Abstract
The roles of long non-coding RNAs (lncRNAs), a class of long non-protein-coding RNAs, in the tumorigenesis of ovarian epithelial cells remain unknown. In this study, we discovered that the expression of long intergenic non-coding RNA 1088 (LINC01088) was clearly reduced in benign epithelial ovarian tumor tissues compared to matched normal ovarian tissues. This was shown by global cDNA gene chip scanning and real-time qPCR, and validated in 42 clinical specimens. Furthermore, we found that LINC01088 inhibited the growth of ovarian cancer xenografts in nude mice. Correlation analysis between LINC01088 and mircoRNAs (miRNAs) conducted using primary clinical samples and RNA co-precipitation experiments revealed that miR-24-1-5p was one of the targets of LINC01088. Overexpression of miR-24-1-5p facilitated cell proliferation both in vitro and in vivo, however, LINC01088 could partially reverse the cell proliferation induced by miR-24-1-5p. Finally, we demonstrated that p21 activated kinase 4 (PAK4) was one of the downstream key targets of miR-24-1-5p by luciferase reporter assay and Western blotting; and our results showed a remarkable decrease in cell proliferation after overexpression of PAK4. We conclude that LINC01088 might function as a tumor suppressor, inhibiting the tumorigenesis of ovarian epithelial cells through LINC01088/ miR-24-1-5p/ PAK4 axis.
Similar content being viewed by others
Introduction
Ovarian neoplasm is one of the most common gynecological tumors. The benign ovarian tumors can be cured by excision. Epithelial ovarian cancer is among the most lethal gynecological malignancies, with the five-year survival rate of less than 30%1. Despite constant improvement in surgical techniques and chemotherapy regimens for epithelial ovarian cancers in the past few years, the survival rates have not improved significantly1. Thus, it is essential to decipher the mechanism of ovarian epithelial tumorigenesis, in order to suppress tumor progression efficiently.
Long non-coding RNAs (LncRNAs) and mircoRNAs (miRNAs) are two classes of common non-protein-coding RNAs. LncRNAs are transcripts longer than 200 nucleotides2 that are involved in various biological processes, such as transcription regulation, cell proliferation and differentiation3. Also, lncRNAs play a crucial role in cancer development and progression4,5,6,7. MiRNAs are composed of approximately 18 to 22 nucleotides, and they play a role in biological processes like cell proliferation, apoptosis and cell differentiation8,9. It has been confirmed that aberrant expression of miRNAs is not only closely related to tumor progression, but also to tumorigenesis and neoplasm metastasis10,11. Studies have shown that lncRNAs contain miRNA response elements (MRE), which can bind to their target miRNAs to inhibit their biological functions (molecule “sponges”)12. Amanda et al.13 confirmed that lncRNA-H19 bound to endogenous let-7 by acting as a molecular sponge, which de-repressed the effect of endogenous let-7 on targeting high mobility group A2 (Hmga2) and promoted embryonic carcinoma cell proliferation and invasion. To identify the genes involved in tumorigenesis of ovarian epithelial cells, we conducted the global cDNA gene chip scanning and found that the expression of long intergenic non-coding RNA 1088 (LINC01088) in benign epithelial ovarian tumor tissues was reduced in comparison to normal ovarian epithelial tissues. Further exploration indicated that miR-24-1-5p was a target of LINC01088, which provided experimental support for the role of LINC01088 in effective suppression of the occurrence of epithelial ovarian cancers.
Results
LINC01088 is down-regulated in benign epithelial ovarian tumors
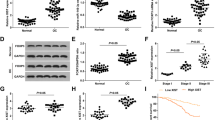
To evaluate the main lincRNAs involved in tumorigenesis of ovarian epithelial cells, we analysed lincRNA expression in 3 benign epithelial ovarian tumor tissue specimens and 3 normal ovarian epithelial tissue specimens using gene chip scanning. Compared with the normal group, the expression levels of the most significant four lincRNAs (LINC01088, LINC01018, LINC01436 and LINC00243) were remarkably down-regulated in the tumor group (Fig. 1A), and the down-regulation of LINC01088 was the most marked. Also, RT-qPCR on the same tissue specimens gave similar results with the gene chip analyses (Fig. 1B). Next, we compared the expression levels of these four lincRNAs in normal ovarian epithelial tissue specimens, and found that the expression level of LINC01088 was the highest (Fig. 1C). Thus, we chose LINC01088 for further study. Moreover, in an experiment with increased sample size, we discovered once again that the expression of LINC01088 was prominently decreased in benign epithelial ovarian tumor tissues compared to the normal tissues. Additionally, LINC01088 expression declined even more in borderline and malignant epithelial ovarian tumors (Fig. 1D). The above results suggested that LINC01088 was a molecule that was negatively associated with tumorigenesis and might be a tumor suppressor gene.
