Abstract
Protein-protein interactions (PPIs) underlie the molecular mechanisms of most biological processes. Mitogen-activated protein kinases (MAPKs) can be dephosphorylated by MAPK-specific phosphatases such as PP2C, which are critical to transduce extracellular signals into adaptive and programmed responses. However, the experimental approaches for identifying PPIs are expensive, time-consuming, laborious and challenging. In response, many computational methods have been developed to predict PPIs. Yet, these methods have inherent disadvantages such as high false positive and negative results. Thus, it is crucial to develop in silico approaches for predicting PPIs efficiently and accurately. In this study, we identified PPIs among 16 BdMAPKs and 86 BdPP2Cs in B. distachyon using a novel docking approach. Further, we systematically investigated the docking site (D-site) of BdPP2C which plays a vital role for recognition and docking of BdMAPKs. D-site analysis revealed that there were 96 pairs of PPIs including all BdMAPKs and most BdPP2Cs, which indicated that BdPP2C may play roles in other signaling networks. Moreover, most BdPP2Cs have a D-site for BdMAPKs in our prediction results, which suggested that our method can effectively predict PPIs, as confirmed by their 3D structure. In addition, we validated this methodology with known Arabidopsis and yeast phosphatase-MAPK interactions from the STRING database. The results obtained provide a vital research resource for exploring an accurate network of PPIs between BdMAPKs and BdPP2Cs.
Similar content being viewed by others
Introduction
Protein kinases or phosphatases in eukaryotic cells are main basics of signal transduction mechanisms by catalyzing covalent addition or subtraction of phosphate groups to serine and threonine/tyrosine residues in their substrates, which causes the quick and reversible modification of proteins and thus regulates plant living situations to adapt to their environment rapidly and precisely1. In particular, mitogen-activated protein kinase (MAPK, also called MPK) is a crucial protein kinase plays vital role in converting environmental and developmental signals into distinct nuclear responses2,3. MAPKs are activated by their specific activator MAPK kinases (MAPKKs, MKKs) through double phosphorylation on both conserved threonine (T) and tyrosine(Y) residues located in the kinase activation loop4,5. In contrast, inactivation of MAPKs is induced by dephosphorylation of cognate residues by various tyrosine phosphatases6, serine/threonine phosphatases(e.g.PP2C)7 and/or dual-specificity phosphatases (DUSPs)8.
The formation of complex between MAPK and its connate activator, substrate, scaffold or inactivator is normally achieved through specific docking interactions9. The docking interactions increase the efficiency of all the enzymatic reactions and may help to regulate the specificity of molecular recognition10. It was discovered that MAPKs contain a common docking domain (CD domain) that is featured by a cluster of negatively charged amino acids in the C-terminal region outside the catalytic domain that binds the basic residues at the N terminus of the docking site (D-site) in MAPK-interaction proteins11,12. Such D-sites has more consecutive positively charged amino acids and promote binding specificity and high affinity interactions with cognate MAPKs. D-sites are also found in MAPK regulating proteins such as MKKs13,14, scaffold proteins and MAPK phosphatases and substrates10. The D-sites in these proteins consist of a cluster of basic residues followed by a hydrophobic sub-motif containing Leu, Ile or Val separated by one residue (R/K1–3-X1–6- φ-X-φ) (φ is any hydrophobic residue)15,16. The specificity of MAPK docking interactions may be determined by the two hydrophobic residues at the distal end of the D-site in MAPKKs17. However, by comparing the current and previous structures, we found that MAPK have actually three (not two) hydrophobic pockets on this surface, which collectively form a “docking groove”, that bind the motif LxLxL/I17. Of course, it is least critical for MAPK binding about the most C-terminal hydrophobic residue in the LxLxL/I motif18,19. Furthermore, MAPK phosphatase MKP-1 harbor another type of MAPK-docking site named the DEF motif 20. Notably, D-sites bind to acidic residues in the CD domain of MAPKs, while DEF motifs interact with a hydrophobic pocket that is only exposed upon MAPK activation21. Moreover, the other one atypical MAPK-docking site within Msg5 named the IYT motif mediates the interaction of Slt2 and Mlp122,23.
