Abstract
Auxin response factors (ARFs) encode transcriptional factors that function in the regulation of plant development processes. A tomato ARF gene, SlARF5, was observed to be expressed at high levels in emasculated ovaries but maintained low expression levels in pollinated ovaries. The amiRNA SlARF5 lines exhibited ovary growth and formed seedless fruits following emasculation. These parthenocarpic fruits developed fewer locular tissues, and the fruit size and weight were decreased in transgenic lines compared to those of wild-type fruits. Gene expression analysis demonstrated that several genes involved in the auxin-signaling pathway were downregulated, whereas some genes involved in the gibberellin-signaling pathway were enhanced by the decreased SlARF5 mRNA levels in transgenic plants, indicating that SlARF5 may play an important role in regulating both the auxin- and gibberellin-signaling pathways during fruit set and development.
Similar content being viewed by others
Introduction
The plant hormone auxin, indole-3-acetic acid (IAA), plays an important role in various aspects of plant development, such as cell extension, division, and differentiation, as well as in organ and tissue development, and tropism. Auxin response factors (ARFs) bind specifically to a TGTCTC motif found in auxin-responsive promoter elements (AuxREs) and mediate auxin responses. A typical ARF protein contains three parts: an N-terminal DNA-binding domain (DBD), a carboxyl-terminal dimerization domain (CTD) that is similar to motifs III and IV of Aux/IAA proteins, and a middle region (MR) that may activate or repress the expression of early auxin response genes, including small auxin up RNA (SAUR), Gretchen Hagen-3 (GH3) and lateral organ boundaries-domain (LBD)1.
Among the components of the auxin signal transduction pathway, ARF proteins participate in transcriptional regulation of a variety of biological processes related to plant growth and development. Studies of arf mutants have indicated that different ARFs possess diverse functions, which is due to the differences in temporal and spatial expression and affinities with promoters of target genes. Recently, functional studies have demonstrated that ARF genes play an essential role in signal transduction during plant organ development. In Arabidopsis thaliana, ARF2 regulates leaf senescence2, root formation and flower organ senescence3. Moreover, heterologous expression of mango MiARF2 in Arabidopsis inhibits root and hypocotyl growth4. ARF3 plays an important role in lateral root development of Arabidopsis and regulates epidermal cells and trichome formation in tomato5,6. S1ARF4 is involved in hypocotyl development, cotyledon growth and fruit pericarp and sugar metabolism during tomato fruit development7,8. Inhibition of ARF2, ARF3, and ARF4 expression by some tasiRNAs may release the repression of Arabidopsis lateral root growth9. Arabidopsis ARF10 and ARF16 have essential roles in the process of root cap development, and the auxin signal cannot bypass them to initiate columella cell production10. Furthermore, ARF7 and ARF19 may regulate Arabidopsis LR formation initiation by interacting with IAA1411.
Several ARFs also play an essential role in reproductive development. Tomato SlARF2 is involved in the regulatory mechanism of the fruit ripening process12. In Arabidopsis, ARF6 and ARF8 promote jasmonic acid production and flower maturation13 and influence the developing floral organs14. Downregulation of SlARF6 and SlARF8 in transgenic tomato plants by the overexpression of MIR167a may lead to floral development defects and female sterility15. Furthermore, ARF8 is a negative regulator of fruit initiation and development in Arabidopsis, tomato and eggplant16,17. Similarly, SlARF7 functions as a negative regulator of fruit set until pollination and fertilization have occurred18. Models have been proposed for the effects of ARF819, SlARF7 and SlARF918,20 on plant fruit development through the interactions between the auxin- and GA-signaling pathways during tomato fruit initiation and development.
Tomato–a fleshy climacteric fruit–is a model plant for studying fruit development and ripening21. In general, tomato fruit development is divided into four major phases: fruit setting, cell division, cell expansion, and a fruit ripening period. Phytohormones have been demonstrated to participate in the regulation of every aspect of fruit development; however, each hormone plays a specific role in the fruit set, expansion, and ripening stages. Biochemical analyses have identified two peaks in auxin levels during tomato fruit development: the first at 8 d after pollination at the end of active cell division, and the other at 30 d after pollination22, corresponding to cell expansion (stage III) and the fruit maturation stage (stage IV), thereby suggesting that auxin has an important role in promoting fruit cell expansion22,23,24 and initiating and enhancing climacteric ripening25. On the other hand, the levels of endogenous GAs peak in two stages: first at 8 d after flowering and then approximately 15 d before ripening22,26, which corresponds to cell division (stage II) and the cell expansion stage (stage III) of fruit development, thereby indicating that GA is an important factor in fruit cell cycle and expansion27,28. GA activity regulates the expression of cell division and the expansion genes in tomato29. Furthermore, previous studies have demonstrated that the expansion of tomato locular cells coinciding with the expression of genes encoding for water flow, organic acid synthesis, sugar storage, and photosynthesis is regulated by auxin and GA30. SlIAA17 in the auxin-signaling pathway may affect fruit cell size in tomato31. Additionally, Matsuo et al.32 suggested that cytokinins regulate fruit development by modulation of auxin biosynthesis and/or polar auxin transport. Certain cell cycle genes, such as cyclin-dependent kinases (CDKs)33 and cell cycle-associated protein kinase WEE134, affect tomato fruit cell division. Meanwhile, certain other genes, including TAGL1 (SHATTERPROOF)35, MYB factors36, and SlPPC2 (phosphoenolpyruvate carboxylase)37, may regulate fruit cell expansion in tomato.
In a previous study, we identified 21 SlARFs in the tomato genome; their structures, chromosome locations, and phylogeny were predicted38. However, to date, their functions in fruit development have not been studied (beyond SlARF6, SlARF7, SlARF8 and SlARF9). Our data indicated that certain other ARFs also exhibited preferential expression in young ovaries and their mRNA levels were significantly altered during fruit development. Their roles in fruit development remain unclear. To explore the function of SlARF5 during fruit development, SlARF5-silenced tomato plants were generated by the amiRNA method. SlARF5 amiRNA lines form seedless fruits without pollination after emasculation. These parthenocarpic fruits developed fewer locular tissues with reduced fruit size and weight compared to those of wild-type plants. Downregulation of SlARF5 resulted in reduced expression of auxin-related genes and increased the expression levels of certain GA-signaling pathway genes. According to these data, we hypothesized that SlARF5 may participate in the regulation of early fruit set and development by mediating the crosstalk between IAA and GA hormones during tomato fruit development.