LINC01088 is down-regulated in benign epithelial ovarian tumors. (A) LincRNA screening involved in tumorigensis. LincRNA gene chip was performed in 3 benign epithelial ovarian tumor tissue specimens and the matched normal tissues. (B) Validation of LINC01088. RNA was isolated from the clinical specimens using Trizol. After reverse transcription, real-time qPCR was conducted to determine the LINC01088 level in 3 benign epithelial ovarian tumor tissue specimens and the matched normal tissues. Data are represented as mean +/− SD, *means P < 0.05 vs normal group (ANOVA). (C) The expressive abundance of four lincRNAs. Normalized signals of LINC01088, LINC00243, LINC01436 and LINC01018 in normal ovarian epithelial tissues showed by gene chip above. (D) LINC01088 in primary epithelial ovarian tumors. RNA was isolated from the clinical specimens using Trizol. After reverse transcription, real-time qPCR was conducted to determine the LINC01088 level in 12 benign epithelial ovarian tumor tissue specimens, 8 borderline epithelial ovarian tumor tissue specimens, 12 malignant epithelial ovarian tumor tissue specimens and the matched normal tissues. Data are represented as mean +/− SD, **means P < 0.01 vs normal group (ANOVA).
miR-24-1-5p is a target of LINC01088
To determine the role of LINC01088 in tumorigenesis of ovarian epithelial cells, we constructed LINC01088-lentiviral vectors and transfected them into A2780 cells. RT-qPCR verified that we have establish a cell line highly expressing LINC01088 (Fig. 2A). The tumor growth assay showed that LINC01088 significantly inhibited tumor growth (Fig. 2B), implying that LINC01088 was a tumor suppressor gene, although its mechanism remained unclear. Based on the role of lncRNAs as molecular “sponges”, we speculated that LINC01088 had target miRNAs and served as a molecular “sponge”. Thus, we used the BLAST function on www.ncbi.nlm.nih.gov and found out that miR-24-1-5p had two binding sites for LINC01088 (Fig. 2C). To verify the interaction between LINC01088 and miR-24-1-5p, we conducted RT-qPCR analysis using a total of 22 clinical tissue samples, and found a negative correlation between the expression levels of LINC01088 and miR-24-1-5p (spearman rank correlation was −0.568, P < 0.01) (Fig. 2D,E). Furthermore, we transfected A2780 cells with miR-24-1-5p expressing plasmids labeled with biotin and tested LINC01088 by PCR using RNA sediments “pulled-down” by co-precipitation, and the results proved the existence of LINC01088 (Fig. 2F). For further verification, pMIR-LINC01088 luciferase reporter plasmids were constructed and transfected into A2780 cells. The luciferase activity assay showed that miR-24-1-5p could significantly inhibited firefly luciferase activity (Fig. 2G), indicating that LINC01088 could target miR-24-1-5p. The above experiments demonstrated that LINC01088 might suppress cell growth by targeting miR-24-1-5p.
miR-24-1-5p is a target of LINC01088. (A) Determination of LINC01088 in transfected cells. Total RNA was extracted from A2780 cells stably expressing LINC01088. Real-time qPCR was performed for detecting LINC01088. Data are represented as mean +/− SD, **means P < 0.01 vs control group (ANOVA). (B) Tumorigenicity of LINC01088 in vivo. Female Balb/c nude mice were subcutaneously implanted with A2780 cells transfected with LINC01088-lentiviral vector or blank-lentiviral vector, respectively. 28 days after implantation, the nude mice in each group were sacrificed. The tumor volume was calculated. Data are represented as mean +/−SD, **means P < 0.01 vs control (Repeated Measure ANOVA). (C) Prediction of binding sites between LINC01088 and miR-24-1-5p. Gene Blasting was performed and found two potential binding sites between LINC01088 and miR-24-1-5p. (D,E) Level of miR-24-1-5p in primary epithelial ovarian tumors. RNA was isolated from the clinical specimens using Trizol. After reverse transcription, real-time qPCR was conducted to determine the miR-24-1-5p level in 12 benign epithelial ovarian tumor tissue specimens and the matched normal tissues. Data are represented as mean +/− SD, **means P < 0.01 vs normal group (ANOVA) and showed a negative relation between the expression LINC01088 and miR-24-1-5p (spearman rank correlation was −0.568, P < 0.01). (F) RNA co-precipitation. A2780 cells were transiently transfected with miR-24-1-5p expression plasmids labeled with biotin, PCR was performed to detect LINC01088 after reverse transcription through Oligo dT15. Results showed that there was an interaction between LINC01088 and miR-24-1-5p. (G) Validating the interaction between LINC01088 and miR-24-1-5p. The relative luciferase activity was notably decreased in A2780 cells co-transfected with pcDNA6.2-GW/EmGFP-miR-24-1-5p, PRL-TK and pMIR-LINC01088 reporter plasmids. miR-LacZ was taken as the control. Data are represented as mean +/−SD, **means P < 0.01 vs control (ANOVA).
