Abstract
Interleukin-13 (IL-13) has important functions in atherosclerosis, but its role in coronary artery disease (CAD) is unclear. Here, we studied the genetic role of IL-13 in CAD in a Chinese Han population using tag SNPs covering the whole IL13 gene (i.e., rs1881457, rs2069744 and rs20541) and a two-stage cohort containing 1863 CAD cases and 1841 controls. Traditional risk factors for CAD, such as age, BMI, and other factors, were used as covariates in logistic regression analysis. In the total population, we found that two haplotypes of IL13 (ATG and ATA, ordered rs1881457C-rs2069744T-rs20541A) significantly contributed to the risk of CAD with adjusted p values less than 0.05 (padj = 0.019 and padj = 0.042, respectively). In subgroup population analyses, the variant rs1881457C was found to significantly contribute to a nearly two fold increase in the risk of CAD in men (padj = 0.023, OR = 1.91, 95% CI: 1.09-3.33). The variant rs1881457C also significantly contributed to a nearly twofold risk of late-onset CAD (padj = 0.024, OR = 1.93, 95% CI: 1.09-3.42). In conclusion, IL13 might be involved in CAD via different mechanisms under different conditions in the Chinese Han population.
Similar content being viewed by others
Introduction
Previous meta-analyses and genome-wide association studies (GWAS) have detected approximately 95 risk variants for CAD, but the substantial heritability of CAD remains unclear1,2,3.
Interleukin-13 (IL-13) was co-discovered by Minty et al. and McKenzie et al. as a new inflammatory cytokine in 1993. IL-13 is expressed in activated human T lymphocytes and has been reported to play important roles in inflammatory and immune responses4,5. In 2003, Gordon demonstrated that IL-13 could alternatively activate macrophages, which have different roles in humoral immunity and repair6. Early studies reported that IL-13 can increase CD36 expression through PPARγ activation and increase macrophage foam cell formation, which might then lead the development of atherosclerosis7,8,9. However, later studies demonstrated that IL-13 can maintain metabolic homeostasis by inducing macrophage PPARδ or PPARβ expression in adipose tissue, which might then promote the development of atherosclerosis10,11. Additionally, in 2012, Cardilo-Reis et al. performed in vivo studies and found that IL-13 inhibits the progression of atherosclerosis through a new mechanism that is not related to cholesterol levels12. In 2013, Stanya et al. demonstrated that IL-13 deficiency in mice leads to increased weight gain, hyperglycemia, and hepatic insulin resistance, which indicates that IL-13 might inhibit atherogenesis13. Jiang et al. found that IL-13 is involved in glucose uptake and metabolism in skeletal muscle, which regulates glucose homeostasis in metabolic diseases14. These findings indicate that IL-13 might be involved in atherosclerosis and has complex functions. Most recently, Amit et al. studied the function of IL-13Rα1 in myocardial homeostasis and heart failure and discovered that IL-13 is up-regulated in diabetic hearts15. Concordantly, Stanya et al. reported that IL-13 can control hepatic glucose production13, which might cause the reactivity in up-regulation of IL-13 in the diabetic heart.
In previous GWASs, such as analyses from the CARDIoGRAMplusC4D Consortium, two SNPs localized in the IL13 region were found to have no association with CAD and/or myocardial infarction (p = 0.260 for rs20541 and p = 0.975 for rs2069744). However, these data sets were mainly based on a European population, and the reported SNPs did not cover the whole IL13 region (UCSC). Additionally, a promoter variant, i.e., rs1881457, of IL13, which has not been studied in previous CAD GWASs, has been reported to regulate IL13 expression and might be an important functional variant in terms of the risks of various diseases16,17.
Therefore, here, we first selected tag variants covering the whole IL13 gene that included both common coding and regulatory variants and then tested the genetic associations between the selected tag variants of IL13 and CAD in a two-stage case-control cohort from a large Chinese Han population. Next, we classified the total studied population into different subgroups and tested the associations between the selected variants and CAD under different conditions. Subsequently, we assessed CAD severity using the Gensini score and examined whether the selected variants influenced this parameter. These data may elucidate the genetic function of IL-13 in CAD, which may help to determine the exact role of IL-13 in CAD.
