Abstract
Apomixis (asexual reproduction through seeds) is considered a deviation of the sexual reproductive pathway leading to the development of clonal progenies genetically identical to the mother plant. Here we used the Methylation-Sensitive Amplification Polymorphism (MSAP) technique to characterize cytosine methylation patterns occurring in florets of sexual and aposporous Paspalum notatum genotypes, in order to identify epigenetically-controlled genes putatively involved in apomixis development. From twelve polymorphic MSAP-derived sequences, one (PN_6.6, later renamed PN_SCD1) was selected due to its relevant annotation and differential representation in apomictic and sexual floral transcriptome libraries. PN_SCD1 encodes the DENN domain/WD repeat-containing protein SCD1, which interacts with RAB GTPases- and/or MAPKs to promote specialized cell division, functions in clathrin-mediated membrane transport and acts as potential substrate receptor of CUL4 E3 ubiquitin ligases. Quantitative RT-PCR and comparative RNAseq analyses of laser microdissected nucellar cells confirmed PN_SCD1 upregulation in florets of apomictic plants and revealed that overexpression takes place just before the onset of apospory initials. Moreover, we found that several SCD1 molecular partners are expressed in P. notatum florets and upregulated in apomictic plants. Our results disclosed a specific vesicle trafficking molecular pathway epigenetically modulated during apomixis.
Similar content being viewed by others
Introduction
Seed formation in flowering plants can follow variable mechanisms, which have evolved over millions of years to secure persistence and diffusion of adapted genotypes. In this regard, the sexual pathway is far from being the only possible operative route. Seeds can be formed through alternative asexual mechanisms, which are classified under the general term ‘apomixis’1. These asexual reproductive pathways lead to the formation of a clonal progeny genetically identical to the mother plant. Apomixis was described in more than 400 flowering plant species belonging to 35 families2, but it is absent from gymnosperms3, with the exception of a male-type odd case described in Cupressus4. Despite the complexity and diversity of these alternative reproductive routes, it is widely accepted that apomictic behaviours can be regarded as deregulations of sexual developmental processes in time and space, involving both genetic and epigenetic mechanisms5. Often, sexuality and apomixis coexist in the same organism, a condition known as facultative apomixis1.
Apomictic developmental pathways are subclassified into two different subcategories: sporophytic and gametophytic1. In sporophytic apomicts, somatic (2n) cells from the ovule nucellus or the integuments differentiate to generate multiple globular-shaped embryos, which coexist with the legitimate sexually-formed sibling and share its endosperm for nutrition6. In gametophytic apomicts, one or several unreduced (2n) embryo sacs are formed in the ovule nucellus after a series of mitosis6. Gametophytic apomixis is further classified into diplospory and apospory, depending on the cell giving origin to the unreduced embryo sacs. They originate from the megaspore mother cell itself in diplosporous plants and from somatic nucellar companion cells in aposporous ones. Embryo development is fertilization independent i.e. embryos arise through parthenogenesis, whereas endosperm formation may or may not require fertilization6.
Apomixis represents a useful tool for agriculture, due to its intrinsic capacity to immediately fix heterotic combinations7. The absence of female meiosis and subsequent egg cell fertilization leads to the emergence of clonal progenies from genetically maternal seeds. However, when used as male parents in experimental crosses with sexual plants, apomictic individuals are able to transfer the trait and produce hybrid progenies segregating for the mode of reproduction. Natural apomictic species, like perennial forage grasses included in the genera Brachiaria and Paspalum, are currently being improved through novel apomixis-based breeding mechanisms8,9,10,11,12. These programs have accelerated the production of adapted hybrid cultivars and are rapidly boosting livestock farming in areas that had traditionally been marginal for cattle production. Harnessing apomixis into major crops like maize, sorghum or rice has proved to be more difficult, since projects aimed at introgressing the trait from wild relatives through sexual crosses were unsuccessful so far13. Genetic engineering could provide a possible way out, but the genetic determinants that control the expression of the trait remain unknown. Therefore, a full characterization of the molecular pathways governing apomixis is crucial to overcome barriers banning introduction from wild relatives and/or to allow the modulation of the trait through genetic engineering14. Moreover, a deep knowledge of the apomixis genetic determinants will allow a better prediction, avoidance and/or control of potential environmental risks derived from its use in agriculture7.
Evidence originated from sexual model species indicated that epigenetic mechanisms are involved in the expression of apomixis, since the disruption of RdDM (RNA-directed DNA methylation) pathways operating in ovules promoted the appearance of phenotypes mimicking the trait. In Arabidopsis plants, a loss of function of an ARGONAUTE (AGO) protein family member, AGO9, results in the development of supernumerary unreduced megaspores in the ovular nucellus15. In wild-type ovules, the AGO9 protein is not localized in the nucellar gametic cells, but in nucellus layer 1 (L1) cells15. Since AGO9 is involved in the silencing mechanisms mediated by small RNAs, these data suggest the existence of trans-acting small interference RNA (tasiRNA) pathways operating in the nucellus in order to stipulate the legitimate female gametophytic lineage15. Moreover, ago104 defective maize mutants produce up to 70% of functional unreduced female gametes16. Such unreduced gametes arise from mitosis-like divisions of the megaspore mother cells themselves. Thus, whereas AGO9 mutations produce phenotypes resembling apospory, AGO104 disruptions ends up in phenotypes mimicking diplospory15,16. Several other epigenetic regulators were identified in differential expression analyses of sexual maize and apomictic maize-Tripsacum hybrids, including several DNA methyltransferases (DMT102, DMT103 and DMT105), a SWI2/SNF2-like chromatin remodeler (CHR106), a plant-specific histone deacetylase (HDT104) and a histone H1 linker (HON101)17. Further characterization of DMT102 and DMT103 revealed that they are both expressed in germ cells as well as surrounding nucellar cells, and that maize mutants are able to form unreduced gametes and/or extra embryo sacs, and show a release of repressive chromatin states17.
