Abstract
This study was conducted to explore the effects of DNMT1 and DNMT3A polymorphisms on susceptibility to noise-induced hearing loss (NIHL) in Chinese workers. A total of 2689 industrial workers from a single textile factory were recruited. Venous blood was collected, as were questionnaire and pure-tone audiometry (PTA) data by specialist physicians. Four selected SNPs (rs7578575, rs749131, rs1550117, and rs2228611) in DNMT1 and DNMT3A were genotyped in 527 NIHL patients and 527 controls. Then, main effects of the genotypes and their interactions were evaluated. Results revealed that the GG genotype at rs749131 and the AG/GG genotypes at rs1550117 and rs2228611 [odds ratio (OR) = 1.87, 2.57, and 1.98 respectively], as well as the haplotypes AGGG and TGGA (rs7578578-rs749131-rs1550117-rs2228611) (OR = 1.35 and 1.56, respectively) were associated with an increased risk of NIHL in the Chinese population. Multifactor dimensionality reduction analysis indicated that rs7578575, rs749131, and rs2228611 interact and are related to increased NIHL risk (OR = 1.63). The genetic polymorphisms rs749131 G, rs1550117 G, and rs2228611 G within the DNMT1 and DNMT3A genes are associated with an increased risk of NIHL in the Chinese population and have the potential to act as biomarkers for noise-exposed workers.
Similar content being viewed by others
Introduction
Occupational noise is one of the most common occupational hazards affecting the health of industrial workers, and noise-induced hearing loss (NIHL) is the second most frequent form of sensorineural hearing impairment besides age-related hearing loss (ARHL) worldwide1. NIHL is a multifactorial disease resulting from the interaction of genetic and environmental factors2. Currently, the mechanism of NIHL is not completely understood. The possible etiopathogenesis may involve inner ear cell apoptosis or necrosis caused by oxidative stress or metabolic products generated during signal transduction and direct mechanical injury to the structures of the cochlea3,4,5. However, numerous population studies have indicated that individuals exhibit various degrees of NIHL risk, even when they are exposed to equal noise intensity levels1,6. Animal experiments also have demonstrated that genetic variants contribute to the incidence of NIHL7,8. Previous studies have found that single nucleotide polymorphisms (SNPs) in the HSP70, EYA4, CDH23, GRHL2, and DFNA5 genes are associated with genetic susceptibility to NIHL in humans and may increase or decrease the risk of NIHL through interactions with occupational noise9,10,11. The prevailing evidence indicates that genetic susceptibility and interactions with environmental factors may play an important role in the occurrence and development of NIHL.
DNA methylation represses gene expression through the allosteric inhibition of transcription factors and the transcriptional machinery while serving as a recognition site for a host of repressive factors12. This modification is applied by DNA methyltransferase (DNMT) proteins. DNA methyltransferase 1 (DNMT1) is a maintenance methyltransferase, while DNMT3A and DNMT3B are de novo methyltransferases13,14. DNA methylation is recognized as an essential epigenetic mark that regulates chromatin structure and gene expression throughout the genome. Recently, hereditary sensory neuropathy with dementia and hearing loss (HSAN1E) has been shown to be caused by mutations in the maintenance methyltransferase DNMT115. Moreover, a family has been identified with adult-onset autosomal dominant cerebellar ataxia with deafness and narcolepsy (ADCA-DN) caused by mutations in DNMT1, and the DNA methylation profiles of these individuals have been assessed16. An epigenome-wide DNA methylation study investigating the association between hearing ability with age and DNA methylation identified strong associations with DNA methylation in the promoters of 10 genes in humans17. Furthermore, oxidative stress has been shown to be associated with DNA hypermethylation, which is mediated by the deregulation of epigenetic-related enzymes such as DNMT3A, H2AFZ, and H3F3B18.
Associations between NIHL and DNMT SNPs and their functional significance in the DNMT3A genes have not yet been reported and previous study by Hu et al. showed no significant association between three DNMT1 SNPs (rs12984523, rs16999593, and rs2228612) and NIHL susceptibility in the Chinese population19. However, considering the vital roles of DNMT1 in hearing disorders and of DNMT3A in oxidative stress, we speculated that polymorphisms in DNMT1, DNMT3A and their interactions could be associated with genetic susceptibility to NIHL. Therefore, we performed a case-control study to elucidate the associations between four DNMT1 and DNMT3A SNPs, namely rs7578575, rs749131, rs1550117, and rs2228611, and genetic susceptibility to NIHL.