Naturally, the precondition of kinase-substrate reaction is the interaction between MAPK and cognate target proteins. However, it is difficult to identify the PPIs about these proteins, primarily because MAPK-substrate interactions are much transient and unstable. The identification of PPIs about their critical roles is a great challenge of unravel many interactomes for deciphering the molecular mechanisms and further providing insight into numerous physiological and pathological processes24. Consequently, the construction of dynamic PPI networks is a critical step towards better understanding biological function of relevant proteins. Recently, many experimental technologies have been proposed for the large-scale PPIs detection, for example, two-hybrid-based screens25, protein chips26, mass spectrometry27, bimolecular fluorescent complementation (BiFC)28 and phage display. However, these methods have itself inherent limitations such as high false positive rate and very low coverage29,30. Hence, more and more computational methods, such as evolutionary information and physicochemical characteristics method31, alignments of multiple sequences32 or weighted sparse representation model33, and so on, have been proposed to predict PPIs efficiently and accurately. While in silico-prediction of PPIs using “Docking” strategy about MAPK has been developed in this study, which can avoid these disadvantages mentioned above.
The above observations clearly demonstrate that MAPK-PP2C docking is crucial for efficient signal transmission. However, less is known about the role of MAPK-PP2C docking in specificity and the identification of D-site in PP2C that is only found in subgroup B PP2C34,35,36. In order to better understand the PPIs about MAPKs and PP2Cs, we have generated a PPI network based on docking approach to predict the interactions about MAPKs and PP2Cs in Brachypodium distachyon. And further analysis also has performed to study the key basis about MAPK-docking on PP2C proteins. In general, the network of PPIs can provide novel insights into the molecular mechanism and the clues of function about MAPKs and PP2Cs in B. distachyon.
Results
Prediction of interaction pairs
Networks of PPIs supply a framework for understanding the biological processes and can give insights into the molecular mechanism inside the cell. Thus the scholarship of PPIs promotes the understanding of biological mechanisms. MAPK is regulated through gene expression between cell receptor and cell response by transcription factions. To better elucidate the molecular mechanism, our knowledge no work in the literature has been executed regarding the interaction prediction of PP2Cs with MAPKs in B. distachyon. MAPKs are prominent components of protein phosphorylation cascades transduce extracellular signals to plant defense responses. Thus this paper identifying the interacting MAPKs with PP2Cs of B. distachyon which is relate to disease signaling process resorting to docking approach. Docking studies is a method which predicts two proteins (PP2C and MAPK) when bound to each other to a complex, the more stable structure (less global energy) higher the probability of their interaction.
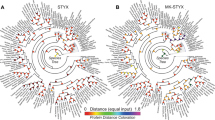
We defined that the absolute value of the lowest energy (AVLE) of two proteins is higher than 65.74 that is the maximum value of the interaction between all subgroup B PP2Cs and MPKs, which means them interacting with each other. Docking studies revealed that 96 from 1376 pairs of PPIs, that is 6.98℅, exhibit higher possibility of interaction including all MAPKs and 49 PP2Cs (Fig. 1, Table S1). The results suggested that PP2C may be connected with other signaling pathway in which PP2C also plays vital roles about negative regulator36,37. Moreover, not all of BdMAPKs and BdPP2Cs are only connected into a single network component (Fig. 1). Some proteins show highly correlated while others do not. For example, BdMAPK20-1, BdMAPK20-2, BdMAPK20-4 and BdMAPK20-5 interact with more BdPP2Cs than others. Of course, there were also some BdPP2Cs showed stronger interaction with BdMAPKs in the chart, including BdPP2C22, BdPP2C31, BdPP2C34, BdPP2C39, BdPP2C41, BdPP2C45, BdPP2C49, BdPP2C50, BdPP2C71 and BdPP2C84(Fig. 1, Table S1). These results indicated that our method could be used to predict the PPIs between BdPP2Cs and BdMAPKs, which were effortless and timesaving in silico “Docking” strategy.
Visualization for the predicted results of PPIs between BdPP2Cs and BdMAPKs was used by the Cytoscape tool. Each node represents a protein and each line refers an interaction. Line thickness reflects the strength of PPIs. Font size and circle color indicated the number of PPIs. Our results showed that there are 143 pairs of potential PPIs. The network is generated using the Cytoscape tool.