Results
Populations
In stage 1, i.e., the discovery cohort, the CAD cases were found to have higher BMI, TG, LDL-c, and Tch level and to be older than the control subjects (Table 1). Smoking, male gender, diabetes mellitus and hypertension were more prevalent among the CAD cases than the controls. The CAD patients had lower levels of HDL-c compared with the control individuals. The validation and combined populations had the same population characteristics. Our sample size provided more than 80% statistical power for the genetic association studies conducted here.
Association analysis of the variants in IL13 with CAD in a Chinese Han population
The p values from Hardy-Weinberg disequilibrium tests in the control subjects were more than 0.001 for all of the studied variants. In the allelic association analysis, we found the following: in the discovery cohort, none of the three variants (i.e., rs1881457, rs2069744 and rs20541) in IL13 exhibited a significant association with CAD before or after adjustment for the traditional risk factors for CAD (p > 0.05); in the validation cohort, the three variants also exhibited no associations with CAD (p > 0.05); and in the combined cohort, the association results between all three SNPs and CAD were non-significant (padj > 0.05; Table 2).
Based on the genotypic association analysis, rs2069744 was significantly associated with CAD in the recessive model in the discovery cohort (padj = 0.044, OR = 0.18, 95% CI: 0.04-0.96). However, in the validation and combined cohorts, the association with rs2069744 did not remain significant (padj > 0.05; see Supplementary Table S1). The other two variants also exhibited the same non-significant association with CAD in all of the studied cohorts (padj > 0.05; see Supplementary Table S1).
Additionally, we analyzed the haplotypic association between IL13 and CAD in the combined population. Two haplotypes (ATG and ATA, ordered rs1881457C-rs2069744T-rs20541A in IL13) were significantly associated with CAD (padj = 0.019 for ATG and padj = 0.042 for ATA; see Supplementary Table S2). We further performed an association analysis of the haplotypes and Gensini scores in the CAD patients but found no significant association result (padj = 0.818). Unfortunately, the numbers of these haplotypes were too small to reach a statistical power of 80%.
Association analysis of the selected variants in IL13 with CAD in the subgroup populations
First, we separated the CAD patients into two subgroups, i.e., an anatomical-CAD group (CAD patients with anatomical disease i.e., severe coronary stenosis) and a clinical-CAD group (patients with clinical disease, i.e., MI or revascularization). In both the anatomical-CAD and clinical-CAD subgroups, the allelic association analyses revealed no significant associations of any of the SNPs with CAD after adjustment for the traditional risk factors (padj > 0.05; see Supplementary Table S3). Genotypic association analysis also demonstrated that the three SNPs were not associated with CAD in either subgroup (see Supplementary Table S3).
Second, we divided our sample into two subgroups by gender to investigate the associations of the three SNPs (rs1881457, rs2069744 and rs20541) with CAD in the different genders. In the female subgroup, none of the three SNPs exhibited a significant association with CAD in the allelic or genotypic association analyses (Table 3). In the male subgroup, the allelic association analysis revealed no significant associations of any of the SNPs with CAD before or after adjustment for the traditional risk factors (p > 0.05; Table 3). However, the genotypic association analysis found a significant association between rs1881457C and CAD in the recessive model (padj = 0.023, OR = 1.91, 95% CI: 1.09-3.33; Table 3).
Finally, we divided the CAD populations according to the age of onset of CAD. The patients with an age of onset of CAD of less than 55 years for males and less than 65 years for females were defined as the early-onset CAD subgroup. The other patients were defined as the late-onset CAD subgroup. No significant association results were discovered between the selected variants and CAD in the early-onset CAD group (padj > 0.05). However, in the late-onset CAD subgroup, rs1881457C resulted in a nearly twofold increase in the risk of CAD (padj = 0.024, OR = 1.93, 95% CI: 1.09-3.42; Table 4).