Materials and Methods
Subjects
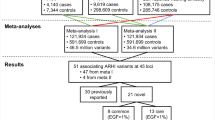
This research has been approved by the Jiangsu Provincial Center for Disease Prevention and Control ethics committee (Nanjing, Jiangsu province, China) and all the experiments were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from each participant before beginning the study. Industrial employees from a single textile factory in eastern China who received annual health examinations performed by the Jiangsu Provincial Center for Disease Prevention and Control were recruited in 2015. A total of 2689 people participated in the health examinations. Occupational health examination items mainly included venous blood collection, general physical examination, and pure-tone audiometry (PTA). During the health examination, personal medical history, smoking and drinking habits, and habitual use of drugs were assessed. In this study, subjects who had quitted smoking or drinking for at least one year were defined as ever smokers and drinkers. Workers who drank a bottle of beer or fifty grams of wine per day for at least one year were defined as “now drinkers”, and subjects who had one cigarette per day for at least one year were defined as “now smokers”. The following subjects were excluded from this study: workers with diseases that may affect hearing thresholds (e.g. otitis media, cholesteatoma, and ear canal stenosis) and workers who have used or are using ototoxic drugs (e.g. aspirin, quinolones, and aminoglycosides). A total of 2477 subjects meet our criteria and were included.
PTA and NIHL assessment
PTA was performed for each participant after a break in noise exposure for up to 12 hours. Using a Madsen Voyager 522 audiometer (Madsen, Taastrup, Denmark), audiometry was conducted by qualified doctors in a soundproof room.
Defining NIHL and control subjects
Hearing loss and normal hearing were defined according to Chinese diagnostic criteria for occupational noise-induced deafness (GBZ 49–2007). In this study, occupational noise exposure was defined as levels of noise exposure (Lex) of at least 85 dB (A) during a nominal 8-hour working day. Hearing loss was identified using binaural hearing thresholds that exceed 25 dB at both high (3000, 4000, 6000 Hz) and speech (500, 1000, 2000 Hz) frequencies. Correspondingly, normal hearing was identified by binaural hearing thresholds below 25 dB both at high and speech frequencies. Hearing thresholds were obtained from the results of PTA. The subjects were divided into two groups: NIHL patients (noise-exposed individuals with hearing loss) and control individuals (noise-exposed individuals with normal hearing). We first selected the NIHL patients, and controls were frequency-matched to the patients by sex, age, and intensity of noise exposure20. As a result, 527 NIHL patients and 527 controls were selected from all eligible subjects.
DNA extraction
Peripheral blood (3 mL) was collected in ethylene diamine tetra acetic acid (EDTA) and used for DNA isolation and genotyping. DNA was extracted from blood samples of subjects using the QIAcube HT and QIAamp 96 DNA QIAcube HT Kit (Qiagen, Dusseldorf, Germany) following the manufacturer’s protocol and was then stored at −20 °C until use.
SNP selection and genotyping
Target SNPs in the DNMT1 and DNMT3A genes were selected on the basis of data from the 1000 Genomes Project and previous findings from the literature21. The criteria for identifying SNPs included a minor allele frequency (MAF) in the Chinese Han population (CHB) >0.10 and a linkage disequilibrium r2 value > 0.8. In the end, twelve candidate SNPs were selected using these criteria and Haploview software. Then, we searched PubMed using these candidate SNPs and found that rs7578575, rs749131, and rs1550117 of DNMT3A and rs2228611 of DNMT1 were the most commonly reported; these SNPs were therefore used in the subsequent analysis.
Genotypes for the selected individuals at each polymorphic site were determined using ABI TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA, USA) and pre-designed commercial genotyping probes and primers. The extracted DNA and genotyping probes and primers were added to TaqMan universal PCR master mix (Roche, Branchburg, NJ, USA) according to the manufacturer’s instructions. Genotyping was then performed on an ABI 7900 real-time PCR system (Applied Biosystems). The results were analyzed using the ABI 7900 system sequence detection software version 1.2.3 (Applied Biosystems).
