Abstract
The detection of germline mutations in BRCA1 and BRCA2 is essential to the formulation of clinical management strategies, and in Brazil, there is limited access to these services, mainly due to the costs/availability of genetic testing. Aiming at the identification of recurrent mutations that could be included in a low-cost mutation panel, used as a first screening approach, we compiled the testing reports of 649 probands with pathogenic/likely pathogenic variants referred to 28 public and private health care centers distributed across 11 Brazilian States. Overall, 126 and 103 distinct mutations were identified in BRCA1 and BRCA2, respectively. Twenty-six novel variants were reported from both genes, and BRCA2 showed higher mutational heterogeneity. Some recurrent mutations were reported exclusively in certain geographic regions, suggesting a founder effect. Our findings confirm that there is significant molecular heterogeneity in these genes among Brazilian carriers, while also suggesting that this heterogeneity precludes the use of screening protocols that include recurrent mutation testing only. This is the first study to show that profiles of recurrent mutations may be unique to different Brazilian regions. These data should be explored in larger regional cohorts to determine if screening with a panel of recurrent mutations would be effective.
Similar content being viewed by others
Introduction
BRCA1 and BRCA2 are tumor suppressor genes and their protein products play an important role in the repair of DNA double-strand breaks through homologous recombination (HR)1. Individuals harboring germline pathogenic variants in BRCA1 and BRCA2 (BRCA) are strongly predisposed to the development of breast (BC; lifetime risk up to 85% and 45%, respectively) and ovarian cancers (OC; lifetime risk up to 39% and 11%, respectively)2 as well as other solid tumors3. As bona fide tumor-suppressor genes, the wild-type allele is frequently lost (mainly through loss of heterozygosity) during tumorigenesis, thereby becoming completely inactivated in the tumor4. Tumors arising within the context of a complete loss of BRCA function are amenable to treatment with agents targeting HR DNA repair deficiency, such as poly(ADP) ribose polymerase (PARP) inhibitors (PARPi) and platinum-based chemotherapy5. In the past few years, these drugs have shown to be effective in the treatment of advanced ovarian cancer in patients harboring somatic and/or germline BRCA mutations6,7, and more recently, the FDA extended the approval of PARPi in the BC BRCA-related metastatic clinical setting8,9. These approvals will benefit many patients since BRCA mutations are identified in 8–13% of all ovarian cancer cases10, and approximately 10% of breast cancers11, which is the most common cancer type among women worldwide and also in Brazil12,13.
Considering this scenario, the identification of a BRCA mutation is of paramount importance not only for providing appropriate genetic counseling and discussing risk-reducing interventions, but also for determining treatment options in patients with metastatic disease. A challenge in the identification of carriers in Latin America, however, is the limited availability of cancer risk evaluation programs and genetic testing per se. In Brazil, 70–80% of the population relies on the public health care system14, which does not provide genetic testing. Consequently, the mutational profile of BRCA remains largely unknown. Systematic testing of at-risk individuals brings knowledge of the genetic background of a population, and may enable the identification of multiple recurrent and/or founder mutations, which, in turn, would support the use of mutation panels as a first-line screening tool.
In this study, we aimed to describe the landscape of BRCA germline mutations in Brazil and investigate if the use of a panel of recurrent mutations would be useful in this population.
Results
A total of 649 reports of pathogenic/likely pathogenic variants were retrieved from 28 centers in 11 different Brazilian States. As shown in Fig. 1, the majority of reports was obtained from the State of São Paulo (60.1%), which is also the State with the largest number of participating centers (N = 7). The second largest number of reports was obtained from the State of Rio Grande do Sul (16.5%).
Geographical distribution of HBOC patients with pathogenic and likely pathogenic BRCA1 and BRCA2 variants in Brazil (N = 649). Legends represent the Brazilian States of Amazonas (AM), Pará (PA), Ceará (CE), Rio Grande do Norte (RN), Bahia (BA), Minas Gerais (MG), Espírito Santo (ES), Rio de Janeiro (RJ), São Paulo (SP), Santa Catarina (SC) and Rio Grande do Sul (RS) and the numbers indicate the number of cases reported from each State. The five Brazilian regions are depicted in different colors and the number between parentheses indicate the approximated population of each region.