Results
Expression pattern of SlARF5 in tomato
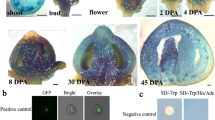
Expression analyses using real time-quantitative PCR (qRT-PCR) demonstrated that SlARF5 was expressed in all tested organs or tissues, with relatively high levels in leaves and stems and significantly low levels in young ovaries before anthesis (Fig. 1A). At different developmental stages of the ovaries and in early-developing fruits, SlARF5 expression decreased with flower development and reached its lowest levels in 6.0–7.5 mm ovaries just before anthesis (Fig. 1B). After pollination and fertilization, SlARF5 expression was maintained at low levels, increasing only slightly at 6 days after anthesis (with hand-pollination) and then decreasing again at 9 days. However, when the flowers were emasculated at 2 days before anthesis, transcription levels increased markedly at anthesis, peaked at 3 days after anthesis (without hand pollination), and then decreased significantly from 6 to 9 days after anthesis. In general, SlARF5 expression was downregulated in ovaries that were pollinated compared to that in ovaries from plants subjected to emasculation. Similar to SlARF5, pollination also suppressed the expression of ARF8 in Arabidopsis and tomato16 and SlARF7 in tomato39.
Real-time PCR analysis of relative gene expression of SlARF5 mRNA in tomato. (A) SlARF5 transcripts in different organs and tissues. (B) Expression levels of SlARF5 at different stages during flower and fruit development. Flower bud samples were at lengths of 3.0–4.5 mm, 4.5–6.0 mm, and 6.0–7.5 mm. Anthesis represents fully opening flowers. Three DAA-U, 6 DAA-U, and 9 DAA-U represent flower samples collected at 3, 6 and 9 d after anthesis, respectively, which were emasculated 2 days before opening. Three DAA-P, 6 DAA-P, and 9 DAA-P represent tomato flowers collected 3, 6, and 9 d after anthesis with hand pollination. ANOVA statistical analyses were performed using SPSS 15.0. Significant differences (p < 0.05) between treatments, as determined by Tukey’s tests, are indicated with different letters. Data are expressed as the mean ± standard errors for three replicates.
Suppression of SlARF5 induces parthenocarpy
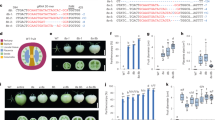
To explore the function of SlARF5 during fruit set and development, amiSlARF5 transgenic plants were generated using A. thaliana miRNA160a as a backbone to express an artificial miRNA (amiRNA) using a 125 bp SlARF5-specific fragment, which was cloned into the pCAMBIA1301-35S vector. The amiRNA binary vector was transferred into tomato using Agrobacterium tumefaciens (Fig. 2A). A total of 11 independent transgenic lines were generated and identified based on PCR and GUS screening (Supplementary Fig. S1). Among them, five lines exhibited significantly lower SlARF5 expression levels compared to those in wild-type plants. In two amiSlARF5 lines, namely, SlARF5-6 and SlARF5-9, the SlARF5 transcription levels were reduced by 67.8% and 68.5%, respectively (Fig. 2B). In addition, although SlARF6, SlARF7, SlARF8, and SlARF19 were closely related with SlARF5 in molecular phylogenetic tree (Supplementary Fig. S2), their expression levels were not evidently affected in these two lines (Supplementary Fig. S3). Thus, these two amiSlARF5 lines were used for further analysis.
Construction of the amiSlARF5 vector and identification of amiRNA transgenic tomato plants. (A) Schematic diagram of the amiSlARF5 vector. The construct is under the control of the CaMV 35 S promoter and terminator and contains a selective marker for hygromycin antibiotic resistance. (B) Relative SlARF5 mRNA levels in amiSlARF5 and wild-type plants analyzed by qRT-PCR. ANOVA statistical analyses were performed using SPSS 15.0. Significant differences (P < 0.05) between treatments, as analyzed by Tukey’s test, are indicated as different letters. Data are expressed as the mean ± standard errors for three replicates.
No evident differences in vegetative growth or floral morphology were observed between amiSlARF5 and wild-type plants (Supplementary Fig. S4). Furthermore, the fruit set rates in the amiSlARF5-6 and amiSlARF5-9 lines, 65.0% and 68.7%, respectively, were comparable to those of wild-type plants (74.2%) (Table 1). However, the self-pollinated amiSlARF5 fruits were significantly smaller than wild-type fruits (Fig. 3A–F, Table 1). Both the polar and equatorial diameter of the transgenic fruits were much smaller compared to those obtained from wild-type and empty vector plants. Similarly, the weights of pollinated amiSlARF5 fruits (1.83 ± 0.27 g for SlARF5-6 and 1.84 ± 0.28 g for SlARF5-9) were much lower than those of the wild-type and empty vector transgenic fruits, at 3.40 ± 0.97 g and 3.32 ± 0.59 g, respectively.
Phenotypic characteristics of amiSlARF5 transgenic lines and control plants. (A–C) Self-pollination fruits of pCAMBIA1301 empty control (A), amiSlARF5-6 (B), and amiSlARF5-9 (C) transgenic plants. (D–F) Vertical section of the corresponding fruits from A, B, C, respectively; Bar = 1 cm. (G–L) Parthenocarpic fruits of amiSlARF5 transgenic plants after emasculation compared to the pollinated control. (G and J) wild-type fruit and section; (H and K) AmiSlARF5-6 fruit and section; (I and L) AmiSlARF5-9 fruit and section. Bar = 1 cm.
Emasculation of untransformed ‘Micro-Tom’ plants 2 d before anthesis negatively affected fruit growth. Emasculated ovaries stopped growing 6 d after anthesis, became abortive and ultimately the flowers abscised within 6–9 days (Table 1). However, when the transgenic flowers were emasculated, the parthenocarpic fruit set and growth occurred in SlARF5-6 and SlARF5-9 lines with fruit set rates of approximately 20.5% and 22.4% in these two transgenic lines, respectively. Although the fruit set rate in emasculated transgenic plants was lower than that in self-pollinated plants, suppression of SlARF5 evidently initiated fruit development and enhanced parthenocarpic capability. However, the average size and weight of the parthenocarpic fruits were significantly lower than those of self-pollinated fruits. Consistently, Mapelli et al.22 also revealed that parthenocarpic fruits were generally smaller than seeded fruits. Furthermore, the smaller fruits in the SlARF5 amiRNA transgenic lines formed fewer locular tissues with reduced pulp and enlarged central columella compared to those observed in the wild-type fruits, resulting in parts of the locular cavities being empty (Fig. 3J–L). Interestingly, in parthenocarpic SlARF7 RNAi fruits39 and in the fruits treated with GA340, the locular tissue was barely developed, resulting in almost empty locular cavities.