Identification of putative D-site in BdPP2Cs
Docking interaction is the precondition of protein molecular recognition and enzymatic reaction. Except subgroup B PP2C, no MAPK-docking sites have been reported. To investigate the correlation of docking site for MAPK with the prediction results, we further analyzed the sequence features of PP2C such as the existence or position of putative docking site. In order to search for the sequence that conform the rules of D-site in PP2C using hidden Markov model, we have derived a list of potential novel D-site (PP2C) from B. distachyon which also showed interaction with BdMAPK through docking studies (Table 1). There are 34 BdPP2C proteins which also show 73.96% PPIs harboring D-site in identified interaction with BdMAPK (Table 1). D-finder assigned a Viterbi probability score of 2.85E-22 to a putative D-site (core sequence: RRGRRRPDILTI) that it found in residues 29–40 of BdPP2C55 (Table 1). It could be hypothesized that the amino acids surrounding the minimal D-site motif contribute to MAPK function. The results indicated that the prediction PPIs have the structural basis of recognition each other and also clearly revealed the complexity of BdMAPK interaction with several PP2C triggered in response to diverse upstream stimuli. Of course, number of novel candidate BdMAPK negative regulators was predicted and need to be confirmed by in vitro assay.
Model Validation
In order to assess the reliability of prediction results and docking site, we investigated the 3D protein structure of BdMAPK6 in complex with docking peptides from BdPP2C25 (high affinity site, BdPP2C25: RRSLSCKA; BdMAPK6, KMLTFDPRQRITVEGAL-visible regions marked in yellow; Fig. 2). It has reported that AtMAPK6 and AtMAPK4 can be dephosphorylated by AtPP2C1 which is orthologous genes with BdPP2C638. Therefore, we choose them to evaluate the possibility of PPIs between BdPP2Cs and BdMAPKs, which was also used as the criterion of cutoff. It is found that the position of BdPP2C6 in “φ-X-φ” locate in protuberant edge which is easier to insert into the hydrophobic pockets. Moreover, BdMAPK6 in common docking motif has three side chain docking pockets which are consistent with the interaction situation (Fig. 2). The results suggested that the PPI of BdMAPK6 and BdPP2C25 have structural evidence support. Furthermore, other BdPP2C proteins also show similar structure in docking site for BdMAPKs (Fig. 3). For example, the 3D structure of BdPP2C47 harbors a D-site (consensus sequence: RRRRRLEMRRFRL, Table 1), which are consistent with the situation of interacting BdMPKs (Fig. 3).
Three-dimensional structural model of the docking site on BdPP2C25 and the CD domain of BdMAPK6. The D-site of BdPP2C25 and CD domain of BdMAPK6 was marked in yellow and connected with the line. Hydrophobic amino acid residues and the basic residues were depicted in blue font; Negative charged amino acid residues are shown in blue type as well.
Database Validation
To better verify our prediction method, the AVLE of more PPIs with reported in public STRING database were performed. Because these PPIs about PP2C and MAPK have confirmed through experiments, instead of pure untested computational procedure. We can assess the reliability of our method through comparing the difference of AVLE about phosphatase-MAPK interaction between B. distachyon and other species. Interestingly, the results show that the AVLE of most (35 pairs) Arabidopsis and yeast PPIs less than 65.74, except 4 pairs PPIs (Table 2). For example, the AVLE of AP2C4-AtMPK6 and AP2C1-AtMPK3 is 73.9 and 70.22, respectively (Table 2). These results suggested that the PPI of BdMAPKs and BdPP2Cs have more favorable evidence support. Because we have already chosen the max AVLE between subgroup B BdPP2Cs and BdMPK3, BdMPK4 or BdMPK6, when the AVLE of most Arabidopsis and yeast PPIs in public database less than 65.74, which indicated that the bigger AVLE of BdPP2C and BdMAPK should even more have protein-protein interaction. That is, these results indirectly suggested that our prediction method have low false positive rate and dependability. Furthermore, in spite of having difference in other species, the same values remain have certain reference value for predicting the PPIs between PP2Cs and MAPKs on other species.