Paspalum became a model genus for the analysis of aposporous apomixis mechanisms, due to its dual nature as a useful system for mining candidate gene(s) and an important target crop18. A wealth of information has been produced regarding the biology, genetic and reproductive modes of many Paspalum species including: (1) analysis of cytoembryological aspects of sexual and asexual development; (2) detailed molecular mapping of apomixis loci; (3) availability of candidate genes for the control of apospory; and (4) development of transformation systems for gene delivery18. The best-characterized species within the genus are Paspalum notatum and Paspalum simplex, both represented by two major cytotypes: sexual diploids and aposporous pseudogamous apomictic tetraploids. In both species, aposporous apomixis is controlled by the single genomic superlocus named Apomixis Controlling Locus (ACL), which is blocked in terms of recombination and shows synteny with a subtelomeric region of the rice chromosome 12 long arm, and for P. notatum only, also with segments of rice chromosome 2 long arm19,20,21,22. In the particular case of P. notatum, the ACL also includes segments syntenic to the rice chromosome 2 long arm19,20,21,22. Partial sequence analysis of the ACL revealed structural features of heterochromatin, namely the presence of repetitive elements, gene degeneration23 and deregulation24. After examining the cytosine methylation status of the ACL in combination with apomixis-linked BAC-FISH analysis, a high level of methylation was detected25. Moreover, DNA methylation is essential for parthenogenesis, since the demethylating agent 5′-azacytidine causes a significant increase of hybrid progenies of a higher ploidy level (BIII hybrids) in the offspring of treated apomictic mother plants25.
Investigating the epigenetic-mediated control of asexual development is substantial not only to projects dealing with reproductive biology, but also to apomixis-based breeding programs, since the latter are strongly dependent on the generation of obligate apomictic hybrids with full capacity to form clonal progeny. Due to the highly heterozygous, polyploid and poorly characterized nature of the genome of natural apomictic grasses22, the technique chosen to detect epigenetic marks must be independent from any prior sequence information, particularly if a genome-wide analysis is required. The Methylation-Sensitive Amplified Polymorphism (MSAP) methodology enables the evaluation of levels and patterns of DNA methylation from an open and broad perspective, without previous sequence knowledge26. It is based on the use of isoschizomers HpaII and MspI, a pair of restriction enzymes that recognize the same target site (5′-CCGG-3′) but have different sensitivity to methylated cytosines26. Although revealing only a fraction of the global methylation changes, MSAP can be very useful to investigate epigenetic variations occurring between different genotypes, since differentially methylated sites lacking genetic polymorphisms can be easily discriminated27. Among many other applications, MSAP has already been used to study methylation changes during ploidy level conversions in apomictic and sexual Eragrostis curvula28 and to estimate the cytosine methylation pattern frequencies and variation in diploid and tetraploid cytotypes of sexual and apomictic P. notatum29.
The objective of this research was to detect and validate epigenetically-controlled apomixis candidate genes. Through the use of MSAP, we found that the vesicle trafficking regulator PN_SCD1 displays gene body hypomethylation and transcript upregulation in the ovule nucellus of Paspalum apomictic plants just before the onset of apospory. Moreover, several PN_SCD1 biological partners showed de-regulated expression in apomictic genotypes.
Results
Methylation-sensitive amplified polymorphism analysis
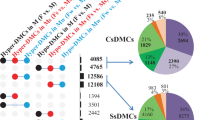
Twelve MSAP primer combinations were used to compare the cytosine methylation status in genomic DNA samples extracted from florets of four sexual (#36, #58, #76, #83) and five apomictic (#Q4117, #Q4086, #9, #40, #112) P. notatum individuals. Overall, these experiments generated 547 clearly detectable and reproducible fragments, which were resolved by capillary electrophoresis linked to an automated detection system. An example of the experimental output is shown in Fig. 1. The frequency of occurrence of the different methylation patterns in the analysed genotypes is shown in Table 1. Sexual and apomictic plants displayed, respectively, an average of: (1) 205 ± 7 and 216 ± 16 unmethylated sites; (2) 62 ± 8 and 46 ± 17 externally hemimethylated sites; (3) 114 ± 8 and 92 ± 13 internally methylated sites; (4) 166 ± 3 and 192 ± 32 methylated at both cytosines or absent sites (the latter due to genetic variation). The average distribution of different methylation patterns in sexual and apomictic samples is summarized in Fig. 2. Tests of Newcombe for 95% confidence intervals of proportions estimations30 showed that the different methylation patterns occurred in equivalent ratios in sexual and apomictic genotypes. However, a locus by locus analysis across the total 547 loci revealed significant variation in the methylation status among the different genotypes. Moreover, clustering analysis based on an epigenetic similarity matrix27 underlined a higher similarity among sexual plants (Supplementary Fig. S1) (see also Discussion).
MSAP output and DNA methylation patterns. Methylation patterns corresponding to a single sexual and a single apomictic individual are shown as an example. Arrows and letters indicate each DNA methylation pattern55. Sexual and apomictic samples were digested with either HpaII or MspI. Patterns A–C represent monomorphic classes, in which the methylation pattern is the same in the sexual and the apomictic genotype. Patterns E–P are indicative of possible cytosine methylation alterations in the apomictic plant with respect to the sexual one.
Average proportion of methylation patterns in flowers of sexual and apomictic P. notatum genotypes. Both plant types (apomictic and sexual) showed identical general methylation patterns proportions, as indicated by Newcombe tests30.