Effect of rs1881457 in IL13 on the severity of CAD
We also tested the association between rs1881457 in IL13 and CAD severity as assessed by the Gensini scoring system. The 1st (lowest) and 4th (highest) quartiles of the LN of the Gensini scores were selected for the case-control association analysis. According to the allelic association analysis, rs1881457 was not associated with CAD severity (quantitative trait association, padj = 0.594; case-control association by the 4th vs 1st, padj = 0.777, OR = 1.04, 95% CI: 0.80–1.35; see Supplementary Table S4). Additionally, we performed Mann-Whitney U tests to assess the influence of the rs1881457 genotypes on the severity of CAD. There were no significant associations between these variants and the Gensini scores, which was consistent with the above association results (p > 0.05; see Supplementary Fig. S1).
Discussion
Here, in a Chinese Han population, we explored the genetic role of IL-13 in CAD. We found the following results: two haplotypes in IL13 (ATG and ATA, ordered rs1881457C-rs2069744T-rs20541A) significantly contributed the risk of CAD; the promoter variant rs1881457C of IL13 not only contributed to the risk of CAD in male patients but was also involved in the development of late-onset CAD; and, in contrast, rs1881457C did not influence the severity of CAD as determined by the Gensini scores.
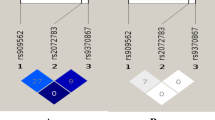
IL13 is located in chromosomal region 5p31.1 near IL4 and has a length of 2,938 bp as illustrated in Fig. 1a. Two variants, rs2706399 in IL5 and rs273909 in SLC22A4-SLC22A5, which have been associated with CAD susceptibility in previous studies, are located in the same region near IL1318. Previous data have confirmed that IL-13, IL-4 and IL-5 have important functions in autoimmune and inflammatory diseases19,20,21,22,23,24. Studies have also reported that IL-13, IL-5 and IL-4 all participate in the pathological process of atherosclerosis, which is the major pathogenesis of CAD12,25,26,27. Additionally, Newland et al. found that the genetic ablation of ILC2 in Ldlr-/- mice accelerates atherosclerosis progression, which might be due to the loss of IL-5 and IL-13 expression by ILC2 cells28. Therefore, IL13 might be involved in CAD and might be a valuable susceptibility locus for CAD.
The location of IL13 and the LD block of IL13. (a) Location of IL13 in the Ensembl database. IL13 is located in the chromosome 5p31.1 region with a length of 2,938 bp. (b) The LD block of IL13 (HapMap CHB and JPT data sets, v.3, release 2). Each diamond indicates the LD degree between the SNPs. The number in each diamond indicates the r2 value. The color indicates the D’. Dark red regions represent a high LD. White regions represent a low LD. The black boxes show the selected tag SNPs in this study.
In this study, we selected three tag SNPs in IL13, including rs1881457, rs2069744 and rs20541, to test the associations of these risk variants with CAD. Among these variants, rs20541 in IL13 has been reported to be a critical genetic risk variant in asthma29. Therefore, we concluded that this variant might also be involved in CAD. However, we did not replicate the association between rs20541 and CAD, and we also found that one of the other SNPs, i.e., rs2069744, was not a risk factor for CAD, which is consistent with previous GWAS results from the CARDIoGRAMplusC4D Consortium. However, we found other interesting results: two haplotypes in IL13 (ATG and ATA, ordered rs1881457C-rs2069744T-rs20541A) might influence the risk of CAD, and rs1881457C might increase the CAD risk in the male population and might also increase the risk of late-onset CAD in the Chinese Han population. Rs1881457, which is located in the promoter region of IL13, was not in the same linkage disequilibrium (LD) region as rs20541 and rs2069744 and was not detected in a previous large GWAS or meta-analysis of CAD that included both European and Asian populations. Therefore, we hypothesized that rs1881457 is involved in CAD via its regulation of IL13 expression. Recently, researchers have demonstrated that variants that regulate the expressions of various genes, which are also known as expression quantitative trait loci (e-QTL), exhibit population heterogeneity and tissue cell specificity and influence the risks of diseases in different conditions30,31. Rs1881457, the promoter variant of IL13, might be an e-QTL of IL13. Additionally, previous studies have reported different roles of IL-13 in vascular tissue, adipose tissue, hepatic tissue, skeletal tissue and the heart7,8,9,10,11. These findings might be due to the different e-QTLs of IL13, especially in different tissues under different disease conditions, which lead to the complex role of IL-13 in atherosclerosis by conditionally regulating IL13 expression. In 2000, Patricia et al. found that HS4 marked the promoter region of IL13 and then enhanced IL13 promoter activity32. Moreover, these researchers also found that rs1881457 is located in the central portion of HS4 and provided a binding site for the transcription factor Oct-1, which enhances IL13 expression17. Here, we discovered that rs1881457C might increase the risk of late-onset CAD risk in the Chinese Han population. In 2014, Li et al. found that the serum IL-13 level is higher in aged healthy individuals than in young individuals33. These findings indicate that the minor allele “C” of rs1881457 might down-regulate IL13 expression in the elderly population. Furthermore, Wouters et al. demonstrated that rs1881457 regulates the mRNA expression of IL13 in both irritable bowel syndrome patients and controls16.