Unexpectedly, self-pollinated transgenic fruits formed well-developed seeds of normal size and with germination potential; however, each fruit contained much lower numbers of fully developed seeds, 8.0 and 8.5 in SlARF5-6 and SlARF5-9 per fruit, respectively, compared to 20.5 and 18.5 in wild-type and empty-vector fruits, respectively (Table 1). The parthenocarpic fruits in the amiSlARF5 lines had no seeds (Fig. 3K,L).
Suppression of SlARF5 expression affects cell division and expansion during early fruit development
In tomato, the cell division period lasts for 10–14 d. In the next 6–7 weeks, cell division activity is weakened and fruit growth set is predominantly dependent on cell expansion22,24,41. Our study demonstrated that transgenic plants of the SlARF5-6 and SlARF5-9 lines, in which SlARF5 transcript levels were downregulated, produced smaller fruits than those of wild-type plants after pollination.
To understand the cause of different fruit phenotypes, histological cross-sections of wild-type and SlARF5 transgenic plants were analyzed using the ovaries and developing fruits collected at diverse developmental stages, ranging from unpollinated ovaries to fruits 10 mm in diameter (Fig. 4A–O). In general, the pericarps of both the tomato fruits differentiated into the three classic layers: exocarp, mesocarp, and endocarp24. At anthesis, cell size in the pericarps of both wild-type and transgenic ovaries showed no obvious differences (Fig. 4A,F,K). In 3–4 mm fruits, the exocarp cells were at the stage of cell division, whereas the mesocarp and the endocarp cells began to expand. At this stage, the cell size in the mesocarps and endocarps of wild-type and transgenic lines were not significantly different. However, in 5–6 mm fruits, the exocarp and mesocarp cells in transgenic lines were significantly larger than those in wild-type plants. The mean cell area of the exocarp in RNAi fruits was nearly 2-fold greater than that of the wild-type fruits, while mesocarp cells were three times larger than those of wild-type cells (Fig. 4C,H,M, Table 2). The differences in exocarp features of RNAi and wild-type fruits were more clearly observed in the 7–8 mm and 9–10 mm fruits.
Microscopic analysis of the pericarp in the fruits of wild-type and transgenic lines during early tomato fruit development. (A–E) Wild-type fruit; (F–J) Transgenic line amiRNA SlARF5-6 fruit; (K–O) Transgenic line amiRNA SlARF5-9 fruit. (A,F,K) Unpollinated ovary at the anthesis stage; (B,G,L) Pericarp of a 3–4 mm diameter fruit; (C,H,M) Pericarp of a 5–6 mm diameter fruit; (D,I,N) Pericarp of a 7–8 mm diameter fruit; (E,J,O) Pericarp of a 9–10 mm diameter fruit. Bars = 50 μm.
On the other hand, there are significant differences in the number of cell layers between transgenic amiSlARF5 lines and wild-type plants. In general, the exocarp cells of tomato fruit comprise 4–5 cell layers outside the fruit, and the endocarp cells comprise 1–2 cell layers inside the fruit; therefore, the mesocarp cell number was the major contributor towards the differences in fruit equatorial diameter among the plants with different genotypes. In 3–4 mm fruits, pollinated transgenic fruits formed fewer cell layers (approximately 16 cell layers) than those in the wild-type plants (at least 23 cell layers), a reduction of 30% compared to wild-type plants. Similarly, in the 5–6 mm and 7–8 mm fruits, the pollinated transgenic fruit formed fewer cell layers than those in the wild-type fruit, with levels being 32% and 25–31% lower, respectively (Fig. 5A–I, Table 3). This indicates that inhibition of SlARF5 leads to a significant reduction in the number of cell layers. The small size of mature transgenic fruits may be primarily attributed to the reduced number of cell layers in transgenic fruits, probably as a result of a reduction in the period of cell division that promotes cell layer formation.
Micrograph of the number of cell layers in wild-type and transgenic line fruits during early fruit development. (A,D,G) 3–4 mm diameter fruits of wild-type (A), amiRNA SlARF5-6 (D), and amiSlARF5-9 plants (G). (B,E,H) 5–6 mm diameter fruits of wild-type (B), amiSlARF5-6 (E), and amiSlARF5-9 plants (H). (C,F,I) 7–8 mm diameter fruits of wild-type (C), amiSlARF5-6 (F) and amiSlARF5-9 plants (I). Bars = 50 μm.
Transcriptomic analysis of the amiSlARF5 and WT fruits
To evaluate expression changes during fruit development in amiSlARF5-9 transgenic and wild-type plants, massively parallel RNA sequencing (RNA-Seq) analyses were carried out using the RNA samples from 3–4 mm young fruits with two replicates. At this stage, SlARF5 was highly expressed in the wild type and no phenotypic differences were observed between amiSlARF5 and wild-type fruits. We generated four cDNA libraries from each genotype for sequencing using the Illumina Hiseq. 2000/2500 system at LC Sciences (Hangzhou, China). Each cDNA library yielded more than 45 million sequence reads (Supplementary Table S1), representing >4 Gb sequence data per sample. RNA data from the two biological replicates exhibited good correlations and were used for further analysis (Supplementary Fig. S5). A summary of RNA sequencing, mapping, assembly and annotation is provided in Supplementary Table S1.
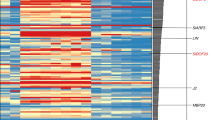
Compared to wild-type fruits, a total of 3152 genes were differentially expressed in amiARF5 transgenic plants, with 1719 genes upregulated and 1433 genes downregulated (p < 0.05, |log2(fold changes)| ≥2) (Fig. 6). As expected, SlARF5 was downregulated by approximately 16-fold in transgenic plants. Seventeen genes from the RNA-seq-derived results were validated by qRT-PCR. The results demonstrated that almost all tested genes were in agreement with those from the RNA-seq data (Supplementary Fig. S6), confirming the reliability of RNA-seq analysis.
Analysis of genes regulated by SlARF5 during fruit development. (A) The numbers of differentially expressed genes in tomato fruit after SlARF5 expression was inhibited. (B) Analysis of biological functions of GO-enriched genes. (C) The primarily enriched KEGG pathway for up- and downregulated differentially expressed genes.
To further evaluate the potential functions of differentially expressed genes (DEGs) in the transgenic fruits, functional enrichment analysis was performed. GO analysis indicated that these DEGs were primarily enriched in ATP, protein, or DNA binding (383), protein kinase activity (147), oxidoreductase activity (131), and catalytic activity (60), which is consistent with kinase genes associated with clusters that are highly expressed in young fruit42. Regarding the biological processes, a high proportion of DEGs was associated with oxidation-reduction processes, regulation of transcription, protein phosphorylation and metabolic metabolism. Among them, transcription regulation, signal transduction, and metabolic processes accounted for more than half of all annotated DEGs, implying that signal transduction pathways and metabolic activities are strongly affected by inhibition of the SlARF5 gene. In addition, in terms of the cellular components, most DEGs were primarily enriched in the membrane, nucleus, or the cell wall (Fig. 6), which is consistent with reports demonstrating that the cell wall plays essential roles in fruit set and fruit size43. Similarly, previous transcriptome analysis indicated that the cell-cycle and cell-wall genes were enriched in ovule/seed-associated expression clusters42.