Identification of differentially methylated DNA fragments expressed in florets
A total of 26 genomic DNA fragments showing statistically significant differential methylation between sexual and apomictic samples were detected (Supplementary Fig. S2). Amplicons were then resolved in polyacrylamide gels and twelve candidate fragments (PN_1.7; PN_1.8; PN_2.2; PN_2.10; PN_4.8; PN_4.10; PN_6.5; PN_6.6; PN_7.6; PN_7.7; PN_8.5; PN_8.8) were successfully retrieved from the gels, cloned and sequenced. These sequences were used as queries for in silico mapping analyses onto the rice genome and transcriptome surveys onto 454 floral transcriptome libraries of sexual and apomictic P. notatum31. Rice in silico mapping results are displayed in Supplementary Table S1. Two pairs of fragments (PN_1.7/PN_1.8 and PN_7.6/PN_7.7) matched to variants of the same sequence and could have been originated from different alleles. Only one fragment (PN_6.6) mapped with top identity onto a functionally characterized gene (BGIOSGA001363, SCD1-like). The rest of the sequences mapped either onto unknown sequences or functionally uncharacterized genes. Regarding the Paspalum floral transcriptome survey, three of the sequences were found expressed in the libraries (PN_2.10, PN_6.6 and PN_8.5). The remaining ones did not find a transcribed match. Therefore, they could be included in promoter, intronic, or intergenic regions, as well as belonged to transcribed regions of genes not expressed in flowers. One of the candidates expressed in flowers (PN_6.6) showed a de-methylated pattern in apomictic plants (11) (with the exception of the Q4117 genotype) and an internally methylated pattern in sexual plants (01) (Table 2). The remaining two candidates (PN_2.10 and PN_8.5) displayed a de-methylated pattern is apomictic plants (11) (with the exception of F1 112 genotype) and a fully-methylated pattern (or absence of the site) in sexual plants (00) (Table 2). The PN_6.6, PN_2.10 and PN_8.5 full sequences were recovered from the 454 floral transcriptome databases31. Annotations were determined from BLASTX at NCBI and confirmed by comparison with ontology classification analysis performed by INDEAR (Instituto de Agrobiotecnología de Rosario) during the construction of the 454/Roche floral transcriptome libraries. Two of the sequences corresponded to protein-coding genes (PN_6.6 and PN_8.5) and the remaining one was unknown (PN_2.10) (Table 2).
One of the sequences (PN_6.6) showed differential expression between sexual and apomictic plants in the 454/Roche libraries (Table 2). NCBI BLASTX searches identified the DENN domain and WD repeat-containing protein SCD1 of Setaria italica (XP_004969033.1; E-value = 0.0; ID = 1000/1036, 97%) as the best match for this sequence (Table 2). PN_6.6 was accordingly renamed PN_SCD1. The Arabidopsis putative ortholog to this gene is At1G49040 (SCD1, STOMATAL CYTOKINESIS-DEFECTIVE 1 SCD1)32,33,34. The original MSAP fragment mapped onto the WD40 domain of apoisotig16740 GFMI02016795.1, with E-value: 9 e−30, ID 99% and 1% gaps (Fig. 3). Due to the relevant annotation of PN_SCD1 concerning specialized cell division, and the putative differential expression detected in the 454/Roche libraries, we decided to extend the characterization of this particular epigenetic candidate to evaluate its potential as an apomixis associated gene.
Structural scheme of PN_SCD1. The scheme was constructed from sequence of P. notatum apoisotig16740 (GFMI02016795.1). White boxes: CDR regions with non-described domains. Black boxes: CDR regions encoding known domains (indicated below). Lines: UTR regions. Numbers indicate the nucleotide positions assigned to each feature. The original fragment isolated from MSAP analysis was represented on top of the scheme, between positions 2993 and 3073. The methylation status of the CCGG site (subjected to MSAP analysis) in apomictic and sexual plants was indicated on top.
PN_SCD1 is upregulated in florets of apomictic P. notatum plants
The relative quantitative expression of PN_SCD1 was investigated in florets collected at anthesis, by conducting qPCR experiments on cDNA samples originated from three sexual (C4-4x, #43, #83) and three apomictic (#40, #74, Q4117) plants. The REST RG relative expression quantitation analysis revealed significant upregulation in all apomictic plants tested. Results are displayed in Fig. 4 and Supplementary Table S2.
qPCR experiments showed highly significant upregulation of PN_6.6 in flowers of apomictic plants. Comparisons were carried out in three sexual (light blue) and three apomictic (red) plants. Sexual genotype C4-4x was used as control. An expression value = 1 (standard deviation = 0) was automatically assigned to C4-4x by the data processing program (REST RG). The software predicted significant upregulation in apomictic plants, in agreement with the methylation patterns detected by MSAP (Supplementary Table S2). Same letters indicate non-significant statistical differences (overlapping standard errors intervals).
PN_SCD1 is upregulated in the ovule nucellus of apomictic P. simplex plants
We investigated the expression in ovules at early developmental stages by using data of P. simplex Illumina RNAseq laser capture microdissection (LCM) libraries available for sexual and apomictic genotypes. These libraries were constructed from groups of cells of the nucellar tissues of ovules sampled at premeiotic stage in sexual and apomictic plants, just before the differentiation of aposporous initials (Galla G., Barcaccia G., Bellucci M., Pupilli F., unpublished). Although P. simplex and P. notatum are not closely related within the Paspalum genus, they display nearly identical apospory mechanisms (see Discussion). Two P. simplex sequences (PS_6.6.1 and PS_6.6.2) scored the best hits with PN_SCD1 (Table 3). As expected, the similarity parameters pointed to a high degree of sequence conservation with P. notatum (Table 3). By using any of the detected P. simplex sequences (PS_6.6 0.1 OM_Locus_1_Transcript_365164/578620 or PS_6.6.2 OM_Locus_82107_Transcript_4/4) as query in BLAST searches onto the P. notatum apomictic and sexual floral transcriptome libraries, a single match was detected in both the apo libraries (isotig16740, gene = isogroup 06295, length = 4008, num Contigs = 1, E-val 0.0) and the sexual library (isotig13672 gene = isogroup 05214 length = 3775 numContigs = 1, E-value 0.0). Transcript PS_6.6.1 resulted significantly upregulated with 97% confidence in the apomictic LCM libraries (Table 3).
SCD1 partners’ orthologs are upregulated in florets of apomictic P. notatum plants
We controlled the representation of genes related to the SCD1 role as vesicle trafficking regulator and CUL4 E3 ubiquitin ligase substrate receptor in the 454/Roche transcriptome libraries of sexual and apomictic P. notatum genotypes. Several candidates were significantly upregulated in apomictic plants (Table 4). Moreover, a PANTHER test showed that biological processes Single Organism Transport (GO:0044765) and Localization (GO:0051179) were overrepresented at highly significant p-values (1.29 E−06 and 1.76 E−07, respectively) in the list of apomictic vs. sexual differentially expressed candidate genes reported from the P. notatum 454/Roche floral transcriptome libraries31. These results point to an integral membrane protein turnover and ubiquitination pathway operating specifically during apomixis development.