Detecting promoter activity of SNP rs1550117
As rs1550117 is located within the 2 kb region upstream of the DNMT3A gene, transcription factor binding sites were predicted by TRANSFAC, and a luciferase reporter assay was conducted to detect the effects of SNP rs1550117 on DNMT3A promoter activity. The target intron containing the rs1550117 A or G allele was synthesized and then cloned into the XhoI/HindIII restriction sites of the pGL3-enhancer plasmid (Promega, Madison, WI, USA). The accuracy of the plasmid sequence was confirmed by DNA sequencing. House Ear Institute-Organ of Corti 1 (HEI-OC1) cells were cultured in 24-well plates and transfected with 800 ng of each allele-specific plasmid using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA). As an internal control, the pRL-SV40 plasmid (Promega) was co-transfected into cells. After 48 h of transfection, dual luciferase activities were determined using a Luciferase Reporter Assay System (Promega). All assays were conducted as three independent replicates.
Statistical analyses
Statistical analyses were performed using SPSS 23.0 software (Chicago, IL, USA). Goodness-of-fit χ2 tests were conducted to determine whether the SNPs were in Hardy-Weinberg equilibrium among the control subjects. Categorical variables are presented as percentages, and continuous variables are presented as the mean ± SD. Odds ratios (ORs) and 95% confidence intervals (95% CI) for genotypes were determined using conditional logistic regression models adjusted for age, sex, smoking, and drinking. Differences in allele-specific promoter activity and gene expression were compared using Student’s t-tests or paired t-tests. Haplotype analysis was performed using the SHEsis platform22. All P-values were corrected (Pc) using the Bonferroni method, and P < 0.05 was used to indicate statistical significance.
Results
Demographic characteristics of study subjects and Hardy-Weinberg tests of selected SNPs
The general characteristics (age, sex, smoking, drinking, work time with noise, and noise exposure level) and high-frequency hearing thresholds of the NIHL cases and controls are shown in Table 1. No significant differences were observed between NIHL cases and controls in terms of the general characteristics (P > 0.05). However, there was a significant difference between NIHL cases and controls in terms of the high-frequency hearing threshold. The average high-frequency hearing threshold was higher for NIHL patients (35.94 ± 9.96) than for controls (14.09 ± 4.14; P < 0.001). General information for the selected SNPs and the results of Hardy-Weinberg tests are shown in Table 2. rs7578575 and rs749131 are located in DNMT3A introns, rs1550117 is located in the 2 kb region upstream of DNMT3A, and rs2228611 is a synonymous SNP in DNMT1. The χ2 tests revealed that all selected SNPs were in Hardy-Weinberg equilibrium (P > 0.05).
Multivariate analyses of DNMT1 and DNMT3A SNPs and the risk of NIHL
Four DNMT1 and DNMT3A SNPs were selected for genotyping in the 1054 noise-exposed workers (527 NIHL patients and 527 controls). Table 3 presents the genotype and allele distributions of rs7578575, rs749131, rs1550117, and rs2228611 under the co-dominant, dominant, recessive, and allelic models. Results showed that the genotype frequencies of rs749131, rs1550117, and rs2228611 under the co-dominant model were significantly different between cases and controls (P = 0.009, 0.034, and 0.017, respectively). Under the recessive model, the rs749131 GG, rs1550117 AG + GG, and rs2228611 AG + GG genotypes were significantly associated with increased NIHL risk (P = 0.002, 0.010, and 0.007, respectively). Subsequent logistic regression analysis to adjust for age, sex, smoking, and drinking showed that individuals with the rs749131 GG, rs1550117 AG + GG, and rs2228611 AG + GG genotypes had increased NIHL risk, with ORs of 1.84 (95% CI = 1.24–2.74), 2.57 (95% CI = 1.22–5.41), and 1.98 (95% CI = 1.19–3.20). Under the allelic model, the rs749131 G (OR = 1.24, 95% CI = 1.03–1.48) allele conferred a significantly increased risk for NIHL (P = 0.020). Thus, our data revealed that the DNMT3A SNPs rs749131 G and rs1550117 G and the DNMT1 SNP rs2228611 G may be significantly associated with increased NIHL susceptibility.
Stratified analysis of rs2802292, rs10457180, and rs12206094 polymorphisms and NIHL risk
The impacts of the rs7578575, rs749131, rs1550117, and rs2228611 genotypes on a series of NIHL risk characteristics were analyzed under a recessive model, and the results are shown in Table 4. We selected 16 years as work time standard as approximately half of the subjects were ≤6 work years, and the other were >16 work years. Significant differences between cases and controls were observed in the genotype distributions of rs2228611 in those with noise exposure time ≤16 years (OR = 4.05, 95% CI = 1.71–9.60) and in the genotype distributions of rs749131 in those with noise exposure time >16 years (OR = 2.34, 95% CI = 1.36–4.03). For persons exposed to ≤85 dB, those carrying the rs749131 GG (OR = 1.97, 95% CI = 1.03–3.84) or rs1550117 AG + GG genotypes (OR = 3.22, 95% CI = 1.02–10.16) exhibited an increased risk for NIHL. Workers exposed to >92 dB carrying the rs2228611 AG + GG genotypes (OR = 4.30, 95% CI = 1.58–11.70) also exhibited an increased risk for NIHL.
