Abstract
Rice, a major food crop, is grown in a wide range of ecological conditions and suffers significant yield losses as it is constantly exposed to a wide range of environmental and biotic stresses. The prevalence of different biotypes/strains has necessitated assembling of numerous resistance genes/QTLs into elite genotypes to confer a broader scale of resistance. The current study reports successful pyramiding of genes/QTLs that confer tolerance/resistance to submergence (Sub1), salinity (Saltol), blast (Pi2, Pi9) and gall midge (Gm1, Gm4) to supplement the four bacterial blight resistance genes (Xa 4, xa5, xa13, Xa21) present in Improved Tapaswini, an elite cultivar. The precise transfer of genes/QTLs was accomplished through effective foreground selection and suitable gene pyramids were identified. Background selection was practiced using morphological and grain quality traits to enhance the recovery of the recurrent parental genome. In the bioassays, the pyramids exhibited higher levels of resistance/ tolerance against the target stresses. The novel feature of the study was successful pyramidization and demonstration of the function of ten genes/QTLs in a new genotype. This success can stimulate several such studies to realize the full potential of molecular plant breeding as the foundation for rice improvement.
Similar content being viewed by others
Introduction
Rice (Oryza sativa L.), is the primary source of food for more than half of the world’s population1 and is being cultivated under a wide range of environments, from arid highlands to flooded lowlands, high humidity to high temperature climates and also in acid, saline and alkaline soils. The rice plant growth and productivity, exposed to a wide range of environmental stresses such as submergence and salinity, suffer significant crop losses globally year after year. A range of biotic factors, which can be insect pests such as rice gall midge or diseases like bacterial leaf blight and blast, severely affect rice grown throughout tropical, subtropical and temperate areas of Asia.
Bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the oldest and most devastating diseases of rice throughout the world2 and significant yield losses up to 80–100% from bacterial blight infection were known to occur3 and when infection occurs during panicle initiation or earlier, the grain development will also be severely affected. As chemical control of the disease is not effective and reliable, genetic enhancement of host plant resistance is a viable and practical option against bacterial blight4.
In rice, the genetics of resistance to the pathogen has been well characterized and till date, thirty nine BB resistance genes, series from Xa1 to Xa39 have been identified from diverse sources5,6 and of these, nine genes (Xa1, Xa3/Xa26, xa5, xa13, Xa10, Xa21, Xa23, xa25 and Xa27) have been isolated and characterized7,8 while seven genes (Xa4, Xa7, Xa22, Xa30, Xa31, Xa33, and xa34) have been fine-mapped9.
Breakdown of resistance in varieties having a single resistance gene has been reported in rice10,11,12 after 2 or 3 years as a result of shifts in the frequency of pathotypes or the emergence of new ones through mutation or other mechanisms. Multiple resistance genes confer durable broad spectrum resistance through synergistic and complementary gene action to a wide range of races compared to one, two and three gene combinations13,14,15.
Among the several fungal diseases infecting the rice plant, the rice blast disease, caused by an ascomycete fungus Magnaporthe grisea Barr. (Telomorph Pyricularia oryzae Sacc), is one of the major destructive disease on rice. Also known as rice fever disease, it was reported in all the rice growing countries of the globe16,17,18 and it infects panicles, culm and leaves of the rice plants, consequently reducing photosynthetic efficiency and yield of rice grain19,20 and causes 40–70% loss of rice grain21,22. For management of blast disease, use of chemical pesticides is not considered as an ecologically sound approach and fortification of host plant was a highly reasonable method22.
So far, more than 100 blast resistance (R) genes and 350 QTLs associated with resistance to blast have been reported. Between them, 26 blast resistance genes (Pib, Pita, Pi54, Pid2, Pi9, Pi2, Pizt, Pi36, Pi37, Pikm, Pi5, Pit, Pid3, pi21, Pish, Pb1, Pik, Pikp, Pia, Pi25, Pid3A4, Pi35, NLS1, Pikh, Pi54 and Pi54rh) have been cloned and functionally authenticated22,23,24,25,26,27,28,29,30. Stacking blast resistance genes into a single cultivar has been recommended approach to broaden the resistance spectrum of genotypes as well as to achieve strong resistance17,22,31.
Asian rice gall midge (Orseolia oryzae), a major insect pest of rice occur mainly in wet season, in specific provinces of central, south, and east India, resulting major yield loss, mostly throughout the kharif season. Seven biotypes of the gall midge have been reported on rice so far32. Genetic studies have identified 11 major genes conferring resistance to gall midge33,34. Of these, eight genes (Gm1, Gm2, Gm4, Gm5, Gm6, Gm7, Gm8 and Gm11) have been tagged and mapped34,35.