PN_SCD1 in silico mapping
The possibility that PN_SCD1 was located within the ACL of Paspalum was explored by mapping its sequence in silico onto the rice genome and checking the Paspalum-rice syntenic relationship of the target regions19,22. PN_SCD1 mapped in silico onto a single rice genome site (overlapping gene BGIOSGA001363), placed at chromosome 1 long arm, in segments scattered between positions 24,525,345 and 24,508,528. The total length of the similarity region was 3,494 nt, with E-values ranging between 2.0 e73 and 0.023 depending on the length of the fragment aligned. The mapped region was found not syntenic to the ACL19,22. Moreover, both P. simplex orthologous sequences (PS_6.6 0.1 and PS_6.6.2) mapped in silico onto the same rice locus (BGIOSGA001363). No other region showing significant similarity was found in the rice genome. This evidence suggests that PN_SCD1 is represented by a single copy in the rice genome, that the different sequences detected in both Paspalum species are alleles/splice variants of the same gene and that PN_SCD1 is most probably one of the downstream participants of the molecular cascade involved in the expression of apomixis rather than its trigger. However, we could not assess the representation of PN_SCD1 directly in the Paspalum genome, since a full genome sequence assembly is still not available.
Discussion
A thorough characterization of the molecular pathways underlying apomixis is a prerequisite for its effective use in breeding. Although in the past few years considerable progress was made on the identification of genes possibly associated to the trait35 only one of them (BABYBOOM) was able to trigger a specific apomictic step (parthenogenesis) in a sexual background36, while no candidate or candidate combination was found capable to govern the whole process involved in the trait. In the genus Paspalum, comprehensive lists of candidate genes showing putative positional22,23 or expression24,37 association with apomixis were produced. However, characterization studies revealed the possible function for only a few of these candidates, like LORELEI, SERK or ORC338,39,40. Moreover, in spite of all efforts, the triggers of the apospory developmental cascade still remain elusive.
Since an epigenetic control was predicted for some apomixis crucial steps, i.e. AI onset15, MMC reduction16 and parthenogenesis25, we focused in the detection of candidate genes showing differential methylation patterns in flowers of apomictic and sexual P. notatum genotypes. MSAP was selected as a reference technique to study cytosine methylation in a wide genome basis, due to its simple procedure, low cost and independence on previous sequence characterization. Major methylation polymorphisms were spotted among genotypes, despite a general conservation of methylation proportions. Similar results were found in a prior work dealing with methylation variations in sexual and apomictic Eragrostis curvula genotypes28. Moreover, an epigenetic similarity clustering analysis revealed that sexual genotypes grouped closer in comparison with apomictic ones. These results are in agreement with a previous work conducted in P. notatum, showing that apomictic tetraploids are epigenetically more variable than sexual diploids29 and suggest an incomplete and rather disordered cytosine methylation reprogramming occurring in association with apomixis.
Our work confirmed the potential of MSAP used in combination with the Cervera et al. matrix construction method27 to detect epigenetic regulation differences even when comparing genetically different plants. Naturally, the identification of consistent epigenetic patterns between different reproductive modes turns difficult when genotypes also differ in sequence. But the technique has the potential to detect at least a bias in the occurrence of particular epigenetic features in genetically well conserved sites, even on this highly variable background. Through MSAP analyses involving 547 loci we were able to identify three differentially-methylated sequences expressed in flowers. One of them (PN_6.6) corresponded to a protein coding gene demethylated in apomictic plants and methylated at the internal cytosine in sexual plants. The remaining two sequences (PN_2.10 and PN_8.5) represented an unknown sequence and a functionally not characterized protein coding gene, respectively. However, they both displayed 11/00 MSAP patterns, which disqualified their unequivocal classification as epigenetic marks. Therefore, according to our results, 0.18% of the genome (1 band over 547) unambiguously corresponds to protein coding genes transcribed in flowers and differentially methylated at CCGG sites in apomictic and sexual genotypes. Due to the rather limited MSAP methylation detection capacity (only CCGG sites were surveyed) and the lack of discrimination between full methylation and sequence mutations for 00 patterns, this number likely represents an underestimation. However, our results demonstrated that, even partial, MSAP exploration retains the capacity to contribute to the characterization of the epigenetic status of relevant candidates.
SCD1 has a reported functional role related with specialized cell division32. It encodes the DENN domain and WD repeat-containing protein SCD1, a RAB GTPase- and/or MAPK-interacting protein32,41. In the leaf epidermis, the gene participates in the symmetric division of the guard mother cells during stomatal development32. SCD1 has specific RAB GEF activity and it might be involved in the regulation of MAPKs-mediated pathways32 as well as the clathrin-mediated membrane transport33. Moreover, it was defined as potential substrate receptor of CUL4 E3 ubiquitin ligases34. Since RAB-encoding transcripts had been found differentially expressed in sexual and apomictic Poa pratensis genotypes42, we considered PN_SCD1 (PN_6.6) worth of further investigation, under the hypothesis that it could represent the second identified member of a RAB-mediated protein localization pathway operative at the onset of apospory. In fact, RNAseq and qRT-PCR analysis revealed PN_SCD1 upregulation in florets of apomictic plants, in agreement with its de-methylated status. To study further the expression profile, we took advantage of P. simplex LCM Illumina RNAseq libraries available in our laboratory.
Although P. notatum and P. simplex belong to different Paspalum subgenera (Anachyris Nees, Chase for P. simplex and Paspalum sensu stricto for P. notatum), these display almost identical aposporous developmental patterns (i.e. several aposporic initials differentiate from nucellar cells giving rise to i) pentanucleated embryo sacs lacking antipodals in P. notatum and, ii) eight-nucleated ones including antipodals in P. simplex). The comparison between these two species allows us to identify similar or identical candidate genes between the apomictic and sexual development in two rather different genetic background once the effect of genotype differences have been eliminated by considering several genotypes for each of the two species. After P. simplex LCM libraries analyses, PN_SCD1 was confirmed to be upregulated in the ovule nucellus of apomictic plants at premeiotic stage, suggesting that overexpression is conserved within the genus and might occur at premature developmental stages.