Discussions
D-sites variants for MAPK on PP2Cs in B. distachyon
It has been demonstrated that MAPK phosphatases can act as negative regulators of multiple distinct MAPK pathways according to many studies39. It is one of key problems to understand their roles on signaling under given conditions how they manage to act specifically on the appropriate substrate. In this context, it is critical to know the mechanisms that mediate the interaction between PP2Cs and MAPKs. In general, MAPK-interaction proteins harbor D-sites at the N-terminus region, which can specific bind cognate CD domain of MAPK(Fig. 4B)11,12. The D-site is characterized by a cluster of positively charged amino acids outside the catalytic domain, whose consensus sequence is (R/K1–3-X1–6-φ-X-φ) (Fig. 4A,C)10. As expected, PP2C-type phosphatases were identified a region matching the consensus MAPK interaction motif (named KIM; [K/R](3–4)-X(1–6)-[L/I]-X-[L/I]) in subgroup B phosphatases38. BdPP2C6, BdPP2C25, BdPP2C73 and BdPP2C78 belong to the subgroup B PP2Cs, which also has a typical KIM40. In addition, it only has reported that the interactions and structural features of PP2Cs with MAPKs in model plant Arabidopsis. So, we also independently investigated the AVLE of PPIs with BdMPK3/4/6 for the clues of their interaction each other (Table 3). However, other PP2Cs maybe interact with MAPK, although the PP2C protein does not apparently contain the KIM on N-terminal extension, commonly used MAPK docking sites. It is possible that these plant PP2C-type phosphatases are also interacted with MAPK through other residues located within or outside the typical D-site (Table 1). Indeed, another type of D-site named DEF motif which has a consensus FXFP of human and yeast PP2C proteins has been reported41. Furthermore, spatial and temporal regulators of PP2C dephosphorylation are probably needed to ensure specific downstream activation of MAPKs. Future identification of all PP2C D-sites and their effects to interaction possibility will be necessary for a detailed understanding of PP2C regulation.
MAP kinases interact with D-sites on substrates and regulators. (A) MAPK and several classes of MAPK-interacting proteins. The D-site on MAPK-binding proteins is shown as a triangle. (B) Cartoon of MAPK docking surface MAPK and its interaction proteins complex. The peptides display similar interactions with the two main regions of the MAPK: the hydrophobic groove, which has three side chain docking pockets (A, B and C; B and C were earlier described as “-x- groove”), and the acidic region, known as the “common docking” (CD) site, which binds the basic residues at the N terminus of the docking motifs. (C) Comparison of D-sites found in the human, yeast and plant MAPK-interacting proteins. Residues comprising the basic submotif (+++) are shown in blue type; residues comprising the hydrophobic sub-motif (-X-) are shown in red type. Gaps have been introduced to maximize alignment of functionally similar residues; spaces are for visual clarity.
The PPI network of BdPP2Cs and BdMAPKs
To date, knowledge about the function of MAPK signaling pathways is rather limited, especially their negative regulator, including PP2Cs. OsMKP1, a dual-specificity phosphatase, can negatively regulate the OsMPK6 via dephosphorylation to coordinate the trade-off between grain number per panicle and grain size42. The expression of AtPTP1, a protein Tyr phosphatase, was manifold increased under salinity stress while decreased obviously in cold stress43. DOG1 can interact with the type 2C protein phosphatases AHG1 and AHG3 regulated the ABA signaling pathway to control seed dormancy36. Furthermore, little or no information is available on the molecular regulation network about MAPK cascades including activators or regulators. In addition, previous searching for MAPK target network using protein microarrays yielded 570 MPK phosphorylation substrates44 and MKKK-MKK-MPK molecular regulation network using co-expression systems showed some signaling pathways45. Moreover, despite a large amount of computational approaches have been developed to effectively and accurately predict protein interactions30,32, to date we still need develop more effective methods to investigate the PPIs situation of them. However, construction of a PPI network is the first step for the systematically study of a given organism, which not only provides clues for the signaling pathway but also contributes to comprehend protein functions46. Therefore, we took a high-throughput approach to understand the PP2C/MAPK signaling pathways. Specially, we identified 96 pairs PPIs and generated a PPI network (Fig. 1). Moreover, our study uncovers a series of putative docking site variants on PP2Cs (Table 1), thus inferring and predicting their functions and roles in MAPK signaling pathways.