Association of DNMT1 and DNMT3A haplotypes with NIHL risk
The haplotype frequencies of the four SNPs were compared between NIHL cases and controls (Table 5). Ten common haplotypes (frequency > 2%) derived from the four SNPs accounting for 95% of the haplotype variation were selected, and the rest of the haplotypes were pooled into the mixed group. The haplotypes AGGG and TGGA (rs7578578-rs749131-rs1550117-rs2228611) were found to be associated with an increased risk for NIHL (OR = 1.346 and 1.557, respectively).
Association between high-frequency hearing threshold shift and SNP genotypes
Figure 1 shows the results of the association analysis of the high-frequency hearing threshold shift and the genotypes at rs7578575, rs749131, rs1550117, and rs2228611 in the 1054 noise-exposed workers. Subjects with the GG genotype at rs749131 exhibited a significantly larger high-frequency hearing threshold shift than those with the GT or TT genotypes (P = 0.018). In addition, those with the GG genotype at rs1550117 and AG + GG genotypes at rs2228611 were found to have higher high-frequency hearing threshold shifts than those with the AA genotype.
Multifactor dimensionality reduction analysis of interactions among SNPs
The results of the multifactor dimensionality analysis of the interactions among the four SNPs are presented in Table 6 and Fig. 2. Figure 2 shows the distribution of high-risk and low-risk genotypes in the best locus model. The interaction results suggested that interactions between rs749131 and rs1550117 and among rs7578575, rs749131, and rs2228611 were associated with an increased NIHL risk (OR = 1.44 and 1.63, P = 0.0024 and P < 0.0001, respectively).
Graph model of the interaction between the four SNPs. Dark gray and light gray boxes presented the high- and low-risk factor combinations, respectively. Left bars within each box represented case while the right bars represented control. The heights of the bars are proportional to the sum of samples in each group.
Identification of promoter activity associated with SNP
Transcription factor binding site prediction by TRANSFAC showed that the version of DNMT3A intron 1 containing the rs1550117 G allele (Fig. 3A) may exhibit higher transcription factor binding activity than that containing the A allele (Fig. 3B). As shown in Fig. 3C, a subsequent luciferase reporter assay showed that the luciferase activity of intron 1 containing the G allele was significantly higher than that of intron 1 carrying the A allele in HEI-OC1 cells (P = 0.0035), indicating that there may be allele-specific modulation of DNMT3A expression, with the allele at rs1550117 potentially determining transcription factor binding affinity.
Identification of promoter activity in SNP rs1550117 region. Transcription factor binding sites prediction of intron 1 region containing rs1550117 G allele (A) and A allele (B). The arrow represents the transcription factor E47 binding site (CCACCAGCAC) from 9 to 18 bp. (C) Luciferase reporter assay of the luciferase activity of rs15550117.
Discussion
SNPs are a common type of genetic variation in the human genome, with about 15 million SNPs among all humans23. Analysis of haplotypes, defined as a specific set of alleles observed on a single chromosome or part of a chromosome, has been an integral part of human genetics for decades24. However, the genomic distribution of SNPs is not homogeneous, with most SNPs occurring in noncoding regions. At present, SNP detection methods include denaturing gradient gel electrophoresis (DGGE), single-strand conformational polymorphism (SSCP), cleaved amplified polymorphic sequence (CAPS), and allele-specific PCR (such as TaqMan genotype-PCR).
In the current study, a genetic association analysis was performed with four selected DNMT1 and DNMT3A SNPs (rs7578575, rs749131, rs1550117, and rs2228611) in 527 NIHL patients and 527 controls using TaqMan genotyping. The results revealed that the rs749131 GG and rs1550117 AG/GG genotypes of DNMT3A and the rs2228611 AG/GG genotype of DNMT1 are associated with a significantly higher NIHL risk. Subsequent haplotype analysis shows that the haplotypes AGGG and TGGA (rs7578578-rs749131-rs1550117-rs2228611) confer increased risk of NIHL. These findings support our hypothesis that DNMT1 and DNMT3A polymorphisms may contribute to NIHL susceptibility in the Chinese population. To our knowledge, this is the first association study showing that DNMT3A genes are correlated with increased NIHL risk in the Chinese population.