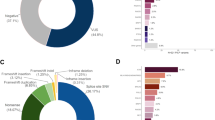
The most common types of pathogenic variants identified in both genes were small deletions and single nucleotide variants (SNVs), and are predicted to result in frameshift and non-sense alterations in the protein sequence (Fig. 2). Synonymous pathogenic variants were identified only once in each gene, in two distinct patients: BRCA1 c.4185G>A and the BRCA2 c.9117G>A. Large genomic rearrangements (LGR) were present in 4.9% of all cases, and among them, the BRCA2 c.156_157insAlu corresponded to 34.3% of all LGRs. The frequency of each type of variant and their molecular consequences are depicted in Fig. 2A,B, respectively.
As shown in Table 1, 126 distinct pathogenic BRCA1 variants were identified among 441 probands, corresponding to 68% (441/649) of all reported mutations. Among these, a subset of 33 distinct BRCA1 mutations corresponded to 73.4% of all variants identified in this gene. The nine most prevalent BRCA1 mutations accounted for 50.3% of all BRCA1 reported mutations, and among these, the European founder mutation c.5266dupC (formerly known as 5382insC) was the most common, corresponding to 20.2% of all variants found in BRCA1. In BRCA2, 103 distinct variants were identified in 208 probands, corresponding to 32% of all individuals tested (Table 2). The mutational profile of BRCA2 was more heterogeneous, since non-recurring mutations (those seen only once) were more common (35.1%) than in BRCA1 (15.4%). Moreover, a higher frequency of novel variants was identified in BRCA2 (17/103) when compared to BRCA1 (9/126). Figures 3 and 4 show all reported BRCA1 and BRCA2 mutations, respectively, including LGR in both genes. Detailed information about BRCA1 and BRCA2 mutations (predicted protein change, rs number and overall frequency) are summarized in the Supplementary Dataset.
Circos plot showing the distribution of all reported BRCA1 mutations. Point mutations and small deletions and insertions are shown around in the outermost ring, which represents the BRCA1 exons. The number between brackets correspond to the number of mutation carriers. Each reported LGR is represented by dashed blocks in the three intermediate rings, while the innermost ring represent the BRCA1 domains.
Circos plot showing the distribution of all reported BRCA2 mutations. Point mutations and small deletions and insertions are shown around in the outermost ring, which represents the BRCA2 exons. The number between brackets correspond to the number of mutation carriers. Each reported LGR is represented by dashed blocks in the intermediate ring, while the innermost ring represent the BRCA2 domains.
Although the most common mutation, BRCA1 c.5266dupC, was reported in all geographical regions, some recurrent BRCA1 mutations (detected in three or more individuals) seem to be unique to a particular Brazilian State. The variants c.188 T>A, c.2405_2406delTG, c.3916_3917delTT, c.689_692delAGAC, c.4287C>A, and c.5123C>A were reported exclusively among individuals recruited from the State of São Paulo (Southeastern region). In addition, the c.1039_1040delCT and c.1039delC variants were reported exclusively in the State of Pará (Northern region), while c.3598C>T and c.5177_5180delGAAA were only reported in pathogenic mutation carriers from the State of Rio Grande do Sul (Southern Region). No similar trends were observed among BRCA2 recurrent mutations.
When considering all BRCA1 and BRCA2 mutations seen in three or more individuals, a subset of 51 variants (33 in BRCA1 and 18 in BRCA2), accounted for 67% of all reports. In a more stringent scenario, mutations seen in four or more individuals, totaling 30 variants (23 in BRCA1 and 7 in BRCA2) corresponded to 57.3% of all mutations.
Discussion
Many factors affect the probability of developing breast or ovarian cancer, but no predictor is as determinant and prevalent as the inheritance of a BRCA mutation. There are several clinical management options for individuals harboring BRCA mutations, including risk reducing surgeries (bilateral risk-reducing mastectomy, salpingo-oophorectomy)15, chemoprevention16 and intensive surveillance with annual breast magnetic resonance imaging17. Several studies have demonstrated that, after identifying a BRCA-mutation carrier, genetic counseling and testing of at-risk individuals results in increased surveillance and use of risk-reduction strategies ultimately leading to primary or secondary prevention of cancer and improved outcomes in carriers18. Despite these benefits there is limited availability of genetic testing in Latin American countries, including Brazil14,19.