Comparison with other computational methods
So far, a variety of computational methods for predicting PPIs have been developed by investigators. They all can accurately predict protein interaction only based on different aspects. Such as, the evolutionary information, protein structure, physicochemical characteristics, weighted sparse representation, and so on. The incorporating evolutionary information and physicochemical characteristics feature extraction method for predicting PPIs using a newly developed discriminative vector machine (DVM) classifier had proposed31. The RVM-BiGP that combines the relevance vector machine (RVM) model and Bi-gram Probabilities (BiGP) for PPIs detection from protein sequences had developed using protein evolutionary information47. Moreover, the combining weighted sparse representation based classifier (WSRC) and global encoding (GE) of amino acid sequence had also performed33. It has reported a method for inference of protein-protein interactions from protein amino acids sequences32. While we also propose a novel computational method for predicting PPIs based on the energy of the protein 3D structure closed. Subsequently, we verified the feasibility of our method through a series of models, including the 3D structure reconstruction, putative protein docking domain prediction and public STRING database validation. For example, BdMAPK6 in common docking motif has three side chain docking pockets which are consistent with the situation of interacting BdPP2C25 which contain a D-site (Fig. 2). The analysis results indicated that our method is feasible for predicting PPIs, and can be used as an effective supplementary tool for future proteomics research in the traditional experimental methods.
Conclusion
In this study, we proposed in silico “Docking” strategy to predict the PPIs network about between 16 BdMAPKs and 86 BdPP2Cs. Docking studies showed 96 pairs of probable PPIs including all BdMAPK and 49 BdPP2C, which indicated that BdPP2C may be involved in other signaling pathway. In addition, 34 BdPP2C proteins harbor D-site in identified interaction with BdMAPK, which means that D-site plays important role in BdPP2C protein bind to BdMAPK. Moreover, we evaluate the reliability of prediction results and docking site by compare the 3D protein structure of BdMAPK6 and BdPP2C25 which can provide the structural support for our study. Furthermore, STRING database also indicated that our prediction results have low false positive rate and dependability. These findings provide insight into docking interactions in MAPK signaling networks and should be related to efforts to predict new MAPK substrates and regulators by in silico-prediction methods.
Methods
Prediction of PPIs between BdPP2Cs and BdMAPKs
The sequences of B. distachyon (BdMAPKs and BdPP2Cs) were downloaded from PLAZA (https://bioinformatics.psb.ugent.be/plaza/)48 in FASTA format. Homology modeling was performed with the help of MODELLER and NCBI BLAST. For reconstructing the 3D conformation of all investigated in this study, a template for homology modeling was searched with PDB search in NCBI BLAST. Six structures were generated in Protein Data Bank proteins for each protein molecule, out of which the minimized average models with maximum score, lowest E-value and with a cut off identity of >30% were selected. The final structures generated by MODELLER 9.17 were used to construct and evaluate 3D models. Structures editing through energy minimization model was implemented using structure minimization tool from enhanced UCSF Chimera (http://www.rbvi.ucsf.edu/chimera/) which was a program for the analysis of molecular structures and related data and interactive visualization using a toolkit Opal49. Therefore, the default parameter value of all structures are retained, in details as follows: Steepest descent steps: 100, Steepest descent step size(A): 0.02, Conjugate gradient steps: 10, Conjugate gradient step size: 0.02, Update interval: 10, Fixed atoms: none, Method: also consider H-bonds(slower), Standard residues: AMBER ff14SB, Other residues: AM1-BCC Protonation states for: histidine, Residue-name-based (HIS/HID/HIE/HIP = unspecified/delta/epsilon/both)50. The minimized energy structures were finally saved as *.pdf files which were checked and validated by PROCHECK, ERRAT and VERIFY_3D. Subsequently, the refined structures of BdMAPKs were taken as receptor and the structures of BdPP2Cs were taken as ligand for the docking studies on the online Patch Dock server which is based on shape complementary principle and keeping parameter value Clustering RMSD:4.0, Complex Type: Default51,52. The results obtained were refined using Fire Dock online server which rearranges the interface side chains and adjusts the relative orientation of the molecules53. The AVLE is in solutions number 1000. At last, we can obtain the AVLE of all phosphatase-MAPK interaction pairs (Table S2). The prediction framework is shown in Fig. 5.
