed to include all variants.
Protein isolation and immunoblot
Protein from granulosa cells was isolated using the AllPrep® DNA/RNA/Protein kit (Qiagen, Mississauga, ON, Canada), followed by a dilution in Laemmli buffer (Bio-Rad Laboratories, Mississauga, ON, Canada) and boiled at 95 °C for 5 min. Protein extracts were resolved by polyacrylamide electrophoresis (10% gel) and transferred to nitrocellulose membranes. Blocking after the transfer was performed for 1.5 h using 5% milk in Tris-buffered saline with 0.1% Tween-20 (TBS-T). All membrane fractions were incubated with their respective primary antibodies (STAR, Phospho-ERK1/2 and EGR1) overnight at 4 °C. Next, membranes were washed with TBS-T (4 × 7 min) followed by incubation with secondary antibody for 1.5 hours at room temperature. The Immun-Star Western Chemi luminescent Kit (Bio-Rad) and Chemidoc Analyzer were used to detect immunoblotted proteins. The membrane was stripped twice for detection of Total-ERK1/2 and β-actin using a stripping buffer (10% SDS, 0.5 M Tris-HCl, milliQ grade water and 2-mercaptoehtanol). We established ERK1/2 activity by determining the abundance of phosphorylated isoform of ERK1/2 relative to its total isoform. All antibodies used in the experiment have been validated in multiple species. The references for the studies that used these antibodies in the bovine ovary are given in Supplementary Table 7 with additional information pertaining to the required concentration and the purchasing information for each antibody. Image Lab Software from Bio-Rad was used to quantify protein abundance. The images of the immunoblots in Figs 2 and 9 were cropped and thus, uncropped images are available in Supplementary Fig. 2.
Follicular fluid analysis
Progesterone concentrations were determined with an electrochemiluminescence immunoassay (Roche, Brazil) in a commercial lab79. Glucose and beta-hydroxybutyric acid (BHBA)/ketone levels were measured with the FreeStyle Optium Neo Blood Glucose and Ketone Monitoring System from Abbott (available from local pharmacies; Abbott Laboratories, Limited).
Statistical analysis for RT-PCR, immunoblot, and follicular fluid analysis
Analysis of data was performed using SAS University Edition (SAS Institute). Data that did not follow a normal distribution (Shapiro-Wilk test) were log-transformed. Differences between means were tested with the Tukey-Kramer honestly significant difference test or by Student’s t-test. Data are presented as the mean ± SEM and significant differences were designated by P < 0.05 and a tendency was marked by P < 0.1.
References
Thatcher, W. W. A 100-Year Review: Historical development of female reproductive physiology in dairy cattle. J Dairy Sci 100, 10272–10291, https://doi.org/10.3168/jds.2017-13399 (2017).
Bearden, H. J., Fuquay, J. W. & Willard, S. T. Applied Animal Reproduction. 6th Edition edn (2004).
Duggavathi, R. & Murphy, B. D. Development. Ovulation signals. Science 324, 890–891, https://doi.org/10.1126/science.1174130 (2009).
Fukuda, S., Orisaka, M., Tajima, K., Hattori, K. & Kotsuji, F. Luteinizing hormone-induced Akt phosphorylation and androgen production are modulated by MAP Kinase in bovine theca cells. J Ovarian Res 2, 17, https://doi.org/10.1186/1757-2215-2-17 (2009).
Tajima, K. et al. Luteinizing hormone-induced extracellular-signal regulated kinase activation differently modulates progesterone and androstenedione production in bovine theca cells. Endocrinology 146, 2903–2910, https://doi.org/10.1210/en.2005-0093 (2005).
Ryan, K. et al. Akt and Erk signal transduction pathways are early markers of differentiation in dominant and subordinate ovarian follicles in cattle. Reproduction (Cambridge, England) 133, 617–626 (2007).
Ryan, K. E. et al. Functional significance of the signal transduction pathways Akt and Erk in ovarian follicles: in vitro and in vivo studies in cattle and sheep. Journal of ovarian research 1, 2 (2008).