LINC01088 reduces cell proliferation promoted by miR-24-1-5p
The experiments described above demonstrated the interaction between LINC01088 and miR-24-1-5p. Next, we went on to explore the effect of miR-24-1-5p on the growth of ovarian epithelial cells. Firstly, we assessed the impact of miR-24-1-5p on cell proliferation. Plasmids expressing miR-24-1-5p were transfected into A2780 cells to establish stable cell line expressing miR-24-1-5p. The MTS assay demonstrated that miR-24-1-5p cloud remarkably facilitated cell proliferation, while LINC01088 distinctly inhibited cell proliferation promoted by miR-24-1-5p (Fig. 3A). Secondly, we investigated the role of miR-24-1-5p in cell migration, and the scratch assay showed that miR-24-1-5p had no regulatory effect on cell migration (Fig. 3B). At last, we explored the influence of miR-24-1-5p on tumorigenicity in animals. Balb/c nude mice were subcutaneously implanted with cells stably overexpressing miR-24-1-5p. Four weeks after implantation, it was evident that not only the tumor cell growth was markedly accelerated, but also the tumor volume and weight were significantly increased in the miR-24-1-5p group, compared with those in the control group (Fig. 3C,D). Moreover, we performed in vivo experiment with cells stably overexpressing miR-24-1-5p + LINC01088, the result showed that LINC01088 inhibited tumor growth facilitated by miR-24-1-5p (Fig. 3E). These findings suggested that miR-24-1-5p promoted cell proliferation but did not affect cell migration, hinting that miR-24-1-5p was an oncogene and LINC01088 could reduce cell proliferation promoted by miR-24-1-5p.
LINC01088 reduces cell proliferation promoted by miR-24-1-5p. (A) Determination of proliferation in LINC01088- and miR-24-1-5p- expressing cells. MTS assay was performed among control group, miR-24-1-5p group, miR-24-1-5p + LINC01088 group and LINC01088 group to determine cell proliferation. Data are represented as mean +/− SD, ** means P < 0.01 vs control group (ANOVA). (B) Determination of migration in miR-24-1-5p-expressing cells. Scratch assay was performed to evaluated cell migration. Data are represented as mean +/− SD, P > 0.05 vs control group (ANOVA). (C,D) Tumorigenicity of miR-24-1-5p in mice. Female Balb/c nude mice were subcutaneously implanted with A2780 cells stably transfected with pcDNA6.2-GW/EmGFP-miR-24-1-5p or pcDNA6.2-GW/EmGFP-miR-LacZ controls, respectively. 28 days after implantation, the nude mice in each group were sacrificed. The tumor volume was calculated (C). Data are represented as mean +/−SD, *means P < 0.05, **means P < 0.01 vs control (Repeated Measure ANOVA). The tumor weights were calculated (D). Data are represented as mean +/−SD, *means P < 0.05 vs Control (ANOVA). (E) Tumorigenicity of miR-24-1-5p + LINC01088 in mice. Female Balb/c nude mice were randomly divided into four groups (control, miR-24-1-5p, LINC01088 and miR-24-1-5p + LINC01088). 28 days after implantation, the nude mice in each group were sacrificed. The tumor volume was calculated. Data are represented as mean +/−SD, *means P < 0.05, **means P < 0.01 vs control (Repeated Measure ANOVA).
PAK4 is the potential target of miR-24-1-5p
It had previously been demonstrated that miR-24-1-5p increased in benign epithelial ovarian tumors and promoted cell proliferation, and was also involved in the development of epithelial ovarian tumors. Hence, we studied the target genes downstream of miR-24-1-5p further. Gene ‘BLAST’ on NCBI suggested that PAK4 was the possible target of miR-24-1-5p (Fig. 4A). To test this assumption, firstly we performed a IHC assay on normal ovarian epithelial tissue sections and benign epithelial ovarian tumor tissue sections. The results showed that PAK4 was expressed in both normal ovarian epithelial tissues and benign epithelial ovarian tumor tissues, but PAK4 expression seemed to be lower in benign epithelial ovarian tumor (Fig. 4B). Next, pMIR-PAK4 3′UTR luciferase reporter plasmids were constructed and transfected into A2780 cells. The luciferase activity assay showed that miR-24-1-5p inhibited firefly luciferase activity dramatically (Fig. 4C). Lastly, we detected PAK4 expression in miR-24-1-5p-expressing cells. The western blotting data indicated that PAK4 expression in A2780 cells stably transfected with miR-24-1-5p recombinant plasmids was indeed lessened (Fig. 4D), while LINC01088 had no direct effect on the expression of PAK4 (data not shown). Taken together, these findings demonstrated that PAK4 was the target of miR-24-1-5p, indicating that miR-24-1-5p might participate in the development and progression of epithelial ovarian tumors by targeting PAK4.