Selection of threshold as a criterion
To distinguish the interacting and non-interacting partners between BdMAPKs and BdPP2Cs, we need chose a reasonable value as a criterion. It has previously reported that phosphatase-MAPK interaction is only found in subgroup B PP2Cs for putative D-site with several kinds of MPKs, including MPK3, MPK4 or MPK635,38. It is more important that these interactions with MPK3, MPK4 and MPK6 depends on D-site in PP2C, which have proved through experiments such as yeast two hybrid screen (Y2H), BiFC assays and so on. According to the principle that homologous genes have similar functions, we therefore analyzed the AVLE of the interaction between all the subgroup B PP2Cs (BdPP2C6, BdPP2C25, BdPP2C73 and BdPP2C78) and BdMAPKs (BdMAPK3, BdMAPK4 and BdMAPK6). Specifically, the AVLE of the interaction between BdPP2C6 and BdMAPKs (BdMAPK3, BdMAPK4 and BdMAPK6) is 60.33, 50.23 and 62.83, respectively (Table 3). Likewise, the AVLE of between BdPP2C25 (43.51, 45.07 and 65.74), BdPP2C73 (41.79, 47.49 and 59.1) and BdPP2C78 (44.1, 49.9 and 53.52) and BdMAPKs are obtained, respectively (Table 3). For maximum assurance of the accuracy of the prediction results, e.g. the false positive rate, etc. we have chosen the max AVLE (that is 65.74) from them was used as a criterion which was distinguished the interacting and non-interacting partners between BdMAPKs and BdPP2Cs. Since there is a smaller AVLE of phosphatase-MPK interaction pairs that are likely to interact, then which has a greater AVLE that is of course more likely to interact.
Prediction of docking site in BdPP2Cs using hidden Markov model
The docking interactions are contributed to the efficiency of all the enzymatic reactions and the specificity of molecular recognition. In order to search for the docking site for BdMAPKs in BdPP2Cs, we aligned the full-sequence of BdPP2Cs with others that contains conserved D-site using hidden Markov model (HMM). A profile HMM architecture, composed of linked main, insert, and delete states was performed in the programming language Java to implement the computational analysis and prediction. An HMM model of length 19 was designed to be a minimal prescreen that simply checks for a basic residue followed after a spacer of 1–5 residues by a hydrophobic-X-hydrophobic (as defined above)54. D-finder is used to select suitable strings and then assigns a standard HMM Viterbi probability score. The code of D-finder is downloadable from http://dfinder.sourceforge.net as a.zip file that contains the Java files along with the original training set file, a sample testing file, and a README file.
Confirmation of docking motif and interolog based protein-protein interaction prediction
The docking-based and interolog method uses domain interaction information and sequence similarity to infer the potential PPIs, respectively. If two proteins contain an interacting domain pair, it is expected that these two proteins may interact with each other. While if two proteins have interacting homolog in another organism such as Arabidopsis, it is also thought that these two proteins are conserved interaction55. Thus, interologs are homologous pairs of protein interactions across different organisms. To assess the PPIs of BdPP2Cs and BdMAPKs, protein 3D structures are used to check their probability of interaction from spatial conformation, especially docking site.
Validation of results against STRING database with our methods
To validate the feasibility and reliability of our methods for phosphatase-MAPK interaction, the appraisement about AVLE of PPIs that have confirmed with experiments in STRING database56 (https://string-db.org/cgi/input.pl) were performed. Subsequently, we compared the AVLE of other species phosphatase-MAPK interaction, including Arabidopsis and yeast.
References
Besteiro, M. A. G. & Ulm, R. Phosphorylation and Stabilization of Arabidopsis MAP Kinase Phosphatase 1 in Response to UV-B Stress. Journal of Biological Chemistry 288, 480–486 (2013).
Xu, J. & Zhang, S. Q. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science 20, 56–64 (2015).
Colcombet, J. & Hirt, H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413, 217–26 (2008).
Samajova, O., Plihal, O., Al-Yousif, M., Hirt, H. & Samaj, J. Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotechnology Advances 31, 118–128 (2013).
Ma, H. et al. MAPK Kinase 10.2 Promotes Disease Resistance and Drought Tolerance by Activating Different MAPKs in rice. Plant J 92, 557–70 (2017).
Alonso, A. et al. Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 (2004).