KEGG analysis further indicated that most of the downregulated genes are involved in the cell cycle (14%), the cell cycle of yeast (12%), and DNA replication (10%) in amiSlARF5 transgenic fruits, suggesting that SlARF5 is positively involved in cell division. The upregulated genes were associated with phenylalanine metabolism (12%), phenylpropanoid biosynthesis (16%), plant-pathogen interactions (13%), and starch and sucrose metabolism (7%) (Fig. 6). Among these DEGs, the expression of several genes related to fruit development was significantly altered in transgenic plants, including those involved in cell division, cell expansion, and hormone-signaling pathways (Tables 4 and 5 and Fig. 6).
Cell division- and cell expansion-related genes
As shown in Table 4, 12 cell cycle-related genes, including cycA1, cycA2, cyclin B1, cycd3C2/C3, and cyclin-dependent protein kinases, and eight cell wall development- related genes, including EXP2, EXP8, xyloglucan, pectinacetylesterase, and endoglucanase, were downregulated in the transgenic lines compared to the levels observed in wild-type plants, suggesting that both fruit cell division and expansion is negatively affected in amiRNA SlARF5 transgenic plants.
To explain the histological data obtained from microscopic analysis in relation to RNA-Seq results, the ovaries and fruits were collected from wild-type and transgenic plants for RNA extraction to analyze expression patterns of cell expansion- and division-related genes. In the wild-type fruits, the transcript levels of cell cycle genes, including SlCyclinB1.1 and SlCDKB2.1, were markedly increased following anthesis, and then decreased continuously during later fruit development. However, in transgenic lines, these cell cycle-related genes (CycB1.1 and CDKB2.1) were significantly downregulated in the early stages of fruit development compared to the levels observed in wild-type fruits (Fig. 7A). In the later stages of fruit development, i.e., 7–8 mm fruits and above, there were no significant differences in the expression patterns of the cell cycle genes between the transgenic and wild-type fruits.
Expression of differentially expressed genes in developing fruits of wild-type and transgenic plants by qPCR. (A) Cell division and cell expansion genes-CycB1.1, CDKB2.1, EXPA5, PEC and XTH1. (B) Auxin-related genes-ARF5, ARF9, GH3-like, IAA1, IAA2 and IAA14. (C) Gibberellin-related genes-GA20ox1, GA3ox1, GA2ox2, GA2ox4, GID1 and GAST1. Ovaries and young fruits of WT and amiRNA plants were sampled at anthesis, and at 3–4 mm, 5–6 mm, 7–8 mm, and 9–10 mm diameter stages. Significant differences (P < 0.05) between treatments, as analyzed by Tukey’s test, are indicated as different letters. Data are expressed as the means ± standard errors for three replicates.
Similarly, in wild-type fruits, the expression of cell expansion-related genes (i.e., SlEXPA5, SlPEC, and SlXTH1) increased significantly following anthesis during early fruit development. In the 3–4 mm and 5–6 mm fruits, the expression of SlEXPA5 in transgenic plants was significantly lower than that in wild-type fruits, and in the 7–8 mm fruits and later stages, the transcript levels of SlEXPA5 in transgenic fruits were similar to those in wild-type fruits. However, the mRNA levels of SlPEC and SlXTH1 in both transgenic and wild-type fruits were maintained at similar levels during early fruit development (Fig. 7A). These results suggest that the microscopic characteristics of the transgenic lines are primarily due to reduced levels of cell division-related genes.
Auxin-signaling pathway genes
Previous studies have confirmed that auxin stimulates cell division40,41. As shown in Table 5, 15 genes involved in auxin homeostasis, transport and signaling, including LAX2, ARF5, SlARF9, IAA1, IAA2, and IAA14, were downregulated in the transgenic lines compared to the levels observed in wild-type plants, suggesting that IAA-related genes are negatively affected in amiRNA SlARF5 transgenic plants. In wild-type plants, the mRNA levels of some auxin response genes (e.g., SlARF9, SlIAA1, SlIAA2, and SlIAA14) increased in 3–4 mm fruits and then decreased to significantly low levels during later stages. However, in transgenic plants, the mRNA levels of SlARF9, SlIAA2, and SlIAA14 were downregulated compared to those in wild-type plants, whereas SlIAA1 was maintained at similar mRNA levels as those in wild-type plants. As expected, the expression of SlARF5 in transgenic plants was significantly lower than that in wild-type plants in the early stages, up to the 5–6 mm fruit stage. Similarly, all detected auxin-responsive genes, including SlARF9, SlGH3, SlIAA1, and SlIAA2, were significantly downregulated in the early development stage compared to the levels observed in wild-type plants, with the exception of a GH3-like gene, which was significantly upregulated in the 3–4 mm and 5–6 mm fruit stages (Fig. 7B); these results are consistent with the RNA-seq data, thereby suggesting that suppression of SlARF5 strongly affects several genes related to the auxin-signaling pathway. Unlike previous microarray data43, in which the expression levels of most auxin-related genes were increased in the pollinated fruit, our RNA-Seq results demonstrated that a majority of the genes involved in the auxin signaling pathway were downregulated, in alignment with the microarray data of GA-treated fruit43.
Gibberellin-related genes
The GA-biosynthesis gene, GA20ox1, was significantly downregulated in the transgenic lines compared to levels observed in wild-type plants (Fig. 7C), whereas four GA-related genes, including SLY1, GID1B, Cell wall protein (GA-induced), and GA-regulated family protein, were upregulated in the transgenic lines compared to the levels in wild-type plants (Table 5), thereby suggesting that GA-biosynthesis genes are negatively affected in amiRNA SlARF5 transgenic plants, whereas certain GA-signaling genes are upregulated. Similarly, qRT-PCR analysis demonstrated that in wild-type plants, the expression levels of SlGA20ox1 and SlGAST1 markedly increased following pollination, and progressively decreased thereafter. However, SlGA2ox2, SlGA3ox1, SlGA2ox4, and SlGID1 continued to decline following anthesis during fruit development. In the amiRNA SlARF5 lines, SlGA3ox1, SlGID1, and SlGAST1 exhibited similar expression patterns as those in wild-type plants, with no significant differences between each successive stage of early fruit development, with the exception of SlGAST1 levels at the 3–4 mm stage. However, the expression level of SlGA20ox1 was significantly downregulated in all successive stages following anthesis, whereas SlGA2ox2 and SlGA20ox4 were significantly downregulated at the anthesis, 3–4 mm and 5–6 mm stages. During later stages, the differences in expression levels became non-significant in both transgenic lines and wild-type plants (Fig. 7C). Previous microarray data43 also demonstrates increased the expression of most GA-signaling genes in GA3-treated ovaries.