Sayasith, K., Bouchard, N., Dore, M. & Sirois, J. Regulation of bovine tumor necrosis factor-alpha-induced protein 6 in ovarian follicles during the ovulatory process and promoter activation in granulosa cells. Endocrinology 149, 6213–6225, https://doi.org/10.1210/en.2008-0459 (2008).
Sayasith, K. & Sirois, J. Molecular characterization of a disintegrin and metalloprotease-17 (ADAM17) in granulosa cells of bovine preovulatory follicles. Molecular and cellular endocrinology 411, 49–57, https://doi.org/10.1016/j.mce.2015.04.010 (2015).
Siddappa, D. et al. Effect of the transient pharmacological inhibition of Mapk3/1 pathway on ovulation in mice. PLoS One 10, e0119387, https://doi.org/10.1371/journal.pone.0119387 (2015).
Su, Y. Q., Nyegaard, M., Overgaard, M. T., Qiao, J. & Giudice, L. C. Participation of mitogen-activated protein kinase in luteinizing hormone-induced differential regulation of steroidogenesis and steroidogenic gene expression in mural and cumulus granulosa cells of mouse preovulatory follicles. Biology of reproduction 75, 859–867, https://doi.org/10.1095/biolreprod.106.052613 (2006).
Fan, H. Y. et al. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 324, 938–941, https://doi.org/10.1126/science.1171396 (2009).
Wauson, E. M., Guerra, M. L., Barylko, B., Albanesi, J. P. & Cobb, M. H. Off-target effects of MEK inhibitors. Biochemistry 52, 5164–5166, https://doi.org/10.1021/bi4007644 (2013).
Su, Y. Q., Wigglesworth, K., Pendola, F. L., O’Brien, M. J. & Eppig, J. J. Mitogen-activated protein kinase activity in cumulus cells is essential for gonadotropin-induced oocyte meiotic resumption and cumulus expansion in the mouse. Endocrinology 143, 2221–2232, https://doi.org/10.1210/endo.143.6.8845 (2002).
Seger, R. et al. The ERK signaling cascade inhibits gonadotropin-stimulated steroidogenesis. Journal of Biological Chemistry 276, 13957–13964 (2001).
Tajima, K. et al. Down-regulation of steroidogenic response to gonadotropins in human and rat preovulatory granulosa cells involves mitogen-activated protein kinase activation and modulation of DAX-1 and steroidogenic factor-1. The Journal of clinical endocrinology and metabolism 88, 2288–2299, https://doi.org/10.1210/jc.2002-020913 (2003).
Henriquez, S. et al. In-vitro study of gonadotrophin signaling pathways in human granulosa cells in relation to progesterone receptor expression. Reproductive biomedicine online 35, 363–371, https://doi.org/10.1016/j.rbmo.2017.06.011 (2017).
Sayasith, K., Sirois, J. & Lussier, J. G. Expression and regulation of regulator of G-protein signaling protein-2 (RGS2) in equine and bovine follicles prior to ovulation: molecular characterization of RGS2 transactivation in bovine granulosa cells. Biology of reproduction 91, 139, https://doi.org/10.1095/biolreprod.114.121186 (2014).
Sayasith, K., Sirois, J. & Lussier, J. G. Expression, regulation, and promoter activation of vanin-2 (VNN2) in bovine follicles prior to ovulation. Biology of reproduction 89, 98, https://doi.org/10.1095/biolreprod.113.111849 (2013).
Sayasith, K. & Sirois, J. Expression and regulation of stromal cell-derived factor-1 (SDF1) and chemokine CXC motif receptor 4 (CXCR4) in equine and bovine preovulatory follicles. Molecular and cellular endocrinology 391, 10–21, https://doi.org/10.1016/j.mce.2014.04.009 (2014).
Sayasith, K., Lussier, J. & Sirois, J. Molecular characterization and transcriptional regulation of a disintegrin and metalloproteinase with thrombospondin motif 1 (ADAMTS1) in bovine preovulatory follicles. Endocrinology 154, 2857–2869, https://doi.org/10.1210/en.2013-1140 (2013).