Low cost screening panels including recurrent BRCA pathogenic variants (e.g. Ashkenazi Jewish Panel) have been used in certain countries/populations as an initial approach to overcome technical and economical restrictions that still exist for comprehensive BRCA1 and BRCA2 testing. Most of the populations where this strategy is used show few mutations occurring at a high frequency, often due to founder effects19,20. Thus, the development of such panels depends on a deep knowledge of the mutational spectrum of the target population and the presence of a relatively small number of recurrent mutations explaining a significant proportion of cases. This strategy has been proposed, for instance, for Hispanic breast and/or ovarian cancer families (with predominantly Mexican origin) where nine recurrent variants account for 53% of all detected BRCA mutations21. For this population, a low-cost multiplex PCR-based panel (HISPANEL) was developed and subsequently estimated to identify up to 75% of all true Mexican BRCA mutations. The pattern of highly recurrent mutations is also seen in other Latin American countries: Bahamas (six recurrent mutations correspond to 89.4% of all carriers), Colombia (three recurrent mutations correspond to 88.9% of all carriers) and Peru (three recurrent mutations correspond to 84.6% of all carriers)19. However, this striking pattern of recurrent mutations seen in several Latin American countries may not be observed in all countries, and specific mutations maybe be shared only by a few populations. In fact, a recent Brazilian study showed that the use of a single screening panel for different Latin American populations will likely not be effective22, because there does not seem to be a significant overlap of recurrent mutations among different Latin American populations19,23. These results are not surprising due to vary distinct population migration waves and therefore genetic admixture background of Brazil in comparison with the other Latin American countries24.
Knowledge about the germline mutational spectrum among Brazilian HBOC patients is limited. Only five studies have performed comprehensive BRCA mutation testing (using gene sequencing and LGR analysis) to date25,26,27,28,29, corresponding to only 1,041 individuals tested, among a Brazilian population of over 207 million people30. Most studies have focused on specific mutations, or screened only a few regions of BRCA1 and/or BRCA2 (summarized in the Supplementary Dataset). To our knowledge, this is the largest comprehensive description of the spectrum of germline BRCA mutations in different geographical Brazilian regions.
Most of the mutations reported previously in smaller Brazilian studies, involving the analysis of only certain gene regions, have also been identified in our cohort, but it is noteworthy that some of the previously reported recurrent mutations are completely absent in this dataset. The most striking example is the BRCA1 ins6Kb rearrangement, which was reported by Esteves et al.31 in seven carriers, five of whom were from Rio Grande do Sul State. However, in our cohort we did not identify this rearrangement in any patient, even considering that Rio Grande do Sul was the second State in terms of the number of reported carriers (N = 107). The BRCA1 6 kb insertion can be detected by routine LGR testing through MLPA, and although we cannot assure that all probands were subjected to MLPA analysis, we can expect that most of them with negative sequencing results were also investigated for LGRs, since patients from the private healthcare setting and also those enrolled in research studies are routinely tested for LGR by MLPA. Indeed, a recent study from Alemar et al. reported LGR data from 351 HBOC probands from the same Brazilian State where the previous cases harboring the 6 kb insertion were reported originally, and the BRCA1 6 kb insertion was not detected, suggesting a very low frequency of this LGR in probands with the HBOC phenotype29.
Among all distinct mutations identified 11.8% were novel, corresponding to 4.6% of all carriers and highlighting the heterogeneity of our population. The identification of novel mutations linked to HBOC is a vital information that should be shared with established mutation databases, in order to become useful for interpreting further tests and to answer questions about the association between a variant and phenotype.