Discussion
In plants, ARFs encode important transcription factors and regulate gene expression in response to auxin by binding specifically to the TGTCTC sequence in the auxin response elements found in the promoters of primary/early auxin response genes. Recently, all ARF family genes were identified in tomato44, an ideal model plant for studying fruit development and ripening. However, only SlARF6, SlARF7, SlARF8, and SlARF9 have been characterized to date and have been demonstrated to be essential for tomato fruits generated through the regulation of auxin signal transduction. In this study, transgenic plants with reduced SlARF5 expression were developed using artificial miRNA silencing methods in order to examine the role of SlARF5 in fruit initiation and development by affecting the IAA and GA signaling pathways.
Suppressing expression of SlARF5 in tomato induces parthenocarpic fruit
Previous studies have demonstrated that SlARF8 is a negative regulator of fruit initiation in Arabidopsis16. SlARF7 and SlARF8 affect fruit initiation and lead to parthenocarpy16,39. SlARF7 functions as a negative regulator of fruit set until pollination and fertilization18. In addition, SlARF9, which is enriched with serine, was reported to negatively regulate cell division during early fruit development20. Interestingly, these ARFs are typical tomato auxin response factors with a glutamine-rich middle region belonging to the class II subfamily, and the others being SlARF5, SlARF6-1, SlARF8-1, SlARF19, and SlARF19-138. ARFs enriched with glutamine are likely to function as transcriptional activators38. It is worth noting that although these genes belong to the same subfamily, their expression patterns during fruit development were quite different. SlARF5 mRNA levels were evidently higher in emasculated fruits than those in self-fertilized fruits. These results suggest that SlARF5 plays a role in fruit initiation when pollination occurs.
In Arabidopsis, ARF5 affects lateral organ development45, primary root initiation46, flower primordium initiation47, and cotyledon development in embryos48. Our data demonstrated that amiRNA SlARF5 transgenic fruits were significantly smaller than wild-type fruits, and their average weight and equatorial and longitudinal diameter were significantly decreased compared to that of wild-type fruits (Table 1). Interestingly, the number of seeds in the self-fertilized transgenic lines was significantly lower than that in wild-type plants. However, when the transgenic flowers were emasculated, the amiSlARF5 plants developed parthenocarpic fruits, and the parthenocarpic fruit set rate was approximately 20.5% and 22.4% in two independent transgenic lines. The average size and weight of the parthenocarpic fruits were significantly lower than those in self-pollinated fruits. In previous studies, downregulation of SlDELLA49, TM2950, CHS51 and AUCSIA genes52, mutations of pat53 and hydra54 and transformation of aberrant ARF819 led to the formation of smaller parthenocarpic fruits, implying that these genes are positive regulators of fruit development in tomato. However, the parthenocarpic tomato fruits developed in rolB55, SlARF7 RNAi18, SlIAA9 knockout56 and SlTIR1 over-expression transgenic plants57 were similar in size to those of wild-type plants. Interestingly, overexpression and inhibition of SlARF9 mRNA levels led to smaller fruits and larger fruits, respectively20, suggesting different functions of ARFs in parthenocarpic fruit development.
SlARF5 modulates the expression of cell division- and expansion-related genes and the crosstalk between auxin- and GA-signaling
The signal transduction mechanism by which auxin regulates cell division and cell expansion during fruit set and growth is largely unknown. Previous studies have demonstrated that suppression of SlARF7 reduces cell division ability and enhances cell expansion ability18, whereas ARF6 may affect epidermal cell differentiation of petals in Arabidopsis14. In the present study, the inhibition of SlARF5 reduced fruit cell division and expansion. RNA-Seq data demonstrated that the expression levels of most cell cycle genes were downregulated in the 3–4 mm diameter fruit stage. GO annotation and KEGG analyses indicated that the cellular components of cell walls and pathways of the cell cycle-related to fruit development were observed to be enriched. The cell cycle genes, cycB1.1 and CDKB2.1, were significantly downregulated in amiSlARF5 plants during early fruit development, compared to the levels in wild-type fruits, indicating that cell division activity in amiSlARF5 fruits was decreased. However, in transgenic tomato plants with inhibition of SlARF7 or SlIAA9, the expression of cell cycle-related genes was lower and higher than that in wild-type plants, respectively39,58.
Cell expansion genes are induced by pollination and GA treatment43. Expression of the cell expansion gene EXPA5, which is induced by pollination rather than by GA application, is downregulated during early fruit development, whereas XTH1 and PEC are upregulated by GA application43. In our study, RNA-Seq data demonstrated that certain cell expansion genes (e.g., EXP2/EXP12, AGPS1, CEL5, and TPX1) in the amiSlARF5 line were downregulated compared to the levels in wild-type plants, suggesting that cell expansion abilities in the amiSlARF5 line were weakened. In a previous study, the mRNA levels of two cell expansion-related genes, namely, SlPEC and SlXTH1 in the SlARF7 RNAi line, were similar at anthesis but became higher than those in the wild-type plants following anthesis during early fruit development39.
Plant hormones, such as auxin and gibberellins, play pivotal roles in tomato fruit development. The IAA and GA pathways regulate the cell cycle and expansion genes and determine the final fruit size24,40,41. In normal fruit, there is a rapid increase in the GA levels induced by successful pollination and fertilization, following which GA may induce an increase in auxin levels during the first 10 DPA22,59. However, in amiSlARF5 plants, a majority of the IAA-related genes were significantly downregulated, as demonstrated by the RNA-Seq data and further verified by qRT-PCR analysis. IAA-related genes, SlARF9, IAA2 and IAA14, which are induced by pollination and auxin treatment43, were downregulated during early fruit development in amiSlARF5 lines. However, expression of GH3, which is induced by auxin application to unpollinated ovaries43, increased in transgenic plants, as demonstrated by the qRT-PCR and RNA-Seq results, thereby suggesting that at least part of the IAA pathway was enhanced.