Carletti, M. Z. & Christenson, L. K. Rapid effects of LH on gene expression in the mural granulosa cells of mouse periovulatory follicles. Reproduction (Cambridge, England) 137, 843–855, https://doi.org/10.1530/REP-08-0457 (2009).
Murphy, M. J., Halow, N. G., Royer, P. A. & Hennebold, J. D. Leukemia Inhibitory Factor Is Necessary for Ovulation in Female Rhesus Macaques. Endocrinology 157, 4378–4387, https://doi.org/10.1210/en.2016-1283 (2016).
Christenson, L. K. et al. Research resource: preovulatory LH surge effects on follicular theca and granulosa transcriptomes. Molecular endocrinology (Baltimore, Md.) 27, 1153–1171, https://doi.org/10.1210/me.2013-1093 (2013).
Shrestha, K. et al. Regulation of ovulatory genes in bovine granulosa cells: lessons from siRNA silencing of PTGS2. Reproduction (Cambridge, England) 149, 21–29, https://doi.org/10.1530/REP-14-0337 (2015).
Murayama, C., Miyazaki, H., Miyamoto, A. & Shimizu, T. Luteinizing hormone (LH) regulates production of androstenedione and progesterone via control of histone acetylation of StAR and CYP17 promoters in ovarian theca cells. Molecular and cellular endocrinology 350, 1–9, https://doi.org/10.1016/j.mce.2011.11.014 (2012).
Pescador, N., Soumano, K., Stocco, D. M., Price, C. A. & Murphy, B. D. Steroidogenic acute regulatory protein in bovine corpora lutea. Biology of reproduction 55, 485–491 (1996).
Bao, B. et al. Expression of steroidogenic acute regulatory protein messenger ribonucleic acid is limited to theca of healthy bovine follicles collected during recruitment, selection, and dominance of follicles of the first follicular wave. Biology of reproduction 59, 953–959 (1998).
Fu, S. P. et al. beta-Hydroxybutyric sodium salt inhibition of growth hormone and prolactin secretion via the cAMP/PKA/CREB and AMPK signaling pathways in dairy cow anterior pituitary cells. Int J Mol Sci 16, 4265–4280, https://doi.org/10.3390/ijms16024265 (2015).
Muller, F. et al. Transport of ketone bodies and lactate in the sheep ruminal epithelium by monocarboxylate transporter 1. Am J Physiol Gastrointest Liver Physiol 283, G1139–1146, https://doi.org/10.1152/ajpgi.00268.2001 (2002).
Vanholder, T. et al. Effects of beta-OH butyrate on bovine granulosa and theca cell function in vitro. Reproduction in domestic animals = Zuchthygiene 41, 39–40, https://doi.org/10.1111/j.1439-0531.2006.00634.x (2006).
Gumen, A., Guenther, J. N. & Wiltbank, M. C. Follicular size and response to Ovsynch versus detection of estrus in anovular and ovular lactating dairy cows. J Dairy Sci 86, 3184–3194, https://doi.org/10.3168/jds.S0022-0302(03)73921-6 (2003).
Walsh, S. W. et al. Effect of the metabolic environment at key stages of follicle development in cattle: focus on steroid biosynthesis. Physiological genomics 44, 504–517, https://doi.org/10.1152/physiolgenomics.00178.2011 (2012).
Li, P. et al. Transcriptome Analysis of Bovine Ovarian Follicles at Predeviation and Onset of Deviation Stages of a Follicular Wave. Int J Genomics 2016, 3472748, https://doi.org/10.1155/2016/3472748 (2016).
Kaplan, M. H. STAT signaling in inflammation. JAKSTAT 2, e24198, https://doi.org/10.4161/jkst.24198 (2013).
Newton, K. & Dixit, V. M. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4, https://doi.org/10.1101/cshperspect.a006049 (2012).