Cardilo-Reis et al. determined that IL-13 inhibits the progression of atherosclerosis in Ldlr−/− mice12. Stanya et al. demonstrated that IL-13 deficiency in mice leads to increased weight gain, hyperglycemia, and hepatic insulin resistance13. Jiang et al. discovered that IL-13 is involved in glucose uptake and metabolism in the skeletal muscle, which regulates glucose homeostasis in metabolic diseases14. Most recently, Newland et al. demonstrated that a major innate cell source of IL-13 is required for its atheroprotective effects against the development of atherosclerosis28. Based on these findings, we concluded that, under specific conditions, the “C” allele of rs1881457 might increase the risk of CAD by reducing IL13 expression. In our study, we found that the prevalence of diabetes mellitus in the late-onset CAD subgroup population was significantly higher than that in the early-onset CAD subgroup population (see Supplementary Table S5). We hypothesized that, in diabetes mellitus, rs1881457C could reduce the IL-13 level and consequently increase the risk of CAD. Decreased IL-13 increased hyperglycemia and hepatic insulin resistance, decreased glucose uptake and then accelerated atherosclerosis, and this might be a slow and cumulative process. Cardilo-Reis et al. found that IL-13 could not reverse the course of atherosclerosis when it was only used in the last period of the experiment12. Consistent with these findings, our study demonstrated that rs1881457 in IL13 was only associated with late-onset CAD, which might be due to the constantly decreasing IL-13 level, which in turn, might lead to the accumulation of glucose and a weakened anti-inflammatory response.
Men are more susceptible CAD than women, but no studies have explained the exact mechanism underlying this difference. In 1991, Bonnin et al. reported that male sex modifies the effect of smoking34. Subsequently, Liu et al. and Sadeghnejad et al. both discovered that the function of variants in IL13 might be influenced by exposure to tobacco smoke35,36. Additionally, Chen et al. first reported that male gender can modify the interactions between variants in IL13 and prenatal exposure to tobacco smoke in atopic diseases37. In our study, we found that rs1881457C in IL13 increased the risk of CAD in male patients. According to these findings, we hypothesized that, in males, rs1881457 in IL13 might enhance the effect of prenatal exposure to tobacco smoke, which could then increase the risk of CAD. Thus, rs1881457 in IL13 is a major cause of CAD in male subjects, and the underlying mechanism is the interaction of rs1881457 in IL13 with prenatal exposure to tobacco smoke.
In summary, our study confirmed that two previous variants in IL13 that were identified in a GWAS (rs2069744 and rs20541) were not associated with CAD in a large Chinese Han population. However, we discovered that a new variant in the regulatory region of IL13, i.e., rs1881457C, increased the CAD risk, and this result was lacking from the findings of the CARDIoGRAMplusC4D Consortium. We conclude that IL13 might be involved in the development of CAD. Further studies with gene-gene and gene-environmental factor interaction analyses and larger sample sizes and functional studies in human tissues or cells are needed to provide more evidence about the exact functional mechanisms of IL-13 in CAD.