Overall our data show a significant molecular heterogeneity among the BRCA1 and BRCA2 mutations identified, and a similar profile of type and molecular consequence of pathogenic variants in both genes. In addition, BRCA1 mutations were more frequent than BRCA2 mutations, which is in agreement with previous data showing this same proportion of mutations between both genes among women from different ethnicities, except Asians32. Also, similar to previous report33, the rate of large genomic rearrangements did not exceed 5% of all mutations. However, it is remarkable that 34.3% of all LGR reported here correspond to the Portuguese founder mutation BRCA2 c.156_157insAlu34. Although significant, this frequency may still be an underestimation, since until very recently the detection of this particular mutation, which requires a specific PCR reaction, was not carried out by most commercial laboratories. Recently (July 2016), MRC-Holland included an extra probe that detects the wild-type sequence of this region in its BRCA2 MLPA kits (P090 version B1 and P45 version C1), allowing the detection of this variant during MLPA testing. This simple modification is expected to increase significantly the detection rate of c.156_157insAlu in Brazilian HBOC patients. The probands with c.156_157insAlu identified here were from the States of Minas Gerais (1), Rio de Janeiro (3) Rio Grande do Sul (2) and São Paulo (5) but screening for this LGR should be done for patients regardless of State of origin.
In the current HBOC genetic testing landscape, where most laboratories have migrated to next generation sequencing (NGS), analysis workflows allow the filtering of many types of mutations, including the exclusion of synonymous variants. In our study, we have identified two pathogenic synonymous mutations, and this finding highlights the importance of careful evaluation of each BRCA variant detected. It is widely known that synonymous substitutions can alter splicing accuracy, creating or destroying a native donor or acceptor splice site, but they can also can modify translation fidelity, mRNA structure and protein folding35. Indeed, both pathogenic synonymous variants identified in this study disrupt splice donor sites, leading to exon skipping. The G nucleotide of BRCA1 c.4185G>A represents the last nucleotide of exon 11 (according to LRG nomenclature, formerly known as exon 12), and is conserved in 86% of the splice sites in mammals36. This variant lead to an aberrant transcript lacking exon 1237. Similarly, the BRCA2 c.9117G>A leads to a complete deletion of exon 2338 and produces a frameshift effect similar to other deleterious mutations39. The process of evaluating variant significance should include multiple databases, as four mutations reported here were not described in ClinVar, although classified as clearly pathogenic in other databases. Finally, even variants described in one database should have their significance confirmed in other databases, especially if classified as variants of uncertain significance (VUS). As an example, using an ex vivo assay based on a splicing reporter minigene, Brandão et al.40 demonstrated that the BRCA1 c.4987-3C>G variant leads to the skipping of exon 17. However, it remains classified as VUS in ClinVar and it is not reported in other databases.
In this study, we have attempted to compile pathogenic and likely pathogenic BRCA1 and BRCA2 variants identified in the main Genetic Cancer Risk Assessment centers in Brazil. Although this report in fact is the most comprehensive to date, both in number of mutations reported, as well as in number of centers/regions of the country included, many limitations must be considered when analyzing the results. We were unable to obtain information on the birthplace for most of the carriers, which would have been more informative than the center where the genetic test was performed. Therefore, data on geographical location should be interpreted with caution. In fact, among some of the individuals tested in the State of São Paulo we were able to identify residents from the Midwest States. This is not unexpected since the paucity of clinical and laboratory personnel trained in clinical cancer genetics in Brazil, results in a pattern of patients with suspected hereditary cancer being refered to testing from different parts of the country to only a few reference centers very distant from their residence place. Moreover, it might be possible that the inclusion of data from point mutation analysis could increase the detection and, consequently, the reporting of a few specific mutations. However, our data regarding the most frequently reported mutations is in agreement with previous Brazilian studies that performed full BRCA1 and BRCA2 sequencing and MLPA. These studies show, for example, that the BRCA1 c.5266dupC is the most prevalent mutation across distinct regions of Brazil, which was also the case in our study26,27,29.
We confirm that there is significant molecular heterogeneity in the BRCA1 and BRCA2 genes among Brazilian carriers. Although our findings suggest that this heterogeneity precludes the use of screening protocols that include recurrent mutation testing only, our results also show that certain mutations occur at a high frequency in some Brazilian regions and not others. These variations could be due to mutation founder effects, which have been described for other genes in Brazil. These findings should be explored in larger cohorts from specific Brazilian regions to assess whether in these areas, screening with a panel of recurrent mutations would be effective.