PAK4 is the potential target of miR-24-1-5p. (A) Prediction of binding sites between miR-24-1-5p and PAK4. Gene Blasting was performed and two potential binding sites between miR-24-1-5p and PAK4 were found. (B) PAK4 in primary epithelial ovarian tumors. IHC staining was performed to measure PAK4 expression in (ii) normal ovarian epithelial tissue, (iii) benign epithelial ovarian tumor tissue, while (i) was shown as the negative control (100X, bar = 50 um). The higher magnification (400X, bar = 50 um) was shown in the upper right corner, correspondingly. (C) Validating the interaction between miR-24-1-5p and PAK4. The relative luciferase activity was notably decreased in A2780 cells co-transfected with pcDNA6.2-GW/EmGFP-miR-24-1-5p, PRL-TK and pMIR-PAK4 3′UTR reporter plasmids. miR-LacZ was taken as the control. Data are represented as mean +/−SD, **means P < 0.01 vs control (ANOVA). (D) Detection of PAK4 in miR-24-1-5p-expressing cells. Western blot was conducted to examine the protein expression of PAK4 in A2780 cells stably transfected with pcDNA6.2-GW/ EmGFP-miR-24-1-5p or control plasmids (cropped; full length blots can be found in Supplementary Fig. S1).
PAK4 inhibits cell proliferation
To further study the biological function of PAK4 in ovarian epithelial cells, we transfected A2780 cells with plasmids expressing PAK4 (Fig. 5A), and evaluated their proliferation and migration abilities respectively. The data showed that overexpressed PAK4 significantly decreased cell proliferation (Fig. 5B), but promoted cell migration (Fig. 5C), which implied that PAK4 was a cell proliferation-inhibiting protein.
PAK4 inhibits cell proliferation. A2780 cells transfected with pCMV6-Entry-PAK4 or control plasmids for 48 h were harvested to perform the experiments as follows. (A) Detection of PAK4 in stable PAK4-expressing cells. Western blot was conducted to examine the protein expression of PAK4 in two groups (cropped; full length blots can be found in Supplementary Fig. S2). (B) Determination of proliferation. MTS assay was performed to determine cell proliferation. Data are represented as mean +/− SD, ** means P < 0.01 vs control group (ANOVA). (C) Determination of migration. Scratch assay was performed to determine cell migration. Data are represented as mean +/− SD, ** means P < 0.01 vs control group (ANOVA).
Discussion
This study demonstrates that the expression of LINC01088 is lowered in benign epithelial ovarian tumors compared to normal ovarian epithelial tissues, and LINC01088 expression has a negative correlation with that of miR-24-1-5p. The study also proves that LINC01088 can target miR-24-1-5p to regulate the expression of PAK4, and influence cell proliferation and migration. The brief diagrammatic representation is made in the Fig. 6. This implies that LINC01088 might inhibit the development of epithelial ovarian tumors through its interaction with miR-24-1-5p and the downstream effector protein PAK4.
LncRNAs perform their biological function through regulation of gene expression. Their interaction with miRNAs is very important in tumorigenesis and tumor progression14,15,16,17, and lncRNA-mediated sponge regulatory network in miRNAs and pri-miRNAs is one of the modes of action. For example, Kumar et al.18 proved that Hmga2 promoted lung cancer progression by competing for let-7 occupancy with TGF-beta type III receptor (Tgfbr3t) through transforming growth factor-β (TGF-β) signaling pathway. Also, lncRNAGAS5 acted as a tumor suppressor in hepatocellular carcinoma through negative regulation of miR-21 to up-regulate its targets programmed cell death 4 (PDCD4) and phosphatase and tensin homologue (PTEN), resulting in inhibition of cancer cell migration and invasion19. In our study, we demonstrated that LINC01088 targeted miR-24-1-5p to regulate the expression of PAK4, but the exact mechanism of the interaction between LINC01088 and miR-24-1-5p required further exploration.
MiR-24 was initially discovered through a research involving invertebrates and vertebrates by Lagos-Quintana et al.20 in 2001. Has-mir-24 has two forms, mir-24-1 and mir-24-2, located on Chromosome 9 and Chromosome 19 respectively21, which includes three mature sequences, miR-24-1-5p, miR-24-3p and miR-24-2-5p. To date, the action mechanism of miR-24-1-5p in carcinomas remains unclear. As reported in Braoudaki’s retrospective study in patients with ependymoma (EP)22, miR-24-1-5p was up-regulated significantly in relapse and progression cases compared to clinical remission and survival cases. Moreover, miR-24-1-5p was considered as an oncogene associated with multiple endocrine neoplasia type1 (MEN1)23. While Goto et al.24 indicated that miR-24-1-5p could clearly inhibit cell proliferation, migration and metastasis in prostate cancer. Inoguchi et al.25 also verified that miR-24-1-5p inhibited bladder cancer cell proliferation by targeting forkhead box protein M1 (FOXM1). However, the possible roles of miR-24-1-5p in epithelial ovarian tumors haven’t been reported yet. This study confirmed that miR-24-1-5p could promote epithelial ovarian tumors by negatively regulating PAK4 expression. In fact, miRNAs have innumerable target molecules, and regulate every aspect of cellular activity, such as cell proliferation, differentiation, apoptosis and so on26,27,28,29. Furthermore, miRNAs play multiple roles in development and progression of tumors, including angiogenesis, metastasis, and also exosome-mediated regulation30,31,32,33,34. Hence, it can be concluded that the results of our study elucidate only one of the countless signaling pathways involved in various biological processes associated with tumors, therefore further studies are required.