SCD1 was initially described in plants as a conditional mutant allele that severely disrupted the formation of the cell plate during guard mother cell cytokinesis32. The SCD1 protein contains a DENN domain, the distinctive mark of vesicle trafficking regulators, which has been shown to have specific RAB GEF activity32,41,43. Moreover, the DENN domain of SCD1 might be involved in the regulation of other signaling routes required for cell expansion and cytokinesis, like MAPKs-mediated pathways32. More recently, it was reported that SCD1 functions in clathrin-mediated membrane transport33, by associating with isolated clathrin coated vesicles and colocalizing with clathrin light chain 2 in dynamic clathrin-coated pits at or near the plasma membrane33.
SCD1 contains also a WD40 domain including a DWD motif, which is defined as the signature of potential substrate receptors of CUL4 E3 ubiquitin ligases34, one of the largest subfamilies of CULLIN-RING E3 ubiquitin ligases (CRLs). The enzyme consists of three core subunits: CULLIN4 (CUL4), a RING finger protein REGULATOR OF CULLIN S1 (ROC1)/RING-BOX 1 (RBX1), and UV DAMAGED DNA BINDING PROTEIN1 (DDB1)44. While the adapter protein DDB1 assemble the substrate receptor complex, the RING finger protein ROC1/RBX1recruits E2 enzyme to form a catalytic core. Therefore, CUL4 forms a rigid packing architecture for the precise positioning of substrate toward the E2 enzyme, facilitating the ubiquitin transfer45,46,47. As a member of the small subgroup of 85 Arabidopsis WD40 proteins containing a DWD motif, SCD1 was reported to act as potential substrate receptor for DDB1-CUL4-ROC1 based E3 ubiquitin ligases34. Interestingly, CUL4 is critical for early embryonic development in mice46. Bearing this in mind, we decided to evaluate the expression of SCD1 molecular partners (RAB proteins, MAP3Ks, clathrin and CUL4-based E3 ubiquitin ligases and its associated proteins) in the closely related P. notatum floral transcriptomes. Interestingly, several of these genes were differentially expressed between apomictic and sexual plants. All of them were upregulated in aposporous plants, suggesting that a vesicle trafficking molecular pathway directing protein localization is operating in particular ways during apomixis, although the path is active in both reproductive modes.
Even when SCD1 was found demethylated and overexpressed in apomictic plants, the gene is present and active also in sexual genotypes, and even in sexual species. Therefore, the particularity of SCD1 expression in apomictic plants do not seem to be related with the occurrence of the sequence itself in this particular reproductive type, but with its tissue-specific regulation. Due to its specific expression in the cell layer originating the aposporous initials (i.e. the somatic cells acting as progenitors of the aposporous embryo sacs) just before differentiation of these cells, as well as its previously reported role in specialized plant cell division, we hypothesized that PN_6.6 might be one of the initial components of the molecular cascade associated with aposporous development. We postulate that the upregulation of PN_SCD1 and its partners in the apospory initials’ precursors might be related with the onset of the cell division processes necessary to generate an unreduced embryo sacs in functionally distinct cell types.
Methods
Plant material
The tetraploid (2n = 4x = 40) P. notatum genotypes used in the MSAP and/or qPCR analysis were selected according to the availability of flowers in the spring/summer season 2015/2016. They were listed below: Q4117 (highly apomictic accession from Brazil, 97% of ovules bearing aposporous embryo sacs, 96% of F1 progenies showing maternal genotypes)48; Q4086 (experimental facultative apomictic genotype, 78.4% aposporous embryo sacs, derived from the duplication of the diploid plant Q4084 by colchicine treatment)49; C4-4x (sexual tetraploids derived from the duplication of a diploid plant C4-2x)50; Q4188 (sexual tetraploid derived from the cross Q3664 x Q3853; parent Q3664 originated from a cross between the sexual tetraploid plant PT-2, induced by colchicine treatment of the sexual diploid Pensacola bahiagrass biotype (P. notatum var. saurae) and the white-stigma bahiagrass strain WSB51; four apomictic (#9, #40, #112, #74) and five sexual (#36, #43, #58, #76, #83) F1 hybrids derived from a cross between Q4188 (sexual) x Q4117 (apomictic)20,21. A scheme showing the pedigree of the material used is presented in Supplementary Fig. S3.
MSAP analysis
The MSAP methodology26 is based on the use of isoschizomers HpaII and MspI, a pair of restriction enzymes which recognize the same target site (5′-CCGG-3′) but show different sensitivity to the first or second methylated cytosine. HpaII is able to cut when the external cytosine is hemi-methylated (single strand), whereas MspI cuts when the internal cytosine is hemi- or fully methylated (double strand)27,52,53. Total genomic DNA was extracted from P. notatum inflorescences at anthesis using the method described by Martínez et al.54. The developmental stages were classified according to the Laspina et al.37 reproductive calendar. Inflorescences were collected at 9:00 a.m. DNA was used for MSAP analyses as described by Marconi et al.55. The list of primer combinations used and their relative sequences is provided in Supplementary Table S3. After amplifications, amplified fragments were separated by capillary electrophoresis in an ABI 3130xl Genetic Analyzer (Life Technologies). Comparisons of the patterns produced from samples digested in parallel with EcoRI/HpaII and EcoRI/MspI allowed identification of genomic alterations in the methylation patterns.
Epigenetic similarity analysis
To graphically represent the epigenetic landscape in each individual plant, a table was constructed by recording the methylation patterns corresponding to the 547 total loci in all analysed plants. The patterns were codified as I: unmethylated; II: externally methylated; III: internally methylated; IV: fully methylated or absent. Then, we produced a linear graphic representation of the methylation status at each locus for each plant. For clustering analysis, a binary matrix was assembled by codifying the presence of a fragment with 1 and the absence with 0. Then, a second derived binary matrix was produced, codifying the HpaII/MspI amplification patterns as indicated by Cervera et al.27, in order to represent methylated sites (10 or 01) as 1 and unmethylated or uninformative sites (11 or 00) as 0 (Table 1). After that, a pairwise epigenetic similarity matrix was generated using the Jaccard coefficient56. This similarity matrix was used to produce an UPGMA dendrogram.