Materials and Methods
Laboratory reports of BRCA1 and BRCA2 testing showing pathogenic or likely pathogenic germline mutations were compiled from 28 public and private health care offices located in 11 Brazilian states, including the main reference centers for Genetic Cancer Risk Assessment (GCRA) in Brazil. Not all probands were subjected to a comprehensive BRCA testing (full BRCA sequencing and multiplex ligation-dependent probe amplification, MLPA). The genetic testing was performed using distinct methodologies, including full gene analysis by Sanger or next generation sequencing, point mutation analysis by Sanger or genotyping methods (as HISPANEL), and MLPA for analysis of large genomic rearrangements. Most data came from institutions participating in the Brazilian Hereditary Cancer Network (BHCN), convened by the Brazilian National Cancer Institute (INCA, Instituto Nacional de Cancer) and partially supported by public funding from the National Council for Scientific and Technical Development (CNPq)41. These centers, mostly public hospitals, are established in the Cities/States of Belém/Pará (in the Northern region, encompassing the Amazon basin), Salvador/Bahia (in the Northeastern region), Vitória/Espírito Santo, Rio de Janeiro/Rio de Janeiro, São Paulo/São Paulo, Ribeirão Preto/São Paulo, Barretos/São Paulo (in the Southeastern region) and Porto Alegre/Rio Grande do Sul (in Southern Brazil). In addition, public or private health care offices from the States of Amazonas (Northern region and the Amazon basin), Minas Gerais (Southeastern region), Rio Grande do Norte, Ceará (Northeastern region) and Santa Catarina (Southern region) also contributed with molecular data from their patients (Fig. 1). All subjects were unrelated and fulfilled HBOC criteria for BRCA testing. Some of the mutations described in this manuscript were also described in prior population/region-specific prevalence studies22,25,26,33,42,43,44,45. This project was approved by the Institutional Review Board from Hospital de Clínicas de Porto Alegre (approval n° 10-0521) and all individuals provided written or verbal consent for BRCA testing. All methods were performed in accordance with the relevant guidelines and regulations, and all data supporting the results are shown in the Supplementary Dataset.
The Human Genome Variation Society (HGVS) nomenclature guidelines (http://varnomen.hgvs.org/) were used to annotate identified variants and the ClinVar database (www.ncbi.nlm.nih.gov/clinvar/) was used to determine the biological significance of all reported variants. For novel variants, BRCA Share (formerly known as UMD, http://www.umd.be/), LOVD (http://www.lovd.nl/3.0/home), ARUP (http://arup.utah.edu/database/BRCA/) and BRCA Exchange (http://brcaexchange.org/) databases were also checked. Current ACMG46 guidelines were also used for further classification. BRCA1 and BRCA2 domains were defined using the boundaries in the Pfam database (http://pfam.xfam.org).
References
Roy, R., Chun, J. & Powell, S. N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12, 68–78 (2012).
Antoniou, A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72, 1117–30 (2003).
Mersch, J. et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 121, 269–75 (2015).
Venkitaraman, A. R. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 343, 1470–5 (2014).
Maxwell, K. N. & Domchek, S. M. Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol 9, 520–8 (2012).
Kim, G. et al. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin Cancer Res 21, 4257–61 (2015).
Syed, Y. Y. Rucaparib: First Global Approval. Drugs 77, 585–592 (2017).
Lord, C. J. & Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
FDA. FDA approves olaparib for germline BRCA-mutated metastatic breast cancer. (U.S Food and Drug Administration, 2018).
Liu, G. et al. Differing clinical impact of BRCA1 and BRCA2 mutations in serous ovarian cancer. Pharmacogenomics 13, 1523–35 (2012).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–86 (2015).
Instituto Nacional de Câncer (INCa) (2016).
Goss, P. E. et al. Planning cancer control in Latin America and the Caribbean. Lancet Oncol 14, 391–436 (2013).
Ludwig, K. K., Neuner, J., Butler, A., Geurts, J. L. & Kong, A. L. Risk reduction and survival benefit of prophylactic surgery in BRCA mutation carriers, a systematic review. Am J Surg 212, 660–669 (2016).
Roukos, D. H. & Briasoulis, E. Individualized preventive and therapeutic management of hereditary breast ovarian cancer syndrome. Nat Clin Pract Oncol 4, 578–90 (2007).
Saslow, D. et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin 57, 75–89 (2007).
Scheuer, L. et al. Outcome of preventive surgery and screening for breast and ovarian cancer in BRCA mutation carriers. J Clin Oncol 20, 1260–8 (2002).