GA20ox1, an enzyme involved in GA biosynthesis, catalyzed the conversion of GA12 and GA53 into GA9 and GA20, respectively, which are precursors of bioactive GAs60. The expression of GA20ox1 was significantly decreased in amiSlARF5 lines during early fruit development compared to that in wild-type fruits, which is consistent with the reduction in parthenocarpic fruit size of SlARF7 RNAi plants18. Similarly, the genes coding GA biosynthetic enzymes, GA2ox2 and GA2ox4, which catalyze active GA1 and GA4 conversion into inactive GA34 and GA8, were significantly downregulated in amiSlARF5 lines. However, GA3ox1, involved in the conversion of GA9 and GA20 into GA1 and GA4, exhibited similar expression levels in transgenic and wild-type plants. On the other hand, GA-signaling transduction genes, such as GID1, were positively induced by GA treatment59. In our RNA-Seq data, the expression levels of GID1 and certain other known GA-signaling and response genes in amiSlARF5 lines increased compared to those in wild-type plants. These results suggest that levels of certain GA-signaling-related genes increased. This may be attributed to the feedback regulation on GA-signaling, which is similar with a previous study in PttGID1 over-expression lines61. The more pronounced responses to GA compared to those of wild-type plants were found in over-expression lines, while PttGA20ox1 expression was reduced61. Although three GA-regulated family proteins were downregulated in the amiSlARF5 lines, their functions were undefined. Two of these genes were probably cell cycle family genes in Arabidopsis (Table 5), which may function as the downstream genes of GA signaling pathway. Consistently, in our RNA-Seq results, mRNA levels of most cell cycle genes were also downregulated (Table 4).
The DEGs between amiSlARF5 transgenic and wild-type plants, identified by RNA-seq, were compared with the data from microarray analyses of GA3-treated and pollinated fruits (RNA from emasculated flowers)43. Our data demonstrated that the expression levels of most cell cycle and expansion genes were downregulated in transgenic plants, unlike their increased expression in GA3-treated and pollinated plants43. Furthermore, the alterations in the expression of some GA and IAA pathway genes in our study were in contrast to those observed by microarray analysis in pollinated fruits43; however, the altered levels of certain GA-related genes, such as SlGA3ox1 and cell wall protein precursor in GA-treated fruits43, were similar with our data. These results suggest that the inhibition of SlARF5 enhanced part of the GA pathway, similar to the results involving GA-treated fruits43.
Previous studies have indicated that the expression of GA metabolism genes is regulated by Aux/IAA and ARF proteins, and changes in GA metabolism partially mediate auxin action during development62. ARF7 and ARF19 serve as crosstalk between auxin signaling and in ethylene responses in Arabidopsis63. The RNA-seq results of SlARF3 RNAi lines indicated that the SlARF3 gene regulates the auxin-dependent transcription of trichome formation by mediating auxin, ethylene, and GA signaling in tomato5. Recently, Breitel et al.64 demonstrated that SlARF2 links the signals of auxin, ethylene, and other hormones and affects the ripening capacity of fruit tissue. In pollination-dependent and parthenocarpic fruit set, ARF2 and IAA9 mediate the crosstalk between auxin and GA during this development process65. In obligatory parthenocarpy caused by SlMIR159-overexpressing tomato cv. Micro-Tom plants, SlGAMYBs potentially contribute to fruit initiation and regulate auxin and gibberellin responses66.
In the amiRNA SlARF5 lines, when the SlARF5 mRNA was silenced, fruit set was evidently different from that observed in previous studies, in which the fruit set may be due to elevation of hormones (especially auxin) by expression of auxin biosynthesis genes in ovaries and ovules55,67,68. However, in our transgenic lines, GA signaling was potentially regulated by GA biosynthesis pathways (downregulated gene expression) through negative feedback, as a result, some genes involved in the GA-signaling pathway were upregulated. SlARF5 may function as SlARF7, which was suggested by De Jong et al.39. When SlARF5 was inhibited, the special downstream genes, such as auxin attenuating genes might fail to form transcripts; therefore, the inhibition imposed on ovary development was relieved, thereby enabling the ovary to expand and form the fruit. Meanwhile, in our transgenic plants, most of the auxin-related genes were not regulated and only GH3 was increased in the amiSlARF5 transgenic plants. The hormone-related gene expression was consistent with the ARF7 RNAi results18. As a result, once SlARF5 was inhibited, increased GA signaling gene expression may facilitate fruit development. On the other hand, the smaller fruit size in amiSlARF5 plants may be attributed to the weakened auxin signaling pathway.
In conclusion, SlARF5 is a essential component of auxin signaling during tomato fruit development. SlARF5 may be critical for the crosstalk between auxin and GA during fruit development. In normal-pollination ovaries of wild-type plants, the mRNA level of SlARF5 was decreased, and the auxin- and GA-signaling pathways converged at fruit set and development. However, in emasculated flowers of wild-type plants, the SlARF5 mRNA levels were increased. Therefore, the SlARF5 mRNA levels potentially negatively regulate the GA-signaling pathway. A partially enhanced GA-signaling pathway may lead to the formation of parthenocarpic fruit.
Materials and Methods
Plant materials and growth conditions
Tomato (S. lycopersicum L.cv. Micro-Tom) plants were grown in a standardized greenhouse at the experimental farm at Zhejiang University, under 14/10 h light/dark periods with 200 μmol m−2s−1 photo flux and 27/22 °C (day/night) temperatures.
For the measurement of early fruit development parameters, the flowers were emasculated 2 days before opening, then ovaries were collected at 0, 3, 6, and 9 DAA-U and DAA-P, and early developing fruits were collected in the following stages: pollinated ovaries at anthesis and 3–4 mm, 5–6 mm, 7–8 mm, and 9–10 mm diameter fruit stages, corresponding to approximately 6, 8, 10, and 12 DAA-P stages of the wild-type, respectively.
Construction of amiSlARF5 vectors
amiRNAs were engineered into a 128 bp fragment containing the miRNA160a stem-loop, synthesized by Invitrogen, and then cloned into the BamH I and Hind III sites of a modified pCAMBIA1301 vector with a 35 S promoter inserted using Kpn I and Sma I, which was used for plant transformation. The detailed protocol for generating amiRNAs is provided in Supplementary Data S1. The pCAMBIA1301 plasmid with a 35SCaMV promoter was used as a control for plant transformation. Transgenic tomato plants were generated using the amiSlARF5 vectors by the Agrobacterium-mediated leaf disk method, as described by Sun et al.69, with the exception that the antibiotics were replaced with 6 mg/L hygromycin and 300 mg/L Timentin.
For the detection of amiSlARF5 lines to select positive transgenic plants, root materials of the transgenics were assayed for GUS activity70. To confirm that the T-DNA sequence of amiRNA SlARF5 had been transformed into tomato plants, a region of 285 bp DNA from leaf materials was amplified by PCR detection (TaKaRa). The specific amiRNA fragment sense primer, (5-ATGAGACGGATTTCGTGTTT-3) was used in combination with a primer from the pCAMBIA1301 vector (5-CGACAGTGGTCCCAAAGAT-3) to amplify the DNA fragment. PCR procedures included an initial denaturation at 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min plus a final extension at 72 °C for 7 min.