During the course of this study, it was demonstrated that PAK4 was the target of miR-24-1-5p. PAK is a class of evolutionarily conserved serine/threonine protein kinases35, and PAK4 is the earliest and most profoundly studied protein of the group II PAKs. The PAK4 gene is located on chromosome 19 at the 19q13 locus; it is 3064 bp in length and encodes 591 amino acids. Recent studies on PAK4 reveal its significant role in controlling cellular activities. It is considered to be a signaling integrator, regulating numerous fundamental cellular processes, such as actin cytoskeletal dynamics, cell morphology and motility, cell survival, embryonic development, immune defense and oncogenic transformation36,37. Besides, it also plays key roles in gallbladder carcinoma38, pancreatic cancer39,40, colorectal cancer41, lung cancer42 and so on.
Studies show that the activation and expression levels of PAK4 plays significant roles in the development and progression of tumors. It is reported that among the 100 types of cancers affecting humans, 78% have increased expression of PAK443. For example, PAK4 expression is significantly increased in breast cancer and is positively correlated with tumor progression44. This is inconsistent with our results which suggest that cells with increased expression of miR-24-1-5p and decreased expression of PAK4 demonstrate enhanced proliferation. Coarfa et al.45 characterized the proteomic footprint of a panel of 8 miRNAs by using reverse phase protein arrays (RPPA) in prostate cancer; and illustrated that for at most 12% of the proteins, the expression level was determined by direct interaction between miRNAs and target mRNAs, but for the majority of them, various factors were involved. Thus, it can be summarized that the proliferation of ovarian cancer cells is regulated by numerous factors, miR-24-1-5p and PAK4 are only the tip of the iceberg.
Methods
Clinical specimens
42 fresh clinical specimens (10 normal ovarian epithelial tissue; 12 benign, 8 borderline and 12 malignant epithelial ovarian tumor tissue) were obtained in the Department of Gynecology at the Second Affiliated Hospital, College of Medicine, Zhejiang University, between 2013 and 2016. 19 formalin-fixed, paraffin-embedded tissue sections (10 normal ovarian epithelial tissue and 9 benign epithelial ovarian tumor tissue) were provided by the Department of Pathology in the same hospital between 2011 and 2015. All patients involved in this study received neither radiation therapy nor chemotherapy before surgical resection.
Cell lines
The human ovarian cancer cell line A2780 was purchased from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 (Gibco, USA) with 10% fetal bovine serum (FBS) (Invitrogen, USA) and 100 IU/ml gentamycin at 37 °C in a humidified atmosphere with 5% CO2.
The gene chip scanning
3 normal ovarian epithelial tissue and 3 benign epithelial ovarian tumor tissue were used for RNA isolation through AMBION kit (Invitrogen, USA). After passing quality inspection, the total RNA was transcribed into double stranded cDNA, synthesized into cRNA and labeled with Cyanine-3-CTP. The labeled cRNAs were hybridized onto the microarray (Agilent SurePrint G3 Human Gene Expression v3, 8*60 K, Design ID: 072363) and scanned by the Agilent Scanner G2505C (Agilent Technologies, USA). The raw data was normalized using Extraction and GeneSpring. Differentially expressed genes were identified through fold change as well as P values calculated using Student’s-test. The threshold set for up- and down- regulated genes was a fold change ≥ 2.0 and a P value ≤ 0.01. The gene chip scanning was conducted in the laboratory of the OE Biotech Company (Shanghai, China).
Real-time qPCR assay
RNA was isolated from the clinical specimens or cells using Trizol (Invitrogen, USA) and reverse transcription was performed using RNA and primers specific for mature miR-24-1-5p or LINC01088. The reverse transcription of LINC01088 was conducted at 42 °C for 60 min and then 95 °C for 5 min (PrimeScript 1st Strand cDNA Synthesis Kit, TaKaRa, Japan). Real-time qPCR of the reverse transcription products of LINC01088 was performed using Premix Ex Taq (TaKaRa, Japan). The reverse transcription of miR-24-1-5p was conducted at 16 °C for 30 min and then 42 °C for 30 min, inactivated by incubating at 85 °C for 5 min (TaqMan, USA). The reverse transcription products of miR-24-1-5p were mixed with TaqMan universal PCR master mix II (ABI, USA) and the mixture was incubated at 95 °C for 10 min, followed by 40 cycles, with an extension time of 15 s each at 95 °C and 60 s each at 60 °C. The relative LINC01088 expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the relative miR-24-1-5p expression was normalized to miR-484, and analysed by the 2−ΔΔCt method.