Shubchynskyy, V. et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses toPseudomonas syringae. Journal of Experimental Botany 68, 1169 (2017).
Owens, D. M. & Keyse, S. M. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213 (2007).
Tanoue, T. & Nishida, E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacology & Therapeutics 93, 193–202 (2002).
Tanoue, T., Adachi, M., Moriguchi, T. & Nishida, E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature Cell Biology 2, 110–116 (2000).
Tanoue, T. J. & Nishida, E. Molecular recognitions in the MAP kinase cascades. Cellular Signalling 15, 455–462 (2003).
Bardwell, L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans 34, 837–41 (2006).
Bardwell, A. J., Frankson, E. & Bardwell, L. Selectivity of Docking Sites in MAPK Kinases. Journal of Biological Chemistry 284, 13165–13173 (2009).
Jiang, M. & Chu, Z. Q. Comparative analysis of plant MKK gene family reveals novel expansion mechanism of the members and sheds new light on functional conservation. Bmc Genomics 19, 407 (2018).
Bardwell, L. & Shah, K. Analysis of mitogen-activated protein kinase activation and interactions with regulators and substrates. Methods 40, 213–223 (2006).
Akella, R., Moon, T. M. & Goldsmith, E. J. Unique MAP kinase binding sites. Biochimica Et Biophysica Acta-Proteins and Proteomics 1784, 48–55 (2008).
Bardwell, A. J. & Bardwell, L. Two Hydrophobic Residues Can Determine the Specificity of Mitogen-activated Protein Kinase Docking Interactions. Journal of Biological Chemistry 290, 26661–26674 (2015).
Remenyi, A., Good, M. C., Bhattacharyya, R. P. & Lim, W. A. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Molecular Cell 20, 951–962 (2005).
Peti, W. & Page, R. Molecular basis of MAP kinase regulation. Protein Science 22, 1698–1710 (2013).
Lin, Y. W. & Yang, J. L. Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. Journal of Biological Chemistry 281, 915–926 (2006).
Lee, T. et al. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Molecular Cell 14, 43–55 (2004).
Palacios, L. et al. Distinct Docking Mechanisms Mediate Interactions between the Msg5 Phosphatase and Mating or Cell Integrity Mitogen-activated Protein Kinases (MAPKs) in Saccharomyces cerevisiae. Journal of Biological Chemistry 286, 42037–42050 (2011).
Sacristan-Reviriego, A., Madrid, M., Cansado, J., Martin, H. & Molina, M. A Conserved Non-Canonical Docking Mechanism Regulates the Binding of Dual Specificity Phosphatases to Cell Integrity Mitogen-Activated Protein Kinases (MAPKs) in Budding and Fission Yeasts. Plos One 9(1), e85390 (2014).
Bosque, G., Folchfortuny, A., Picó, J., Ferrer, A. & Elena, S. F. Topology analysis and visualization of Potyvirus protein-protein interaction network. Bmc Systems Biology 8, 129 (2014).
Gavin, A. C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Zhu, H. et al. Global analysis of protein activities using proteome chips. Science 293, 2101–2105 (2001).
Ho, Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002).
Schweighofer, A., Shubchynskyy, V., Kazanaviciute, V., Djamei, A. & Meskiene, I. Bimolecular fluorescent complementation (BiFC) by MAP kinases and MAPK phosphatases. Methods Mol Biol 1171, 147–58 (2014).
Huang, Y. A., You, Z. H., Chen, X. & Yan, G. Y. Improved protein-protein interactions prediction via weighted sparse representation model combining continuous wavelet descriptor and PseAA composition. Bmc Systems Biology 10, 120 (2016).
Li, J. Q., You, Z. H., Li, X., Ming, Z. & Chen, X. PSPEL: In Silico Prediction of Self-Interacting Proteins from Amino Acids Sequences Using Ensemble Learning. Ieee-Acm Transactions on Computational Biology and Bioinformatics 14, 1165–1172 (2017).
Li, Z. W., You, Z. H., Chen, X., Gui, J. & Nie, R. Highly Accurate Prediction of Protein-Protein Interactions via Incorporating Evolutionary Information and Physicochemical Characteristics. International Journal of Molecular Sciences 17, 1396 (2016).