Microscopy
To estimate the cell area, cell numbers and cell layers in the ovaries and developing fruits of both wild-type and transgenic SlARF5 plants, five fruits per line in each stage were sampled for analysis using fluorescence microscopy. Ovaries and young fruits were fixed with 2.5% glutaraldehyde overnight, rinsed with 0.1 M phosphate buffer three times, then transferred to 1% osmic acid for 1–2 h. The specimens were again washed with 0.1 M phosphate buffer and then dehydrated through a graded series of ethanol solutions. Samples were embedded in Spurr’s resin, and semithin sections were obtained and stained with 1% methylene blue and viewed using fluorescence microscopy (DMLB, Leica, Germany). The micrographs were captured with a Leica DFC 420 C camera using the Leica Application Suite software (Leica Microsystems, Germany).
Quantitative real-time PCR
Total RNA was extracted from tomato ovaries using the TRIzol reagent (Invitrogen, Germany), according to the manufacturer’s instructions. The first cDNA strand was generated using the Improm-TM Reverse Transcription system (Promega, Madison, WI, USA), following the manufacturer’s protocol. The qPCR experiment was carried out using SYBR Premix Ex Taq (Takara) in a BioRad CFX96 Real-time PCR Detection System, as described by Wu et al.38, with three biological replications and three technical replications. The Ubi3 gene (GenBank accession number X58253) was used as an internal control, and the relative expression was calculated using the comparative 2(−ΔΔCt) method71. The primer pairs for qRT-PCR are listed in Supplementary Table S2.
RNA sequencing and data analysis
Tissues were collected from pollinated fruits during the 3–4 mm diameter stage from amiRNA SlARF5 lines and wild-type plants. Total RNA was extracted using TRIzol reagent (Invitrogen, CA, USA), according to the manufacturer’s instructions with two biological replicates. Four RNA libraries were sequenced using a Hiseq. 2000/2500 sequencing system (Illumina), following the manufacturer’s protocol.
Following the removal of low-quality reads, the sequenced RNA-seq reads were aligned to the tomato genome using Tophat version 2.0.8b with modified parameters72. The aligned read files were handled by Cufflinks, and the relative abundances of the transcripts were measured by the normalized RNA-seq fragment counts. Expression levels of each gene in FPKM (fragment per kilobase of exon per million fragments mapped) and the fold change of each gene were calculated. Comparisons with a q-value less than 0.05 were used for screening differentially expressed genes and further analyzed by enrichment of gene ontology (GO) terms and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. GO analysis was performed on differentially expressed genes using the Plant MetGenMAP database. Pathway enrichment analysis was performed using the KEGG database.
Statistical Analysis
ANOVA statistical analyses were performed on all data using SPSS 15.0 (p < 0.01) and data were tested for significant differences (p < 0.05) using Tukey’s test. Means and standard errors were calculated from three biological replicates.
References
Guilfoyle, T. J. & Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460 (2007).
Lim, P. O. et al. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 61, 1419–1430 (2010).
Ren, Z., Liu, R., Gu, W. & Dong, X. The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. 256, 103–111 (2017).
Wu, B. et al. Over-expression of mango (Mangifera indica L.) MiARF2 inhibits root and hypocotyl growth of Arabidopsis. Mol. Biol. Rep. 38, 3189–94 (2010).
Zhang, X. et al. Auxin Response Gene SlARF3 Plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant Cell Physiol. 56, 2110–2124 (2015).
Yoon, E. K., Yang, J. H. & Lee, W. S. Auxin and abscisic acid responses of auxin response factor 3 in Arabidopsis lateral root development. J. Plant Biol. 53, 150–154 (2010).
Sagar, M. et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374 (2013).
Jones, B. et al. Down- regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 32, 603–613 (2002).
Marin, E. et al. miR390, Arabidopsis TAS3 tasiRNAs, and their Auxin Response Factor targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117 (2010).
Wang, J. W. et al. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17, 2204–2216 (2005).
Ito, J. et al. Auxin-dependent compositional change in mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. 113, 6562–6567 (2016).
Hao, Y. W. et al. Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genet. 11, e1005649 (2015).
Punita, N. et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118 (2005).
Tabata, R. et al. Arabidopsis auxin response factor6 and 8 Regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51, 164–175 (2010).
Liu, N. et al. Down-regulation of auxin response factors 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 65, 2507–2520 (2014).
Goetz, M., Vivian-Smith, A., Johnson, S. D. & Koltunow, A. M. Auxin response factor8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18, 1873–1886 (2006).
Du, L. et al. SmARF8, a transcription factor involved in parthenocarpy in eggplant. Mol. Genet. Genomics 291, 93–105 (2016).
De Jong, M., Wolters-Arts, M., García-Martínez, J. L., Mariani, C. & Vriezen, W. H. The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J. Exp. Bot. 62, 617–26 (2011).
Goetz, M. et al. Expression of aberrant forms of auxin response factor 8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol. 145, 351–366 (2007).
De Jong, M. et al. Solanum lycopersicum auxin response factor 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 66, 3405–16 (2015).
Foolad, M. R. Genome mapping and molecular breeding of tomato. Int. J. Plant Genom. 64358, https://doi.org/10.1155/2007/64358 (2007).
Mapelli, S. C., Frova, C., Torti, G. & Soressi, G. P. Relation-ship between set, development and activities of growth regulators in tomato fruits. Plant Cell Physiol. 19, 1281–1288 (1978).
Abdel-Rahman, M. Patterns of hormone respiration and ripening enzymes during development maturation and ripening of cherry tomato fruits. Physiol. Plant 39, 115–118 (1977).
Gillaspy, G., Ben-David, H. & Gruissem, W. Fruits: a developmental perspective. Plant Cell 5, 1439–1451 (1993).
El-Sharkawy, I. et al. Expression of auxin-binding protein1 during plum fruit ontogeny supports the potential role of auxin in initiating and enhancing climacteric ripening. Plant Cell Rep. 31, 1911–21 (2012).
Sjut, V. & Bangerth, F. Induced parthenocarpy: a way of changing the levels of endogenous hormones in tomato fruits (Lycopersicon esculentum Mill.). 1. Extractable hormones. Plant Growth Regul. 1, 243–251 (1983).
Bohner, J., Hedden, P., Bora-Haber, E. & Bangerth, F. Identification and quantitation of gibberellins in fruits of Lycopersicon esculentum and their relationship to fruit size in L. esculentum and L. pimpinellifolium. Physiol. Plant 73, 348–353 (1988).
Srivastava, A. & Handa, A. K. Hormonal regulation of tomato fruit development: a molecular perspective. J. Plant Growth Regul. 24, 67–82 (2005).