Materials
Populations
The subjects (1863 CAD cases and 1841 controls in total) were selected from the Union Hospital in Wuhan (Hubei, China). The criteria for enrollment as a CAD case were a diagnosis with percutaneous coronary intervention, myocardial infarction, coronary artery bypass graft, and/or ≥50% coronary stenosis in any one of the main vessels (i.e., the right coronary artery, left anterior descending, left main and left circumflex artery) on coronary angiography38,39,40. Individuals with coronary artery spasms, congenital heart disease, adolescent hypertension, valvular heart disease, type 1 diabetes, and serious renal or hepatic diseases were not included in this study.
According to previously published guidelines, we selected subjects with major coronary artery stenosis of no more than 30% and without histories of myocardial infarction, CAD, or ischemic stroke as controls41. Individuals who had coronary artery spasms, adolescent hypertension, type 1 diabetes, congenital heart disease, rheumatic autoimmune disease, tumor or stroke were not included. All of the participants’ clinical data, including age, BMI, gender, hypertension, smoking status and diabetes, were collected as detailed in previous studies38,39,40,41. The fasting TG, Tch, LDL-c and HDL-c concentrations were assessed with by standard methods42,43.
The DNA samples were collected using a purification kit (Wizard Genomic, Promega Corporation, Madison, WI)38,39,40,41. Coronary angiograms were available for a total of 1431 CAD individuals, and the coronary artery stenoses were assessed using the Gensini scores44. The coronary segment scores was judged as 32, 16, 8, 4, 2 or 1 for stenosis percentages of 100%, 99–91%, 90–76%, 75–51%, 50–26% or 25–0%, respectively. We then multiplied the score with a specific factor that was based on the vessel importance and size (ranging from 5.0 to 0.5). The Gensini index for each patient was the total sum of the weights of the segments.
This study followed the ethical principles of Declaration of Helsinki and passed a review by the ethics committee of Tongji Medical College, Huazhong University of Science and Technology. All participants signed an informed consent form of their own accord.
Tag SNPs
We selected the tag SNPs as follows according to the following criteria: (1) the r2 value threshold of the LD between the SNPs was more than 0.8 (HapMap CHB and JPT data sets, v.3, release 2); (2) the threshold for the minor allele frequency (MAF) of the variant was greater than 0.05; and (3) the variant was functional or potentially functional (Promoter and Genevar). Three satisfactory variants (rs1881457, rs2069744 and rs20541) in IL13 were selected for this association study (Fig. 1b). Rs1881457 is located in the promoter region of IL13 and has been reported to regulate the expression of IL1316,17. The variants were genotyped using a high-resolution melt system (Rotor-gene 6000, Corbett Life Science, Concorde, NSW, Australia), with a 25-µl volume PCR reaction system as reported in our previous study38,39,40,41 using 0.5 μl of LC Green dye, 10 pmol of each primer (the forward primer was 5′-CGTGGGCCCTCTACTACAGA-3′, and the reverse primer was 5′-CCACACTCGAAGCTTCCCA-3′ for rs1881457; the forward primer was 5′-TGAGGTTAAGTGACAGAGGCT-3′ and the reverse primer was 5′-AAATGCCCTGCCTTCTGATG-3′ for rs2069744; and the forward primer was 5′-GGTGGCCCAGTTTGTAAAGG-3′ and the reverse primer was 5′-CAGGTCCTGTCTCTGCAAAT-3′ for rs20541), 30 ng of genomic DNA, and other contents45. The concordance rate of this genotyping method was 100%, and the results were confirmed by Sanger DNA sequencing.