Dutil, J. et al. The spectrum of BRCA1 and BRCA2 alleles in Latin America and the Caribbean: a clinical perspective. Breast Cancer Res Treat 154, 441–53 (2015).
Weitzel, J. N. et al. Prevalence of BRCA mutations and founder effect in high-risk Hispanic families. Cancer Epidemiol Biomarkers Prev 14, 1666–71 (2005).
Weitzel, J. N. et al. Prevalence and type of BRCA mutations in Hispanics undergoing genetic cancer risk assessment in the southwestern United States: a report from the Clinical Cancer Genetics Community Research Network. J Clin Oncol 31, 210–6 (2013).
Alemar, B. et al. Prevalence of Hispanic BRCA1 and BRCA2 mutations among hereditary breast and ovarian cancer patients from Brazil reveals differences among Latin American populations. Cancer Genet 209, 417–422 (2016).
Ossa, C. A. & Torres, D. Founder and Recurrent Mutations in BRCA1 and BRCA2 Genes in Latin American Countries: State of the Art and Literature Review. Oncologist 21, 832–9 (2016).
Kehdy, F. S. et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci USA 112, 8696–701 (2015).
Carraro, D. M. et al. Comprehensive analysis of BRCA1, BRCA2 and TP53 germline mutation and tumor characterization: a portrait of early-onset breast cancer in Brazil. PLoS One 8, e57581 (2013).
Fernandes, G. C. et al. Prevalence of BRCA1/BRCA2 mutations in a Brazilian population sample at-risk for hereditary breast cancer and characterization of its genetic ancestry. Oncotarget 49, 80465–81 (2016).
Silva, F. C. et al. Hereditary breast and ovarian cancer: assessment of point mutations and copy number variations in Brazilian patients. BMC Med Genet 15, 55 (2014).
Maistro, S. et al. Germline mutations in BRCA1 and BRCA2 in epithelial ovarian cancer patients in Brazil. BMC Cancer 16, 934 (2016).
Alemar, B. et al. BRCA1 and BRCA2 mutational profile and prevalence in hereditary breast and ovarian cancer (HBOC) probands from Southern Brazil: Are international testing criteria appropriate for this specific population? PLoS One 12, e0187630 (2017).
IBGE. Projeção da população do Brasil. Vol. 2017 (Instituto Brasileiro de Geografia e Estatística (IBGE), 2017).
Esteves, V. F. et al. Prevalence of BRCA1 and BRCA2 gene mutations in families with medium and high risk of breast and ovarian cancer in Brazil. Braz J Med Biol Res 42, 453–7 (2009).
Hall, M. J. et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer 115, 2222–33 (2009).
Ewald, I. P. et al. BRCA1 and BRCA2 rearrangements in Brazilian individuals with Hereditary Breast and Ovarian Cancer Syndrome. Genet Mol Biol 39, 223–31 (2016).
Machado, P. M. et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol 25, 2027–34 (2007).
Sauna, Z. E. & Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12, 683–91 (2011).
Shapiro, M. B. & Senapathy, P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15, 7155–74 (1987).
Claes, K. et al. Differentiating pathogenic mutations from polymorphic alterations in the splice sites of BRCA1 and BRCA2. Genes Chromosomes Cancer 37, 314–20 (2003).
Peelen, T. et al. Screening for BRCA2 mutations in 81 Dutch breast-ovarian cancer families. Br J Cancer 82, 151–6 (2000).
Bonatti, F. et al. RNA-based analysis of BRCA1 and BRCA2 gene alterations. Cancer Genet Cytogenet 170, 93–101 (2006).
Brandão, R. D. et al. BRCA1 c.4987-3C>G is a pathogenic mutation. Breast Cancer Res Treat 131, 723–5 (2012).
Ashton-Prolla, P. & Seuanez, H. N. The Brazilian Hereditary Cancer Network: historical aspects and challenges for clinical cancer genetics in the public health care system in Brazil. Genet Mol Biol 39, 163–5 (2016).
Ewald, I. P. et al. Prevalence of the BRCA1 founder mutation c.5266dupin Brazilian individuals at-risk for the hereditary breast and ovarian cancer syndrome. Hered Cancer Clin Pract 9, 12 (2011).