Isolation of DNA fragments showing differential methylation
A chi square test was used to classify any biased polymorphism. Therefore, selected samples were run on polyacrylamide gels and silver stained with the aim of isolating and sequencing them as described by Bocchini et al.57. Recovered polymorphic bands were excised and rehydrated with 100 μl of ultrapure water, by incubating at 4 °C overnight. Samples were centrifuged at maximum speed and the supernatants were transferred into fresh tubes. Five μl aliquots were used as template for re-amplification. They were added to a 20 μl final volume mix containing the same E and M primer combination used in the pre-selective amplification step (Supplementary Table S3). The PCR cycling conditions were: 1 cycle of 94 °C for 1 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min and ending with a 20-min extension step at 72 °C. One μl of the re-amplified DNA was used for cloning into the pCR4-TOPO vector (TOPO TA cloning kit, Invitrogen). After ON culture, plasmids were purified using the GenElute Plasmid Miniprep Kit (Sigma-Aldrich). Both strands were sequenced with the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on an ABI 3130xl Genetic Analyzer (Applied Biosystems).
NGS library resources
Sequences identified through MSAP were used as queries in BLAST searches on P. notatum 454/Roche floral transcript databases produced in former studies31. Sequences are available at the NCBI Sequence Read Archive database under accession numbers SRX1971037 for apomictic and SRX1971038 for sexual P. notatum. The Transcriptome Shotgun Assembly (TSA) projects corresponding to the apomictic (Q4117) and sexual (C4-4x) samples were loaded at DDBJ/ENA/GenBank under the accessions GFMI00000000 and GFNR00000000, respectively. The versions surveyed in this paper are the first ones (GFMI02000000 and GFNR01000000, respectively). Moreover, MSAP fragments were also used for interrogating apomictic and sexual LCM (Laser Capture Microdissection) P. simplex Illumina libraries. Both sequences resources were constructed in triplicate from laser microdissected nucellar cells at late premeiosis stage (Galla, Bellucci, Barcaccia and Pupilli, unpublished). Cells were laser-microdissected from the ovule nucellus just before MMC differentiation, as this is the developmental stage immediately preceding the onset of apospory initials in the ovule nucellus of aposporous genotypes37.
Bioinformatic analysis
Annotations were carried out by doing BLASTX at NCBI and confirmed by comparing with results produced by INDEAR (Instituto de Agrobiotecnología de Rosario, Argentina) using TRINOTATE (https://trinotate.github.io/). In silico mapping predictions were conducted by doing BLASTN searches onto the Oryza sativa subsp. Indica genome at the Gramene website (www.gramene.org). For 454/Roche comparative quantitation, we used the data originated from mapping the reads originated from each 454 library (apo or sex) onto a global (apo + sex) assembly31. For quantification analyses onto the P. simplex Illumina LCM libraries, the isotigs identified in the P. notatum 454/Roche library were used as queries in BLASTN and BLASTX surveys, and statistically significant differential expression was detected by applying Baggerly tests58. PANTHER Gene Ontology (GO) Term Enrichment Analysis (http://pantherdb.org) was used to detect overrepresentation of Single Organism Transport and Localization GO biological processes in the P. notatum 454/Roche libraries59,60.
RNA isolation and cDNA synthesis
Florets at anthesis were collected at 9:00 a.m and classified according to the method recommended by Laspina et al.37. Total RNA samples were extracted by using the SV Total RNA Isolation System (Promega, Madison, WI, USA), according to the manufacturers’ protocol. Total RNA concentrations and quality indexes were determined by spectrophotometry at 260/280 nm. RNA was retro-transcribed using SuperScriptTM II Reverse Transcriptase (Invitrogen Life Tecnologies, California, CA, USA). Briefly, 1 μg RNA, 10 pmol oligo-dT and 1 μl dNTP mix (10 mM each) were mixed in a 12 μl final volume. After 5 min of 65 °C incubation, 4 μl of 5X First-Stand Buffer and 2 μl 0.1 M DTT were added to the mix. All components were incubated at 42 °C for 2 min. Then, 1 μl of SuperScript RT II was added. The reaction was incubated for 50 min at 42 °C and then stopped by heating at 70 °C for 15 min. The cDNA samples were stored at −80 °C until use.
Quantitative RT-PCR analysis
Gene-specific PCR primer pairs were designed by using Primer3 v.0.4.0 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi)61 (Supplementary Table S4). Oligonucleotides were provided by GBT (GeneBiotech, Argentina). RT-PCR reactions were prepared in a 25 µL final volume reaction containing 200 nM gene specific primers, 1X Mezcla Real qPCR (Biodynamics, Argentina) and 20 ng of reverse-transcribed RNA. Beta-tubulin was used as normalizer gene, since it was characterized as an optimal reference in analogous P. notatum apomictic vs. sexual systems38,39. RT (-) and non-template controls were incorporated to the assays. Each biological replicate was processed including three technical replicates. Amplifications were performed in a Rotor-Gene Q thermocycler (Qiagen), programmed as follows: 2 min at 94 °C, 45 cycles of 15 s at 94 °C, 30 s at 57 °C, 40 s at 72 °C. Melting curves (10 s cycles from 72 to 95 °C, where temperature was increased by 0.2 °C after cycle 2) were produced at the end of the cycling to check consistent amplification of a single amplicon. Relative quantitative expression was estimated by using REST-RG (Relative Expression Software Tool V 2.0.7 for Rotor Gene, Corbett Life Sciences), considering the take-off values and amplification efficiencies for each particular reaction. The REST9 software allows relative quantitation by making comparisons between samples in pairs. Sexual plant C4-4x was used as control, referring the expression of each sample to this particular genotype.