RNA co-precipitation
A2780 cells were transiently transfected with 5ug miR-24-1-5p expression plasmids labeled with biotin using Lipofectamine 3000 (Invitrogen, USA) for 48 h. The cells were then lysed in RIPA buffer and centrifuged at 14 800 rpm to remove the precipitate. A total of 30 µl biotin-avidin conjugated agarose was added to the supernatant and mixed via vortexing for 2 h at 4 °C. The mixture was then centrifuged and the supernatant was carefully discarded. 500 µl chloroform was added to the precipitate for nucleic acids extraction after it was washed thrice with RIPA buffer. PCR was performed to detect LINC01088 after reverse transcription using Oligo dT15. miR-155 labeled with biotin was used as negative control and LINC01088 as positive control.
Construction of miR-24-1-5p expression plasmids
The genetic sequence of mature miR-24-1-5p (MIMAT0000079) was identified by using Genbank. The engineered pre-miRNAs were chemically synthesized according to BLOCK-iT™ Pol II miRRNAi Expression Vector Kits (Invitrogen, USA). Top primer:5′-tgctgactgatatcagctcagtaggcagttttggccactgactgactgcctactgctgatatcagt-3′, bottom primer:5′-cctgactgatatcagcagtaggcagtcagtcagtggccaaaactgcctactgagctgatatcagtc-3′. After denaturation at 95 °C for 4 min, 4 μl of 10 nM oligonucleotides were ligated into 2 μl of 5ng/μl linearized pcDNA6.2-GW/EmGFP-miR plasmids (Invitrogen, USA) using T4 DNA ligase. The ligation mixture was transformed into DH5α and selected by 50 μg/ml spectinomycin. Positive colonies were amplified for plasmid extraction and DNA sequencing. Then the correct plasmids were purified to remove endotoxins for subsequent use.
Cell transfection
A2780 cells were planted in a 6-well plate with an inoculum density of 50–60%. 3.75 μl liposomes and 5 μl plasmids with 10 μl P3000 reagent were respectively added into 125 μl OPTI-MEM culture medium (Gibco, USA) according to the instructions of Lipofectamine 3000. They were mixed thoroughly and incubated at room temperature for 5 min. The solution was then added into each well of the 6-well plate and culture for 48 h.
Establishment of stable cell line expressing miR-24-1-5p
A2780 cells transfected with recombinant plasmids of miR-24-1-5p or miR-LacZ for 48 h were harvested for flow cytometry sorting based on green fluorescent protein (GFP) accumulation. Then, the GFP-positive cells were cloned by limiting dilution. After colony formation, blasticidin at 7 μg/ml was used for selecting resistant cells by 14 days of culture. Blasticidin-resistant colonies were picked and expanded for use.
Western blotting
Cells were washed with phosphate-buffered saline (PBS) and lysed in 500 μl RIPA buffer with 5 μl protease inhibitor cocktail (100X, Merck, USA) on ice for 15 min, then centrifugation at 13 000 g for 10 min. The proteins in 20 μl supernatant were separated with 12% SDS-PAGE and transferred onto a nitrocellulose membrane. Then the membrane was incubated with rabbit anti-human primary antibody PAK4 (1:500–1:1000, Abcam, UK), mouse Anti-Flag Tag (1:1000–1:10 000, Proteintech, USA) and mouse anti-human primary antibody GAPDH (1:5000, KangChen Bio-tech Inc. China) at 4 °C overnight, respectively. Next, it was incubated with a horseradish peroxidase-labeled (HRP-labeled) goat anti-rabbit or anti-mouse secondary antibody (1:2000, Jackson Immunotech, UK) at room temperature for 1 h. The membrane was developed using enhanced chemiluminescence (ECL, Millipore, Germany) for testing.
Luciferase reporter gene assay
The intact RNA was extracted using Trizol.LINC01088 (located in Chr4, size 1011 bp) were chemically synthesized. Forward primer: 5′- ctagtccccttgaaggaataggagtagacctgctgaactatcacatgagagaagaggcccaaa-3′, reverse primer: 5′-agcttttgggcctcttctctcatgtgatagttcagcaggtctactcctattccttcaagggga-3′. DNA fragments of the PAK4 3′UTR were chemically synthesized using cDNA as a template. Forward primer: 5′-cagctctactagtccctcaaccaaagagccccc-3′, reverse primer: 5′-cagtgacaagctttgtctccccatccagccaca-3′. The double-stranded oligonucleotides were then ligated into the Spe I/HindIII sites in the pMIR-report Luciferase plasmid (Invitrogen, USA). After DNA sequencing, recombinant pMIR-PAK4 or pMIR-LINC01088 plasmids were co-transfected with pcDNA6.2-GW/EmGFP-miR plasmids containing miR-24-1-5p and PRL-TK plasmids (10:1:0.1) into A2780 cells using Lipofectmine3000 and incubated for 48 h. Then luciferase reporter gene assay was conducted by using the Dual-Luciferase Reporter Assay System (Promega, USA).