To date, defects in DNA methylation have been identified as causative or contributing to numerous cancers, developmental syndromes, genetic disorders, and complex conditions25. DNMT1 maintains methylation patterns, preserving epigenetic information across subsequent cellular generations, whereas DNMT3A establishes new patterns of methylation. Currently, numerous hearing impairment diseases have been shown to be caused by mutations in the maintenance methyltransferase DNMT1 and corresponding defects in the DNA methylation profiles of patients. In our study, mutation from A to G at SNP rs1550117 was significantly associated with increased NIHL risk. In addition, the G allele resulted in increased promoter activity of DMNT3A compared to the A allele. Despite the fact that rs749131 is located in a noncoding region of the DNMT3A gene, numerous studies have demonstrated functional consequences of noncoding SNPs involved in the regulation of protein-coding genes and long noncoding RNAs26,27. rs2228611 is a synonymous mutation located in the DNMT1 gene. Kimchi-Sarfaty reported that a synonymous SNP in the MDR1 gene, part of a haplotype previously linked to the altered function of the MDR1 gene product P-glycoprotein (P-gp), nonetheless resulted in P-gp with altered drug and inhibitor interactions28. This demonstrates that naturally occurring synonymous SNPs can lead to the synthesis of protein products with the same amino acid sequence but different structural and functional properties. Supek et al. also reported that synonymous mutations can be oncogenic by altering transcript splicing, thereby affecting protein function29.
Our current study has several potential limitations. First, while the sample size of our study was relatively large compared to previous research, the power of the statistical tests may not be sufficient to detect the small biological effects of individual SNPs. Therefore, a larger sample size and cohort studies are needed in the future to confirm the effects of the DNMT polymorphisms on NIHL risk. Second, the subjects in this case-control study were limited to those in the Chinese population. Thus, our results may be more applicable to the Chinese Han population, with limited external generalizability. Third, rs749131 and rs2228611 are noncoding SNPs; thus, we assume that these variants are linked with one or more functional variants within the DNMT genes or their regulatory regions. Future fine mapping of these DNMT genes may detect such functional variants.
Conclusions
In conclusion, genotypes at rs749131 G and rs1550117 G of DNMT3A and rs2228611 G of DNMT1 are associated with a significantly higher risk of NIHL. Thus, our findings indicate that DNMT SNPs (rs749131, rs1550117, and rs2228611) may play crucial roles in NIHL and have the potential to serve as biomarkers for noise-exposed workers.
References
Konings, A., Van Laer, L. & Van Camp, G. Genetic studies on noise-induced hearing loss: a review. Ear Hear 30, 151–159, https://doi.org/10.1097/AUD.0b013e3181987080 (2009).
Carlsson, P. I. et al. The influence of genetic variation in oxidative stress genes on human noise susceptibility. Hear Res 202, 87–96, https://doi.org/10.1016/j.heares.2004.09.005 (2005).
Henderson, D., Bielefeld, E. C., Harris, K. C. & Hu, B. H. The role of oxidative stress in noise-induced hearing loss. Ear Hear 27, 1–19, https://doi.org/10.1097/01.aud.0000191942.36672.f3 (2006).
Le Prell, C. G. et al. Pathways for protection from noise induced hearing loss. Noise & health 5, 1–17 (2003).
Le Prell, C. G., Yamashita, D., Minami, S. B., Yamasoba, T. & Miller, J. M. Mechanisms of noise-induced hearing loss indicate multiple methods of prevention. Hear Res 226, 22–43, https://doi.org/10.1016/j.heares.2006.10.006 (2007).
Henderson, D., Subramaniam, M. & Boettcher, F. A. Individual susceptibility to noise-induced hearing loss: an old topic revisited. Ear Hear 14, 152–168 (1993).
White, C. H. et al. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mammalian genome: official journal of the International Mammalian Genome Society 20, 207–213, https://doi.org/10.1007/s00335-009-9178-5 (2009).
Davis, R. R. et al. Genetic basis for susceptibility to noise-induced hearing loss in mice. Hear Res 155, 82–90 (2001).