Availability of materials and data
The plant materials used as progenitors in the current study were collected/developed and characterized by Prof. Camilo L. Quarin at Instituto de Botánica del Nordeste (IBONE), CONICET-UNNE, Corrientes, Argentina and described in prior works of our research group48,50,51. All materials belong to the IBONE’s live germplasm collection. Plants were grown in accordance with the local legislation at IBONE-CONICET-UNNE and IICAR-CONICET-UNR, Rosario, Argentina. Voucher specimens of this material are deposited at the Herbarium CTES-IBONE (publicly available), under deposition numbers: C4-4X (Quarin, C. L. 4260, barcode CTES0541627, cardboard No. 330064); Q4188 (Quarín, C. L. 4188, barcode CTES0542168, cardboard No. 323424; Q4117 (Quarin, C. L. 4117, barcode CTES0541626, cardboard No. 233851); Q4086 (Quarin, C. L. 4086, barcode CTES0542167, cardboard No. 219629). Part of the evidence presented here was retrieved from datasets reported in a prior article31, which are publicly available at: NCBI Sequence Read Archive database, accession numbers SRX1971037 and SRX1971038; Transcriptome Shotgun Assembly (TSA) projects, DDBJ/ENA/GenBank, accessions GFMI00000000 and GFNR00000000 (the versions surveyed in this paper are the first ones, GFMI02000000 and GFNR01000000, respectively).
References
Nogler, G.A. Gametophytic Apomixis in Embryology of Angiosperms (ed. Johri, B.M.) 475-518 (Springer-Verlag, 1984).
Ozias-Akins, P. Apomixis: developmental characteristics and genetics. Crit. Rev. Plant Sci. 25, 199–214 (2006).
Carman, J. G. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 61, 51–94 (1997).
Pichot, C., El Maataoui, M., Raddi, S. & Raddi, P. Surrogate mother for endangered Cupressus. Nature 412, 39 (2001).
Koltunow, A. M. & Grossniklaus, U. Apomixis: a developmental perspective. Annu. Rev. Plant Biol. 54, 547–574 (2003).
Crane, C. F. Classification of apomictic mechanisms in The Flowering of Apomixis: From Mechanisms to Genetic Engineering (ed. Savidan, Y., Carman, J. G. & Dresselhaus, T.) 24–43 (CIMMYT, IRD, European Commission DG VI, 2001).
Pupilli, F. & Barcaccia, G. Cloning plants by seeds: inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J. Biotechnol. 159, 291–311 (2012).
Acuña, C. A., Blount, A. R., Quesenberry, K. H., Hanna, W. W. & Kenworthy, K. E. Reproductive characterization of bahiagrass germplasm. Crop Sci. 47, 1711–1717 (2007).
Acuña, C. A., Blount, A. R., Quesenberry, K. H., Kenworthy, K. E. & Hanna, W. W. Bahiagrass tetraploid germplasm: reproductive and agronomic characterization of segregating progeny. Crop Sci. 49, 581–588 (2009).
Acuña, C. A., Blount, A. R., Quesenberry, K. H., Kenworthy, K. E. & Hanna, W. W. Tetraploid bahiagrass hybrids: breeding technique, genetic variability and proportion of heterotic hybrids. Euphytica 179, 227–235 (2011).
Quesenberry, K. H., Dampier, J. M., Lee, Y. Y., Smith, R. L. & Acuña, C. A. Doubling the chromosome number of bahiagrass via tissue culture. Euphytica 175, 43–50 (2010).
Barrios, S. C. L., Do Valle, C. B., Alves, G. F., Simeão, R. M. & Jank, L. Reciprocal recurrent selection in the breeding of Brachiaria decumbens. Tropical Grasslands-Forrajes tropicales 1, 52–54 (2013).
Leblanc, O. et al. Seed development and inheritance studies in apomictic maize-Tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. International Journal of Developmental Biology 53, 585–96 (2004).
Barcaccia, G. & Albertini, E. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod. 26, 159–179 (2013).
Olmedo-Monfil, V. et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 25, 628–632 (2010).
Singh, M. et al. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23, 443–458 (2011).
García-Aguilar, M., Michaud, C., Leblanc, O. & Grimanelli, D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22, 3249–3267 (2010).
Ortiz, J. P. A. et al. Harnessing apomictic reproduction in grasses: what we have learned from Paspalum. Ann. Bot. 112, 767–787 (2013).
Pupilli, F. et al. Comparative mapping reveals partial conservation of synteny at the apomixis locus in Paspalum spp. Mol. Genet. Genomics 270, 539–548 (2004).
Stein, J., Quarin, C. L., Martínez, E. J., Pessino, S. C. & Ortiz, J. P. A. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor. Appl. Genet. 109, 186–191 (2004).
Stein, J. et al. A genetic map of tetraploid Paspalum notatum Flügge (bahiagrass) based on single-dose molecular markers. Mol. Breed. 20, 153–166 (2007).
Podio, M. et al. Sequence characterization, in silico mapping and cytosine methylation analysis of markers linked to apospory in Paspalum notatum. Genet. Mol. Biol. 35, 827–837 (2012).
Calderini, O. et al. Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcolinearity with rice. Theor. Appl. Genet. 112, 1179–1191 (2006).
Polegri, L., Calderini, O., Arcioni, S. & Pupilli, F. Specific expression of apomixis-linked alleles revealed by comparative transcriptomic analysis of sexual and apomictic Paspalum simplex Morong flowers. J. Exp. Bot. 61, 1869–1883 (2010).
Podio, M. et al. A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control on parthenogenesis. J. Exp. Bot. 65, 6411–6424 (2014).
Xiong, L. Z., Xu, C. G., Saghai Maroof, M. A. & Zhang, Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 261, 439–446 (1999).
Cervera, M. T., Ruiz García, J. M. & Martínez Zapater, J. M. Analysis of DNA methylation in Arabidopsis thaliana based on methylation sensitive AFLP markers. Mol. Genet. Genomics 268, 543–552 (2002).
Ochogavía, A. C., Cervigni, G., Selva, J. P., Echenique, V. C. & Pessino, S. C. Variation in cytosine methylation patterns during ploidy level conversions in Eragrostis curvula. Plant Mol. Biol. 70, 17–29 (2009).
Rodríguez, M. P., Cervigni, G. D. L., Quarin, C. L. & Ortiz, J. P. A. Frequencies and variation in cytosine methylation patterns in diploid and tetraploid cytotypes of Paspalum notatum. Biol. Plantarum 56, 276–282 (2012).
Newcombe, R. G. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat. Med. 17, 857–872 (1998).
Ortiz, J. P. A. et al. A reference floral transcriptome of sexual and apomictic Paspalum notatum. BMC Genomics 18, 318, https://doi.org/10.1186/s12864-017-3700-z (2017).