Wang, L. et al. An ensemble approach for large-scale identification of protein-protein interactions using the alignments of multiple sequences. Oncotarget 8, 5149–5159 (2017).
Huang, Y. A., You, Z. H., Chen, X., Chan, K. & Luo, X. Sequence-based prediction of protein-protein interactions using weighted sparse representation model combined with global encoding. Bmc Bioinformatics 17, 184 (2016).
Schweighofer, A., Hirt, H. & Meskiene, L. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science 9, 236–243 (2004).
Brock, A. K. et al. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol 153, 1098–111 (2010).
Née, G. et al. Delay of Germination1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nature Communications 8, 72 (2017).
Lu, X. et al. A PP2C-1 Allele Underlying a Quantitative Trait Locus Enhances Soybean 100-Seed Weight. Molecular Plant 10, 670–684 (2017).
Schweighofer, A. et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19, 2213–24 (2007).
Martin, H., Flandez, M., Nombela, C. & Molina, M. Protein phosphatases in MAPK signalling: we keep learning from yeast. Molecular Microbiology 58, 6–16 (2005).
Cao, J. M., Jiang, M., Li, P. & Chu, Z. Q. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. Bmc Genomics 17, 175 (2016).
Jacobs, D., Glossip, D., Xing, H. M., Muslin, A. J. & Kornfeld, K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes & Development 13, 163–175 (1999).
Guo, T. et al. Grain Size and Number1 Negatively Regulates the OsMKKK10-OsMKK4-OsMPK6 Cascade to Coordinate the Trade-off between Grain Number per Panicle and Grain Size in Rice. Plant Cell 30, 871–888 (2018).
Xu, Q., Fu, H. H., Gupta, R. & Luan, S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 10, 849–857 (1998).
Popescu, S. C. et al. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes & Development 23, 80–92 (2009).
Jiang, M. et al. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. Bmc Genomics 16, 228 (2015).
Singh, R. et al. Protein interactome analysis of 12 mitogen-activated protein kinase kinase kinase in rice using a yeast two-hybrid system. Proteomics 14, 105–115 (2014).
An, J. Y. et al. Improving protein-protein interactions prediction accuracy using protein evolutionary information and relevance vector machine model. Protein Science 25, 1825–1833 (2016).
Van Bel, M. et al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research 46, D1190–D1196 (2018).
Huang, C. C., Meng, E. C., Morris, J. H., Pettersen, E. F. & Ferrin, T. E. Enhancing UCSF Chimera through web services. Nucleic Acids Research 42, W478–W484 (2014).
Pettersen, E. F. et al. UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612 (2004).
Mashiach, E. et al. An integrated suite of fast docking algorithms. Proteins-Structure Function and Bioinformatics 78, 3197–3204 (2010).
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H. J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Research 33, W363–W367 (2005).
Mashiach, E., Schneidman-Duhovny, D., Andrusier, N., Nussinov, R. & Wolfson, H. J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Research 36, W229–W232 (2008).
Whisenant, T. C. et al. Computational Prediction and Experimental Verification of New MAP Kinase Docking Sites and Substrates Including Gli Transcription Factors. Plos Computational Biology 6(8) (2010).
Yu, H. Y. et al. Annotation transfer between genomes: Protein-protein interologs and protein-DNA regulogs. Genome Research 14, 1107–1118 (2004).
Szklarczyk, D. et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research 43, D447–D452 (2015).
Acknowledgements
This research is supported by the Grant from Chinese Academy of Sciences strategic resource service network planning of plant germplasm resources innovation project ZSZC-013 and Grant from Shanghai Landscaping Administrative Bureau (Grant Nos. G162406 and G162407). These funding bodies had no role in study design, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Z.C. and M.J. conceived and designed the work. C.N. performed most of the data mining and constructed PPI network. M.J. performed data analysis, wrote and revised the manuscript. J.C. and D.N. helped to retrieve gene sequences data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, M., Niu, C., Cao, J. et al. In silico-prediction of protein–protein interactions network about MAPKs and PP2Cs reveals a novel docking site variants in Brachypodium distachyon. Sci Rep 8, 15083 (2018). https://doi.org/10.1038/s41598-018-33428-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33428-5
Keywords
This article is cited by
-
A topological review on protein–protein interactions: the development and promises in the era of omics
Journal of Proteins and Proteomics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.