Espey, L. L. Ovulation as an inflammatory reaction–a hypothesis. Biology of reproduction 22, 73–106 (1980).
Richards, J. S., Russell, D. L., Ochsner, S. & Espey, L. L. Ovulation: new dimensions and new regulators of the inflammatory-like response. Annual review of physiology 64, 69–92, https://doi.org/10.1146/annurev.physiol.64.081501.131029 (2002).
Ndiaye, K., Castonguay, A., Benoit, G., Silversides, D. W. & Lussier, J. G. Differential regulation of Janus kinase 3 (JAK3) in bovine preovulatory follicles and identification of JAK3 interacting proteins in granulosa cells. J Ovarian Res 9, 71, https://doi.org/10.1186/s13048-016-0280-5 (2016).
Argov, N., Moallem, U. & Sklan, D. Lipid transport in the developing bovine follicle: messenger RNA expression increases for selective uptake receptors and decreases for endocytosis receptors. Biology of reproduction 71, 479–485, https://doi.org/10.1095/biolreprod.104.028092 (2004).
Trigatti, B. et al. Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proceedings of the National Academy of Sciences of the United States of America 96, 9322–9327 (1999).
Miettinen, H. E., Rayburn, H. & Krieger, M. Abnormal lipoprotein metabolism and reversible female infertility in HDL receptor (SR-BI)-deficient mice. The Journal of clinical investigation 108, 1717–1722, https://doi.org/10.1172/JCI13288 (2001).
Luo, W., Gumen, A., Haughian, J. M. & Wiltbank, M. C. The role of luteinizing hormone in regulating gene expression during selection of a dominant follicle in cattle. Biology of reproduction 84, 369–378, https://doi.org/10.1095/biolreprod.110.085274 (2011).
Nyegaard, M. et al. Lack of functional pregnancy-associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female fertility. Biology of reproduction 82, 1129–1138, https://doi.org/10.1095/biolreprod.109.079517 (2010).
Topilko, P. et al. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Molecular endocrinology (Baltimore, Md.) 12, 107–122, https://doi.org/10.1210/mend.12.1.0049 (1998).
Sayasith, K., Brown, K. A., Lussier, J. G., Dore, M. & Sirois, J. Characterization of bovine early growth response factor-1 and its gonadotropin-dependent regulation in ovarian follicles prior to ovulation. Journal of molecular endocrinology 37, 239–250, https://doi.org/10.1677/jme.1.02078 (2006).
Shindo, T. et al. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. The Journal of clinical investigation 105, 1345–1352, https://doi.org/10.1172/JCI8635 (2000).
Brown, H. M. et al. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biology of reproduction 83, 549–557, https://doi.org/10.1095/biolreprod.110.084434 (2010).
Yoshioka, S. et al. Expression of tumor necrosis factor-stimulated gene-6 in the rat ovary in response to an ovulatory dose of gonadotropin. Endocrinology 141, 4114–4119, https://doi.org/10.1210/endo.141.11.7784 (2000).
Fulop, C. et al. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development (Cambridge, England) 130, 2253–2261 (2003).
Sharma, S. C. & Richards, J. S. Regulation of AP1 (Jun/Fos) factor expression and activation in ovarian granulosa cells. Relation of JunD and Fra2 to terminal differentiation. The Journal of biological chemistry 275, 33718–33728, https://doi.org/10.1074/jbc.M003555200 (2000).
Nothnick, W. B. Disruption of the tissue inhibitor of metalloproteinase-1 gene results in altered reproductive cyclicity and uterine morphology in reproductive-age female mice. Biology of reproduction 63, 905–912 (2000).
Nothnick, W. B., Soloway, P. & Curry, T. E. Jr. Assessment of the role of tissue inhibitor of metalloproteinase-1 (TIMP-1) during the periovulatory period in female mice lacking a functional TIMP-1 gene. Biology of reproduction 56, 1181–1188 (1997).
Takeda, K. et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proceedings of the National Academy of Sciences of the United States of America 94, 3801–3804 (1997).