Kowalski, T. J., Pawelczyk, M., Rajkowska, E., Dudarewicz, A. & Sliwinska-Kowalska, M. Genetic variants of CDH23 associated with noise-induced hearing loss. Otology & neurotology: official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology 35, 358–365, https://doi.org/10.1097/MAO.0b013e3182a00332 (2014).
Chang, N. C. et al. Association of polymorphisms of heat shock protein 70 with susceptibility to noise-induced hearing loss in the Taiwanese population. Audiology & neuro-otology 16, 168–174, https://doi.org/10.1159/000317119 (2011).
Zhang, X. et al. Associations of genetic variations in EYA4, GRHL2 and DFNA5 with noise-induced hearing loss in Chinese population: a case- control study. Environmental health: a global access science source 14, 77, https://doi.org/10.1186/s12940-015-0063-2 (2015).
Law, J. A. & Jacobsen, S. E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature reviews. Genetics 11, 204–220, https://doi.org/10.1038/nrg2719 (2010).
Hirasawa, R. et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes & development 22, 1607–1616, https://doi.org/10.1101/gad.1667008 (2008).
Uysal, F., Akkoyunlu, G. & Ozturk, S. Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos. Biochimie 116, 103–113, https://doi.org/10.1016/j.biochi.2015.06.019 (2015).
Klein, C. J. et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nature genetics 43, 595–600, https://doi.org/10.1038/ng.830 (2011).
Kernohan, K. D. et al. Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clinical epigenetics 8, 91, https://doi.org/10.1186/s13148-016-0254-x (2016).
Wolber, L. E. et al. Epigenome-wide DNA methylation in hearing ability: new mechanisms for an old problem. PLoS One 9, e105729, https://doi.org/10.1371/journal.pone.0105729 (2014).
Bomfim, M. M. et al. Antioxidant responses and deregulation of epigenetic writers and erasers link oxidative stress and DNA methylation in bovine blastocysts. Molecular reproduction and development. https://doi.org/10.1002/mrd.22929 (2017).
Hu, F. et al. Lack of association between DNMT1 gene polymorphisms and noise-induced hearing loss in a Chinese population. Noise & health 15, 231–236, https://doi.org/10.4103/1463-1741.113517 (2013).
Shen, H. et al. A functional Ser326Cys polymorphism in hOGG1 is associated with noise-induced hearing loss in a Chinese population. PLoS One 9, e89662, https://doi.org/10.1371/journal.pone.0089662 (2014).
Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74, https://doi.org/10.1038/nature15393 (2015).
Shi, Y. Y. & He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15, 97–98, https://doi.org/10.1038/sj.cr.7290272 (2005).
Jiang, R. et al. Genome-wide evaluation of the public SNP databases. Pharmacogenomics 4, 779–789, https://doi.org/10.1517/phgs.4.6.779.22821 (2003).
International HapMap, C. The International HapMap Project. Nature 426, 789–796, https://doi.org/10.1038/nature02168 (2003).
Bergman, Y. & Cedar, H. DNA methylation dynamics in health and disease. Nature structural & molecular biology 20, 274–281, https://doi.org/10.1038/nsmb.2518 (2013).
Meyer, K. B. et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS genetics 7, e1002165, https://doi.org/10.1371/journal.pgen.1002165 (2011).
Guo, H. et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nature genetics 48, 1142–1150, https://doi.org/10.1038/ng.3637 (2016).
Kimchi-Sarfaty, C. et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528, https://doi.org/10.1126/science.1135308 (2007).
Supek, F., Minana, B., Valcarcel, J., Gabaldon, T. & Lehner, B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335, https://doi.org/10.1016/j.cell.2014.01.051 (2014).
Acknowledgements
This research was funded by Jiangsu Province’s Outstanding Medical Academic Leader program (CXTDA2017029), Nanjing Medical Technology Development Funds (YKK16220), Nanjing Medical Science and technique Development Foundation (QRX17197) and Jiangsu Program for Young Medical Talents (QNRC2016528).
Author information
Authors and Affiliations
Contributions
E.D. performed the experiments and wrote the paper. J.L., H.G. and H.S. collected the specimens. H.Z., W.G. and H.S. statistically analyzed the data. B.Z. designed the research and wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ding, E., Liu, J., Guo, H. et al. DNMT1 and DNMT3A haplotypes associated with noise-induced hearing loss in Chinese workers. Sci Rep 8, 12193 (2018). https://doi.org/10.1038/s41598-018-29648-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29648-4
Keywords
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.