Falbel, T. G. et al. SCD1 is required for cytokinesis and polarized cell expansion in Arabidopsis thaliana. Development 130, 4011–4024 (2003) Erratum: Development 130, 4495 (2003).
McMichael, C. M. et al. Mediation of clathrin-dependent trafficking during cytokinesis and cell expansion by Arabidopsis stomatal cytokinesis defective proteins. Plant Cell 25, 3910–3925 (2013).
Lee, J. H. et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20, 152–167 (2008).
Hand, M. & Koltunow, A. The genetic control of apomixis: asexual seed formation. Genetics 197, 441–450 (2014).
Conner, J. A., Mookkan, M., Huo, H., Chae, K. & Ozias-Akins, P. A parthenogenesis gene of apomictic origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl. Acad. Sci. USA 112, 11205–11210 (2015).
Laspina, N. V. et al. Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol. Biol. 67, 615–628 (2008).
Felitti, S. A. et al. Expression of LORELEI-like genes in aposporous and sexual Paspalum notatum plants. Plant Mol. Biol. 77, 337–354 (2011).
Podio, M. et al. Characterization and expression analysis of SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) genes in sexual and apomictic Paspalum notatum. Plant Mol. Biol. 84, 479–495 (2014).
Siena, L. A. et al. An apomixis-linked ORC3-like pseudogene is associated with silencing of its functional homolog in apomictic Paspalum simplex. J. Exp. Bot. 67, 1965–1978 (2016).
Marat, A. L., Dokainish, H. & McPherson, P. S. DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 286, 13791–13800 (2011).
Albertini, E., Marconi, G., Barcaccia, G., Raggi, L. & Falcinelli, M. Isolation of candidate genes for apomixis in Poa pratensis. Plant Mol Biol 56, 879–894 (2004).
Yoshimura, S., Gerondopoulos, A., Linford, A., Rigden, D. J. & Barr, F. A. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 (2010).
Lee, J. & Zhou, P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26, 775–780 (2007).
Zheng, N. et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 (2002b).
Li, B., Ruiz, J. C. & Chun, K. T. CUL-4A is critical for early embryonic development. Mol. Cell. Biol. 22, 4997–5005 (2002).
Angers, S. et al. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443, 590–593 (2006).
Ortiz, J. P., Pessino, S. C., Leblanc, O., Hayward, M. D. & Quarin, C. L. Genetic fingerprint for determining the mode of reproduction in Paspalum notatum, a subtropical apomictic forage grass. Theor. Appl. Genet. 95, 850–856 (1997).
Espinoza, F., Daurelio, L. D., Pessino, S. C., Quarin, C. L. & Valle, E. M. Genetic characterization of Paspalum notatum accessions by AFLP markers. Plant Syst. Evol. 258, 147–159 (2006).
Quarin, C. L., Espinoza, F., Martínez, E. J., Pessino, S. C. & Bovo, O. A. A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sex. Plant Reprod. 13, 243–249 (2001).
Quarin, C. L. et al. Registration of Q4188 and Q4205, sexual tetraploid germplasm lines of Bahiagrass. Crop Sci 43, 243–249 (2003).
McClelland, M., Nelson, M. & Raschke, E. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucl Acids Res 22, 3640–3659 (1994).
Fraga, M. & Esteller, M. DNA methylation: a profile of methods and applications. Biotechniques 33, 632–649 (2002).
Martínez, E. J., Hopp, H. E., Stein, J., Ortiz, J. P. A. & Quarin, C. L. Genetic characterization of apospory in tetraploid Paspalum notatum based on the identification of linked molecular markers. Mol. Breed. 12, 319–327 (2003).
Marconi, G. et al. Use of MSAP markers to analyse the effects of salt stress on DNA methylation in rapeseed (Brassica napus var. oleifera). PloS One 8, e75597 (2013).
Jaccard, P. Distribution de la flore alpine dans le Bassin des Drouces et dans quelques regions voisines. Bulletin de la Société Vaudoise des Sciences Naturelles 37, 241–272 (1901).
Bocchini, M. et al. Iron deficiency in barley plants: phytosiderophore release, iron translocation, and DNA methylation. Front. Plant Sci. 6, 514, https://doi.org/10.3389/fpls.2015.00514 (2015).
Baggerly, K., Deng, L., Morris, J. & Aldaz, C. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19, 1477–1483 (2003).
Thomas, P. D. et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41(Database issue), D377–D386. https://doi.org/10.1093/nar/gks1118 (2013).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers in Bioinformatics Methods and Protocols: Methods in Molecular Biology, (ed. Krawetzvand, S., Misener, S.) 365–386 (Humana Press, 2000).
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreement No 645674; Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina, Project PICT PICT-2014-1080; Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina, Project PIP 11220090100613; Ministero degli Affari Esteri, e della Cooperazione Internazionale, Direzione Generale per la Promozione del Sistema Paese; Academic Research Project of the University of Padova (Transcriptomics of reproductive organs in model species for a comparative analysis of the genetic-molecular factors characterizing sexual and apomictic processes, CPDA128282/12), Principal investigator: G. Barcaccia. J.P.A. Ortiz and S. Pessino are research staff members of CONICET. We are grateful to Dr. Camilo L. Quarin and the IBONE (Instituto de Botánica del Nordeste, Corrientes, Argentina) for kindly providing the plant material.
Author information
Authors and Affiliations
Contributions
M.Bo. carried out the MSAP experimental work, collaborated with the 454 library bioinformatic studies and performed the qPCR analysis. G.G., F.P., M.Be. and G.B. analysed the L.C.M. Illumina libraries. J.P.A.O. classified the samples according to the plants’ reproductive stages and extracted the genomic DNA samples. S.P. designed part of the experimental approach, conducted the survey on the 454 transcriptome library and wrote the paper. E.A. designed part of the experimental approach, carried out the methylation analysis and wrote the paper. All authors critically revised and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bocchini, M., Galla, G., Pupilli, F. et al. The vesicle trafficking regulator PN_SCD1 is demethylated and overexpressed in florets of apomictic Paspalum notatum genotypes. Sci Rep 8, 3030 (2018). https://doi.org/10.1038/s41598-018-21220-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-21220-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.