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections were deparaffinized, hydrated and soaked in 3% H2O2 at room temperature for 10 min. Then the slides were incubated with rabbit anti-human PAK4 antibody (10 µg/ml, Abcam, UK) overnight at 4 °C in a humidified chamber. The negative control was obtained by omitting the primary antibody. The next day, biotin-labeled sheep anti-rabbit IgG antibody was added to sections and incubated at 37 °C for 10 to 30 min. Then, the HRP streptavidin solution was added and the slides were incubated at 37 °C for 10 to 30 min. Finally, the slides were added with DAB solution, incubated for 5–10 min and counterstained with hematoxylin for 2 min and mounted.
Two pathologists, without access to the clinical data, independently scored the tissue staining. Positive staining was indicated by the presence of brown stain. The PAK4 expression was evaluated based on the intensity of staining. The percentage of positive cells was scored as: “0” (<5%, negative), “1” (5–25%, sporadic), “2” (25–50%, focal), or “3” (>50%, diffuse). The staining intensity was scored as “0” (no staining), “1” (weakly stained), “2” (moderately stained), or “3” (strongly stained). The PAK4 immunostaining score was calculated as the percentage positive score × the staining intensity score46.
MTS assay
Cells in the experimental and control groups were plated at a density of 10 000 cells per well (100 µl) in 96-well plates. After being cultured for 24 h, the cells were incubated with 20 µl MTS (Promega, USA) at 37 °Cfor 4 h. The absorbance was read at 490 nm using a microplate reader.
Scratch assay
Cells in two groups were implanted into 3.5 cm2 culture plates with approximately 90% confluence. Wounds were created with a 200 µl pipette tip and washed with PBS, then serum-free medium was reintroduced at the experiment start time point of 0 hour. The diameter of the scratch was recorded under light microscopy at 24 h.
In vivo tumor growth assay
Female Balb/c nude mice (5–6 weeks old) were randomly divided into groups (control, miR-24-1-5p, LINC01088 and miR-24-1-5p + LINC01088), and each group contained five mice. Cells transfected with LINC01088-, miR-24-1-5p- or LINC01088 + miR-24-1-5p- overexpressing plasmids were injected subcutaneously in the dorsal flank of the nude mice. The mice were observed for tumor formation by measuring the tumor major axis (a) and minor axis (b) every 7 days. The tumor volume was calculated: v = ab2/2. Afterwards the mice were sacrificed by cervical dislocation on the 28th day after injection, the tumors were subsequently recovered and the weight of each tumor was determined.
Data availability
All data generated or analysed during this study are included in this published article.
Ethical statement
We solemnly stated that all methods were performed in accordance with the relevant guidelines and regulations. All human and animal studies have been approved by the Ethics Committee of the Second Affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent was obtained from each patient prior to inclusion in the study.
References
Wang, Y., Jin, X., Song, H. & Meng, F. AEG-1 as a predictor of sensitivity to neoadjuvant chemotherapy in advanced epithelial ovarian cancer. Onco Targets Ther 9, 2385–2392 (2016).
Zhang, H. & Zhu, J. K. Emerging roles of RNA processing factors in regulating long non-coding RNAs. RNA Biol 11, 793–797 (2014).
Tsai, M. C., Spitale, R. C. & Chang, H. Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res 71, 3–7 (2011).
Gutschner, T. & Diederichs, S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9, 703–719 (2012).
Li, J. et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem 37, 687–696 (2015).
Kladi-Skandali, A., Michaelidou, K., Scorilas, A. & Mavridis, K. Long Noncoding RNAs in Digestive System Malignancies: A Novel Class of Cancer Biomarkers and Therapeutic Targets? Gastroenterol Res Pract 2015, 319861 (2015).
Lin, R., Maeda, S., Liu, C., Karin, M. & Edgington, T. S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26, 851–858 (2007).
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 (2003).
Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 (2004).
Kinose, Y., Sawada, K., Nakamura, K. & Kimura, T. The role of microRNAs in ovarian cancer. Biomed Res Int 2014, 249393 (2014).
Iorio, M. V. et al. MicroRNA signatures in human ovarian cancer. Cancer Res 67, 8699–8707 (2007).
Sen, R., Ghosal, S., Das, S., Balti, S. & Chakrabarti, J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorld Journal 2014, 896206 (2014).
Kallen, A. N. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52, 101–112 (2013).
Han, Y. et al. Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long noncoding RNA MALAT1. FEBS Lett (2013).
Xia, T. et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep 5, 13445 (2015).
Ke, J. et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 6, 21934–21949 (2015).
Tsang, F. H. et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int 35, 1597–1606 (2015).
Kumar, M. S. et al. & Downward, J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 505, 212–217 (2014).
Hu, L. et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol 37, 2691–2702 (2016).
Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001).
Sun, Q. et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res 36, 2690–2699 (2008).