Statistics
Hardy-Weinberg disequilibrium tests in the control population were performed using PLINK software (version 1.07, Broad institute, USA). Pearson’s chi-square tests with 2 × 2 or 2 × 3 contingency tables were conducted in the allelic and genotypic association analyses (v.21.0, SPSS, Inc., Chicago, IL)38,39. Categorical data, including allele and haplotype frequencies, gender, smoking status, and other data, were tested using the chi-square test. Measurement data, such as BMI, age and blood lipid levels, were tested using t-tests. The traditional risk factors for CAD, including age, BMI, gender, hypertension, smoking, diabetes and lipid levels, were adjusted as covariates by logistic regression analysis (SPSS, v.21.0). The haplotype reconstruction and haplotype analysis were performed using Haploview (v.4.2, Informer Technologies, Inc.) or SPSS (v.21.0, Inc., Chicago, IL). Quartile case-control association analysis and liner regression analysis were utilized to test the associations between the levels of the Gensini scores and the SNPs. The correlations between the variant genotypes and the Gensini scores (CAD severity) was examined with Mann-Whitney U tests.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
McPherson, R. & Tybjaerg-Hansen, A. Genetics of Coronary Artery Disease. Circ Res. 118, 564–78 (2016).
Howson, J. M. M. et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 49, 1113–1119 (2017).
Nelson, C. P. et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 49, 1385–1391 (2017).
Minty, A. et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 362, 248–50 (1993).
McKenzie, A. N. et al. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc Natl Acad Sci USA 90, 3735–9 (1993).
Gordon, S. Alternative activation of macrophages. Nat Rev Immunol. 3, 23–35 (2003).
Rahaman, S. O. et al. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 4, 211–21 (2006).
Silverstein, R. L. & Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2, re3 (2009).
Berry, A. et al. IL-13 induces expression of CD36 in human monocytes through PPARgamma activation. Eur J Immunol. 37, 1642–52 (2007).
Kang, K. et al. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 7, 485–95 (2008).
Wu, D. et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 332, 243–7 (2011).
Cardilo-Reis, L. et al. Interleukin-13 protects from atherosclerosis and modulates plaque composition by skewing the macrophage phenotype. EMBO Mol Med. 4, 1072–86 (2012).
Stanya, K. J. et al. Direct control of hepatic glucose production by interleukin-13 in mice. J Clin Invest. 123, 261–71 (2013).
Jiang, L. Q. et al. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am J Physiol Endocrinol Metab. 305, E1359–66 (2013).
Amit, U. et al. New Role for Interleukin-13 Receptor alpha1 in Myocardial Homeostasis and Heart Failure. J Am Heart Assoc. 6, e005108 (2017).
Wouters, M. M. et al. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut. 63, 1103–11 (2014).
Kiesler, P., Shakya, A., Tantin, D. & Vercelli, D. An allergy-associated polymorphism in a novel regulatory element enhances IL13 expression. Hum Mol Genet. 18, 4513–20 (2009).
Deloukas, P. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 45, 25–33 (2013).
Cabon, Y. et al. Comparison of anti-interleukin-5 therapies in patients with severe asthma: global and indirect meta-analyses of randomized placebo-controlled trials. Clin Exp Allergy. 47, 129–138 (2017).
Wagner, W., Ciszewski, W., Kania, K. & Dastych, J. Lactate Stimulates IL-4 and IL-13 Production in Activated HuT-78 T Lymphocytes Through a Process That Involves Monocarboxylate Transporters and Protein Hyperacetylation. J Interferon Cytokine Res. 36, 317–27 (2016).
Varricchi, G. & Canonica, G. W. The role of interleukin 5 in asthma. Expert Rev Clin Immunol. 12, 903–5 (2016).
Gour, N. & Wills-Karp, M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 75, 68–78 (2015).
Bao, K. & Reinhardt, R. L. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine. 75, 25–37 (2015).
McLeod, J. J., Baker, B. & Ryan, J. J. Mast cell production and response to IL-4 and IL-13. Cytokine. 75, 57–61 (2015).
McLeod, O. et al. Genetic loci on chromosome 5 are associated with circulating levels of interleukin-5 and eosinophil count in a European population with high risk for cardiovascular disease. Cytokine. 81, 1–9 (2016).
Zhao, W. et al. Macrophage-specific overexpression of interleukin-5 attenuates atherosclerosis in LDL receptor-deficient mice. Gene Ther. 22, 645–52 (2015).
Silveira, A. et al. Plasma IL-5 concentration and subclinical carotid atherosclerosis. Atherosclerosis. 239, 125–30 (2015).