Carrera, E., Ruiz-Rivero, O., Peres, L. E. P., Alejandro, A. & Garcia-Martinez, J. L. Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 160, 1581–1596 (2012).
Lemaire-Chamley, M. et al. Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol. 139, 750–769 (2005).
Su, L. Y., Audran, C., Bouzayen, M., Roustan, J. P. & Chervin, C. The Aux/IAA, Sl-IAA17 regulates quality parameters over tomato fruit development. Plant Signal Behav. 10, e1071001 (2016).
Matsuo, S., Kikuchi, K., Fukuda, M., Honda, I. & Imanishi, S. Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 63, 5569–79 (2012).
Czerednik, A., Busscher, M., Bielen, B. A. M., Wolters-Arts, M. & de Maagd, R. A. Regulation of tomato fruit pericarp development by an interplay between CDKB and CDKA1 cell cycle genes. J. Exp. Bot. 63, 2605–2617 (2012).
Gonzalez, N., Gévaudant, F., Hernould, M., Chevalier, C. & Mouras, A. The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J. 51, 642–55 (2007).
Vrebalov, J. et al. Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–62 (2009).
Machemer, K. et al. Interplay of MYB factors in differential cell expansion, and consequences for tomato fruit development. Plant J. 68, 337–50 (2011).
Guillet, C. et al. Regulation of the fruit-specific PEP carboxylase SlPPC2 promoter at early stages of tomato fruit development. PLoS One 7, e36795 (2012).
Wu, J. et al. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 30, 2059–2073 (2011).
De Jong, M., Wolters-Arts, M., Feron, R., Mariani, C. & Vriezen, W. H. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, I60–170 (2009).
Serrani, J. C., Fos, M., Atarés, A. & García-Martínez, J. L. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv. micro-tom of tomato. J. Plant Growth Regul. 26, 211–221 (2007).
Bünger-Kibler, S. & Bangerth, F. Relationship between cell number, cell size and fruit size of seeded fruits of tomato (Lycopersicon esculentum Mill.), and those induced parthenocarpically by the application of plant growth regulators. Plant Growth Regul. 1, 143–154 (1982).
Pattison, R. J. et al. Comprehensive Tissue-Specific Transcriptome Analysis Reveals Distinct Regulatory Programsduring Early Tomato Fruit Development. Plant Physiol. 168, 1684–701 (2015).
Vriezen, W. H., Feron, R., Maretto, F., Keijman, J. & Mariani, C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 177, 60–76 (2008).
Kumar, R., Tyagi, A. K. & Sharma, A. K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genomics 285, 245–260 (2011).
Krogan, N. T. & Berleth, T. A dominant mutation reveals asymmetry in MP/ARF5 function along the adaxial-abaxial axis of shoot lateral organs. Plant Signal Behav. 7, 940–943 (2012).
Herud, O., Weijers, D., Lau, S. & Jüurgens, G. Auxin responsiveness of the MONOPTEROS- BODENLOS module in primary root initiation critically depends on the nuclear import kinetics of the Aux/IAA inhibitor BODENLOS. Plant J. 85, 269–277 (2016).
Wu, M. F. et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4, e09269 (2015).
Cole, M. et al. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651 (2009).
Martí, C. et al. Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876 (2007).
Ampomah-Dwamena, C., Morris, B. A., Sutherl, P., Veit, B. & Yao, J. L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130, 605–617 (2002).
Schijlen, E. G. et al. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 144, 1520–1530 (2007).
Molesini, B., Pandolfini, T., Rotino, G. L., Dani, V. & Spena, A. Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol. 149, 534–48 (2009).
Olimpieri, I. et al. Tomato fruit set driven by pollination or by the parthenocarpic fruit allele are mediated by transcriptionally regulated gibberellin biosynthesis. Planta 226, 877–888 (2007).
Rojas-Gracia, P. et al. The parthenocarpic hydra mutant reveals a new function for a SPOROCYTELESS -like gene in the control of fruit set in tomato. New Phytol. 214, 1198–1212 (2017).
Carmi, N., Salts, Y., Dedicova, B., Shabtai, S. & Barg, R. Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217, 726–735 (2003).
Ueta, R. et al. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507 (2017).
Ren, Z. & Wang, X. SlTIR1 is involved in crosstalk of phytohormones, regulates auxin-induced root growth and stimulates stenospermocarpic fruit formation in tomato. Plant Sci. 253, 13–20 (2016).
Wang, H. et al. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21, 1428–1452 (2009).
Serrani, J. C., Ruiz-Rivero, O., Fos, M. & García-Martínez, J. L. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 56, 922–934 (2008).
Hedden, P. & Phillips, A. L. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–30 (2000).
Mauriat, M. & Moritz, T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinctroles in wood formation. Plant J. 58, 989–1003 (2009).
Frigerio, M. et al. Transcriptional regulation of gibberellins metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142, 553–63 (2006).
Li, J., Dai, X. & Zhao, Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 140, 899–908 (2006).
Breitel, D. A. et al. Auxin response factor 2 intersects hormonal signals in the regulation of tomato fruit ripening. PLoS Genet. 12, e1005903 (2016).
Tang, N., Deng, W., Hu, G., Hu, N. & Li, Z. Transcriptome profiling reveals the regulatory mechanism underlying pollination dependent andparthenocarpic fruit set mainly mediated by auxin and gibberellin. PLoS One 10, e0125355 (2015).
da Silva, E. M. et al. microRNA159-targeted SlGAMYB transcription factors are required for fruit set in tomato. Plant J. 92, 95–109 (2017).
Rotino, G. L., Perri, E., Zottini, M., Sommer, H. & Spena, A. Genetic engineering of parthenocarpic plants. Nat. Biotechnol. 15, 1398–1401 (1997).
Mezzetti, B., Landi, L., Pandolfini, T. & Spena, A. The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnol. 4, 4 (2004).
Sun, H. J., Uchii, S., Watanabe, S. & Ezura, H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 47, 426–31 (2006).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–7 (1987).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–8 (2008).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31471878 and 31272178), Zhejiang province key science and technology innovation team (2013TD05), and the Natural Science Foundation of Zhejiang province, China (LZ17C150002).
Author information
Authors and Affiliations
Contributions
S.L. performed the experiments, analyzed data and wrote the paper. Q.F. performed the qRT-PCR, Y.Z. and L.Q. helped in experiments, C.P. analyzed the RNA-seq data, A.T.L.S. helped to collect the data and revise the manuscript. G.L. designed the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, S., Zhang, Y., Feng, Q. et al. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci Rep 8, 2971 (2018). https://doi.org/10.1038/s41598-018-21315-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-21315-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.