Alemar, B. et al. BRCA1 and BRCA2 mutational profile and prevalence in Hereditary Breast and Ovarian Cancer (HBOC) probands from Southern Brazil: are international testing criteria valid for this specific population?. ((Submitted), 2017).
Timoteo, A. R. et al. Identification of a new BRCA2 large genomic deletion associated with high risk male breast cancer. Hered Cancer Clin Pract 13, 2 (2015).
Moreira, M. A. et al. Portuguese c.156_157insAlu BRCA2 founder mutation: gastrointestinal and tongue neoplasias may be part of the phenotype. Fam Cancer 11, 657–60 (2012).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–24 (2015).
Acknowledgements
We wish to thank members of the Center of Molecular Diagnosis, Oncogenetics Department and Molecular Oncology Research Center of Barretos Cancer Hospital for their contributions to the study. We also thank Amanda de Franca Nobrega, Maristela Taiari Pimenta, Simone Maistro and Ana Rafaela de Souza Timóteo for their support. This work was supported in part by grants from Barretos Cancer Hospital (FINEP - CT-INFRA, 02/2010), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2013/24633-2 and 2103/23277-8), Fundação de Apoio à Pesquisa do Rio Grande do Norte (FAPERN), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Ministério da Saúde, the Breast Cancer Research Foundation (Avon grant #02-2013-044) and National Institute of Health/National Cancer Institute (grant #RC4 CA153828-01) for the Clinical Cancer Genomics Community Research Network. Support in part was provided by grants from Fundo de Incentivo a Pesquisa e Eventos (FIPE) from Hospital de Clínicas de Porto Alegre, by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, BioComputacional 3381/2013, Rede de Pesquisa em Genômica Populacional Humana), Secretaria da Saúde do Estado da Bahia (SESAB), Laboratório de Imunologia e Biologia Molecular (UFBA), INCT pra Controle do Câncer and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). RMR and PAP are recipients of CNPq Productivity Grants, and Bárbara Alemar received a grant from the same agency. Funding agencies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.I.P., D.M.C., B.A. and P.A.P. conceived the project. All authors (E.I.P., D.M.C., B.A., M.A.M.M., A.R.S., K.A.S., H.C.R.G., R.M.R., C.P.S., N.P.C., M.I.A., R.C.B., M.N.C.F., F.B.M., F.R.V., A.C.E.S., H.N.S., K.R.L.S., C.B.O.N., P.S.S., G.S.S., R.M.R.B., S.S., P.P.A., I.M.M.B., T.M.B.M.L., T.F.B., M.B.P.T., I.N., B.G., S.S., S.N., F.T.L., A.C., C.M.B., J.B., O.A., M.D.P.E.D., T.B.P.L., A.C.L., R.G., T.C., I.V.D.S., P.B., D.M., S.N., J.H., J.W., P.A.P.) worked on data curation, investigation and methodology. E.I.P., D.M.C., B.A., M.A.M.M., A.R.S., K.A.S. and P.A.P. performed the formal analysis. E.I.P., D.M.C., B.A., M.A.M.M., A.R.S., K.A.S., R.M.R., J.W. and P.A.P. were responsible for funding acquisition. B.A., P.A.P. wrote the original draft, and E.I.P., D.M.C., B.A., M.A.M.M., A.R.S., K.A.S., R.M.R., J.H. and P.A.P. reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Palmero, E.I., Carraro, D.M., Alemar, B. et al. The germline mutational landscape of BRCA1 and BRCA2 in Brazil. Sci Rep 8, 9188 (2018). https://doi.org/10.1038/s41598-018-27315-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-27315-2
This article is cited by
-
Germline mutations in BRCA1 and BRCA2 among Brazilian women with ovarian cancer treated in the Public Health System
BMC Cancer (2024)
-
Systematic review of the molecular basis of hereditary breast and ovarian cancer syndrome in Brazil: the current scenario
European Journal of Medical Research (2024)
-
Genomic ancestry and cancer among Latin Americans
Clinical and Translational Oncology (2024)
-
BRCA1 and BRCA2 germline mutation analysis from a cohort of 1267 patients at high risk for breast cancer in Brazil
Breast Cancer Research and Treatment (2023)
-
Familial history and prevalence of BRCA1, BRCA2 and TP53 pathogenic variants in HBOC Brazilian patients from a public healthcare service
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.