Akison, L. K. et al. Regulation of the ovarian inflammatory response at ovulation by nuclear progesterone receptor. Am J Reprod Immunol, https://doi.org/10.1111/aji.12835 (2018).
Tian, Z. J. & An, W. ERK1/2 contributes negative regulation to STAT3 activity in HSS-transfected HepG2 cells. Cell research 14, 141–147, https://doi.org/10.1038/sj.cr.7290213 (2004).
Li, Y. J. et al. Crosstalk between ERK1/2 and STAT3 in the modulation of cardiomyocyte hypertrophy induced by cardiotrophin-1. Chinese medical journal 117, 1135–1142 (2004).
Gkouveris, I., Nikitakis, N., Karanikou, M., Rassidakis, G. & Sklavounou, A. Erk1/2 activation and modulation of STAT3 signaling in oral cancer. Oncology reports 32, 2175–2182, https://doi.org/10.3892/or.2014.3440 (2014).
Young, J. M. & McNeilly, A. S. Theca: the forgotten cell of the ovarian follicle. Reproduction (Cambridge, England) 140, 489–504, https://doi.org/10.1530/REP-10-0094 (2010).
Fortune, J. E., Willis, E. L., Bridges, P. J. & Yang, C. S. The periovulatory period in cattle: progesterone, prostaglandins, oxytocin and ADAMTS proteases. Anim Reprod 6, 60–71 (2009).
Willis, E. L., Bridges, P. J. & Fortune, J. E. Progesterone receptor and prostaglandins mediate luteinizing hormone-induced changes in messenger RNAs for ADAMTS proteases in theca cells of bovine periovulatory follicles. Molecular reproduction and development 84, 55–66, https://doi.org/10.1002/mrd.22761 (2017).
Wen, L., Craig, J., Dyce, P. W. & Li, J. Cloning of porcine signal transducer and activator of transcription 3 cDNA and its expression in reproductive tissues. Reproduction (Cambridge, England) 132, 511–518, https://doi.org/10.1530/rep.1.01055 (2006).
Guerrero-Netro, H. M., Estienne, A., Chorfi, Y. & Price, C. A. The mycotoxin metabolite deepoxy- deoxynivalenol increases apoptosis and decreases steroidogenesis in bovine ovarian theca cells. Biology of reproduction 97, 746–757, https://doi.org/10.1093/biolre/iox127 (2017).
Hatzirodos, N. et al. Transcriptomal profiling of bovine ovarian granulosa and theca interna cells in primary culture in comparison with their in vivo counterparts. PLoS One 12, e0173391, https://doi.org/10.1371/journal.pone.0173391 (2017).
Sayasith, K., Dore, M. & Sirois, J. Molecular characterization of tumor necrosis alpha-induced protein 6 and its human chorionic gonadotropin-dependent induction in theca and mural granulosa cells of equine preovulatory follicles. Reproduction (Cambridge, England) 133, 135–145, https://doi.org/10.1530/rep.1.01200 (2007).
Ishii, T. et al. The roles of circulating high-density lipoproteins and trophic hormones in the phenotype of knockout mice lacking the steroidogenic acute regulatory protein. Molecular endocrinology (Baltimore, Md.) 16, 2297–2309, https://doi.org/10.1210/me.2001-0320 (2002).
Stocco, C., Telleria, C. & Gibori, G. The molecular control of corpus luteum formation, function, and regression. Endocrine reviews 28, 117–149, https://doi.org/10.1210/er.2006-0022 (2007).
Fang, L., Yu, Y., Zhang, R., He, J. & Sun, Y. P. Amphiregulin mediates hCG-induced StAR expression and progesterone production in human granulosa cells. Scientific reports 6, 24917, https://doi.org/10.1038/srep24917 (2016).
McArt, J. A. A., Nydam, D. V. & Oetzel, G. R. Epidemiology of subclinical ketosis in early lactation dairy cattle. J Dairy Sci 95, 5056–5066, https://doi.org/10.3168/jds.2012-5443 (2012).