Braoudaki, M. et al. miR-15a and miR-24-1 as putative prognostic microRNA signatures for pediatric pilocytic astrocytomas and ependymomas. Tumour Biol 37, 9887–9897 (2016).
Luzi, E. et al. The negative feedback-loop between the oncomir Mir-24-1 and menin modulates the Men1 tumorigenesis by mimicking the “Knudson’s second hit”. PLoS One 7, e39767 (2012).
Goto, Y. et al. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget 5, 7748–7759 (2014).
Inoguchi, S. et al. Tumour-suppressive microRNA-24-1 inhibits cancer cell proliferation through targeting FOXM1 in bladder cancer. FEBS Lett 588, 3170–3179 (2014).
Cheng, L. C., Tavazoie, M. & Doetsch, F. Stem cells: from epigenetics to microRNAs. Neuron 46, 363–367 (2005).
Reinhart, B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000).
Suh, M. R. et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270, 488–498 (2004).
Cimmino, A. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102, 13944–13949 (2005).
Falcone, G., Felsani, A. & D’Agnano, I. Signaling by exosomal microRNAs in cancer. J Exp Clin Cancer Res 34, 32 (2015).
Kuninty, P. R., Schnittert, J., Storm, G. & Prakash, J. MicroRNA Targeting to Modulate Tumor Microenvironment. Front Oncol 6, 3 (2016).
Zhang, Y., Yang, P. & Wang, X. F. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol 24, 153–160 (2014).
Kohlhapp, F. J., Mitra, A. K., Lengyel, E. & Peter, M. E. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 34, 5857–5868 (2015).
Cortez, M. A. et al. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol 8, 467–477 (2011).
Wells, C. M. & Jones, G. E. The emerging importance of group II PAKs. Biochem J 425, 465–473 (2010).
Dart, A. E. & Wells, C. M. P21-activated kinase 4–not just one of the PAK. Eur J Cell Biol 92, 129–138 (2013).
Li, Z. et al. p21-activated kinase 4 phosphorylation of integrin beta5 Ser-759 and Ser-762 regulates cell migration. J Biol Chem 285, 23699–23710 (2010).
Kim, J. H. et al. Gene expression profiles in gallbladder cancer: the close genetic similarity seen for early and advanced gallbladder cancers may explain the poor prognosis. Tumour Biol 29, 41–49 (2008).
Tyagi, N. et al. p-21 activated kinase 4 promotes proliferation and survival of pancreatic cancer cells through AKT- and ERK-dependent activation of NF-kappaB pathway. Oncotarget 5, 8778–8789 (2014).
Chen, S. et al. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther 7, 1793–1802 (2008).
Parsons, D. W. et al. Colorectal cancer: mutations in a signalling pathway. Nature 436, 792 (2005).
Callow, M. G. et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem 277, 550–558 (2002).
Shibata, T., Niinobu, T., Ogata, N. & Takami, M. Microwave coagulation therapy for multiple hepatic metastases from colorectal carcinoma. Cancer 89, 276–284 (2000).
Bi, Y. et al. Study on the expression of PAK4 and P54 protein in breast cancer. World J Surg Oncol 14, 160 (2016).
Coarfa, C. et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 35, 2345–2356 (2016).
Chen, C. et al. Expression of Lysine-specific demethylase 1 in human epithelial ovarian cancer. J Ovarian Res 8, 28 (2015).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81371881) and the Zhejiang Provincial Natural Science Foundation of China (LY13H160018). The authors wish to thank Professor Xiaoming Zhang for his help with the preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.W.Z. and W.G.L. designed the research. W.J.Z. and J.F. carried out the experiments and drafted the manuscript. S.Q.Y. and J.Y.S. participated in the coordination of the experiments. X.Q.Z. and A.S. analysed the data. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, W., Fei, J., Yu, S. et al. LINC01088 inhibits tumorigenesis of ovarian epithelial cells by targeting miR-24-1-5p. Sci Rep 8, 2876 (2018). https://doi.org/10.1038/s41598-018-21164-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-21164-9
This article is cited by
-
MiR-24-1-5p Hinders Malignant Phenotypes of Clear Cell Renal Cell Carcinoma by Targeting SHOX2
Biochemical Genetics (2023)
-
Upregulated LINC01088 facilitates malignant phenotypes and immune escape of colorectal cancer by regulating microRNAs/G3BP1/PD-L1 axis
Journal of Cancer Research and Clinical Oncology (2022)
-
CXCL14 facilitates the growth and metastasis of ovarian carcinoma cells via activation of the Wnt/β-catenin signaling pathway
Journal of Ovarian Research (2021)
-
LncRNA IGBP1-AS1/miR-24-1/ZIC3 loop regulates the proliferation and invasion ability in breast cancer
Cancer Cell International (2020)
-
ZEB1-AS1 initiates a miRNA-mediated ceRNA network to facilitate gastric cancer progression
Cancer Cell International (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.