Newland, S. A. et al. Type-2 innate lymphoid cells control the development of atherosclerosis in mice. Nat Commun. 8, 15781 (2017).
Li, J. et al. Interleukin-4 and interleukin-13 pathway genetics affect disease susceptibility, serum immunoglobulin E levels, and gene expression in asthma. Ann Allergy Asthma Immunol. 113, 173–179 e1 (2014).
Westra, H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 45, 1238–43 (2013).
Li, X. et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy. 70, 1309–18 (2015).
Loots, G. G. et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 288, 136–40 (2000).
Li, Q., Liu, X. & Wei, J. Ageing related periostin expression increase from cardiac fibroblasts promotes cardiomyocytes senescent. Biochem Biophys Res Commun. 452, 497–502 (2014).
Bonnin, A. J., Montealegre, F. & Gewurz, A. Association of cigarette smoking with elevated serum IgE levels in Hispanic Puerto Rican men. Ann Allergy. 67, 609–611 (1991).
Liu, X. et al. Associations between total serum IgE levels and the 6 potentially functional variants within the genes IL4, IL13, and IL4RA in German children: The German Multicenter Atopy Study. J Allergy Clin Immunol. 112, 382–388 (2003).
Sadeghnejad, A. et al. IL13 gene polymorphisms modify the effect of exposure to tobacco smoke on persistent wheeze and asthma in childhood, a longitudinal study. Respir Res. 9, 2 (2008).
Chen, C. H. et al. Environmental tobacco smoke and male sex modify the influence of IL-13 genetic variants on cord blood IgE levels. Pediatr Allergy Immunol. 23, 456–63 (2012).
Fan, Q. et al. Analysis of the genetic association between IL27 variants and coronary artery disease in a Chinese Han population. Sci Rep. 6, 25782 (2016).
Tu, X. et al. The IL-33-ST2L pathway is associated with coronary artery disease in a Chinese Han population. Am J Hum Genet. 93, 652–60 (2013).
Cheng, X. et al. The same chromosome 9p21.3 locus is associated with type 2 diabetes and coronary artery disease in a Chinese Han population. Diabetes. 60, 680–4 (2011).
Wang, F. et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 43, 345–9 (2011).
Mancia, G. et al. 2013 ESH/ESC Guidelines for the management of arterial hypertension: the Task Force for the management of arterial hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC). J Hypertens. 31, 1281–357 (2013).
Kapur, A. et al. Randomized comparison of percutaneous coronary intervention with coronary artery bypass grafting in diabetic patients. 1-year results of the CARDia (Coronary Artery Revascularization in Diabetes) trial. J Am Coll Cardiol. 55, 432–40 (2010).
Gensini, G. G. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 51, 606 (1983).
Xu, C. et al. Minor allele C of chromosome 1p32 single nucleotide polymorphism rs11206510 confers risk of ischemic stroke in the Chinese Han population. Stroke. 41, 1587–92 (2010).
Acknowledgements
This research was supported by the funds of the National Natural Science Foundation of China [Nos 81525003, 91639301 and 81720108005 to Xiang Cheng; 81500186 to Shaofang Nie; 81270163 and 91439109 and to Xin Tu; 81400364 to Ni Xia; 81670361 and 81200177 to Tingting Tang].
Author information
Authors and Affiliations
Contributions
Designed the research: X.C. and X.T.; carried out the research: L.F.Z. and S.F.N.; contributed materials/reagents/analysis tools: L.F.Z., S.F.N., Q.W.C., Y.H.L., H.S.Z., J.T.D., T. X., F.W., N.X., T.T.T., Y.C.Z., C.Q.X., Z.P. Z., J.J., P.Y.W., Q.K.W., X.T., X.C.; wrote the paper: L.F.Z. and S.F.N.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zha, LF., Nie, SF., Chen, QW. et al. IL-13 may be involved in the development of CAD via different mechanisms under different conditions in a Chinese Han population. Sci Rep 8, 6182 (2018). https://doi.org/10.1038/s41598-018-24592-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-24592-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.