Laeger, T., Metges, C. C. & Kuhla, B. Role of beta-hydroxybutyric acid in the central regulation of energy balance. Appetite 54, 450–455, https://doi.org/10.1016/j.appet.2010.04.005 (2010).
Ferreira, R., Oliveira, J. F., Fernandes, R., Moraes, J. F. & Goncalves, P. B. The role of angiotensin II in the early stages of bovine ovulation. Reproduction (Cambridge, England) 134, 713–719, https://doi.org/10.1530/REP-07-0239 (2007).
Gasperin, B. G. et al. FGF10 inhibits dominant follicle growth and estradiol secretion in vivo in cattle. Reproduction (Cambridge, England) 143, 815–823, https://doi.org/10.1530/REP-11-0483 (2012).
Bioinformatics, B. FastQC: A quality control tool for high throughput sequence data, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
John Hopkins University, C. f. C. B. TopHat: A splicedread mapper for RNA-Seq, https://ccb.jhu.edu/software/tophat/index.shtml (2016).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, https://doi.org/10.1093/bioinformatics/btu638 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biol 15, 550, https://doi.org/10.1186/s13059-014-0550-8 (2014).
Schuermann, Y., Siddappa, D., Pansera, M. & Duggavathi, R. Activated receptor tyrosine kinases in granulosa cells of ovulating follicles in mice. Molecular reproduction and development, https://doi.org/10.1002/mrd.22966 (2018).
de Campos, F. T. et al. The acute effect of intravenous lipopolysaccharide injection on serum and intrafollicular HDL components and gene expression in granulosa cells of the bovine dominant follicle. Theriogenology 89, 244–249, https://doi.org/10.1016/j.theriogenology.2016.11.013 (2017).
Acknowledgements
The authors would like to thank Dr. Sarah Kimmins for the use of facilities. In addition, the authors would like to thank Dr. Tanya Copley for concept teaching of bioinformatics analysis. YS was supported by the Fonds de Recherche du Québec-Nature et Technologies (FRQNT), Réseau Québécois and Reproduction (RQR)-CREAT Scholarship and Department of Animal Science Graduate Excellence Fellowship and Travel Awards. This work was supported by the Natural Science and Engineering Research Council (NSERC) of Canada and FRQNT to RD.
Author information
Authors and Affiliations
Contributions
Y.S. contributed to ideas, designed experiments, took part in the field work, collected, assembled, analyzed, and interpreted data, prepared figures and wrote manuscript; M.T.R. took part in the field work, collected and analyzed data and prepared figures; B.G. & R.F. contributed to ideas, took part in the field work, and collected and assembled data; J.F. took part in the field work; E.M. assembled and interpreted data; P.B.G. & V.B. contributed to ideas, collected and interpreted data; R.D. conceived the study, designed experiments, analyzed data, and edited manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schuermann, Y., Rovani, M.T., Gasperin, B. et al. ERK1/2-dependent gene expression in the bovine ovulating follicle. Sci Rep 8, 16170 (2018). https://doi.org/10.1038/s41598-018-34015-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-34015-4
Keywords
This article is cited by
-
Predicting ketosis during the transition period in Holstein Friesian cows using hematological and serum biochemical parameters on the calving date
Scientific Reports (2022)
-
TNFAIP3 promotes ALDH-positive breast cancer stem cells through FGFR1/MEK/ERK pathway
Medical Oncology (2022)
-
ADAMTS-1: a novel target gene of an estrogen-induced transcription factor, EGR1, critical for embryo implantation in the mouse uterus
Cell & Bioscience (2021)
-
Impact of Dietary Selenium on Modulation of Expression of Several Non-Selenoprotein Genes Related to Key Ovarian Functions, Female Fertility, and Proteostasis: a Transcriptome-Based Analysis of the Aging Mice Ovaries
Biological Trace Element Research (2021)
-
ADAMTS1 and HSPG2 mRNA levels in cumulus cells are related to human oocyte quality and controlled ovarian hyperstimulation outcomes
Journal of Assisted Reproduction and Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.