In the rainfed lowlands of South and South East Asia, submergence stress of rice is a foremost frequent problem for rice farmers36. Flooding is a foremost natural tragedy that has a disadvantageous consequence on growth and health of plant in natural and agricultural environment37,38,39. In submergence stress and later desubmergence, rice plants use to confront several external challenges sequentially, which produce diverse inner stress which influence the growth and survival of rice plant. In submergence stress, it significantly reduces the gas diffusion rate, restricts uptake of oxygen and ineffective anaerobic metabolism40. Muddy floodwaters decreases light accessibility and slows down underwater photosynthesis. Constraint of efficient gas exchange as well highly limits rate of transpiration41. Under long-standing submergence stress, due to these conditions rice plants gets energy hunger and deficiency of nutrient. As a result of which the plant starts decaying and dying39. The yield loss can reach nearly 70% due to submergence alone42. The average yield loss from submergence is estimated about 80 kg/ha43.
Submergence tolerance is an important trait for rice in rainfed lowland conditions and is being evaluated as a weed control strategy for rice seeded directly into standing water. The indica cultivar FR13A, is well known for its tolerance to submergence which can withstand up to 14 days of total submergence owing to submergence1 (Sub1), a major quantitative trait locus mapped close to the centromere of chromosome 915,44,45,46,47,48. Around eight Sub1 varieties were developed and in Asia through incorporation of Sub1 from FR13A into elite varieties.
The most extensive soil problem in the world is salinity and it is estimated that over 150 million hectares of potential rice land is affected by salinity49 and is considered as one of important physical factor influencing rice production. Salt tolerance has evolved and there are naturally occurring salt tolerant trees (mangroves), shrubs, grasses and herbs and in general, none of the crop plants are able to tolerate even a quarter of seawater without yield loss. Rice plant is classified as a salt sensitive crop50 and it is comparatively tolerant of salt stress during germination, active tillering and towards maturity and is sensitive during early seedling and reproductive stages51.
Tolerance to salinity is complex, involving a number of different physiological mechanisms, such as sodium exclusion from roots, controlled sodium transport between root and shoot, and sequestering of sodium in older tissues and in the vacuoles. A few traditional indica rice varieties such as Nona Bokra, Pokkali and Kala-rata are salinity stress tolerant rice varieties52. Even though several genes likely to contribute salinity stress tolerance, but the most important QTL for salinity tolerance designated as Saltol, present on the short arm of chromosome 153, confers seedling stage tolerance. The Saltol QTL was transferred into elite IRRI breeding lines and a breeding line FL478, contains a 1 Mb DNA fragment (10.6–11.5 Mb on chromosome1) that contains the entire Saltol locus of Pokkali and the fragment was flanked by IR29 alleles and the breeding line displays high levels of tolerance to salinity54.
Incidences of different biotypes/strains/stresses have necessitated assembling of numerous resistance/tolerance genes into background of high yielding cultivars to confer a broad scale of resistance. Enhancement of broad spectrum resistance/tolerance capabilities of a genotype can be achieved with precision through introgression of additional resistance genes/QTLs using marker-assisted backcrossing (MAB) approach55 and the same cannot be accomplished through conventional approaches even with many generations of backcrosses56. With the availability of tightly linked PCR based (STS) markers for target genes and a large number of markers for background selection57, the pyramiding of the target genes is possible in minimum number of backcrosses effectively with significant savings in time, space, labor and money. Successful transfer of genes/QTLs into agronomically superior cultivars has been reported earlier in rice but all the reports deals with pyramiding of genes/QTLs of one or two traits only. In this study, an attempt was made to pyramid ten genes/QTLs in Improved Tapaswini to fortify the host plant defenses against several biotic and abiotic stresses simultaneously.
Results
Parental polymorphism survey
Parental polymorphism survey revealed distinct polymorphism for the markers tested for the traits to be introgressed into Improved Tapaswini i.e. RG64 (C1O1A51-Pi2), P28 (WHD-1S-75-1-127- Pi9), RM444 (Kavya-Gm1), RM547 (Abhaya-Gm4), SUB1BC2 (FR13A-Sub1QTL) and RM10745 and (FL478-Saltol QTL) (Fig. 1 and Table 1). In the bioassays conducted against GM, blast and screenings against submergence and salinity, the respective donors showed high levels of resistance/tolerance for the stresses while Improved Tapaswini was susceptible for all the stresses tested. However, in the bioassays conducted against eight isolates of Xoo, Improved Tapaswini, the recurrent parent expressed high levels of resistance and the resistance levels are similar to the level expressed by IRBB 60, the line used as donor in developing Improved Tapaswini and both exhibited very short lesion lengths (data not shown).
Parental polymorphism survey through PCR assays (cropped gels displayed). (A) Amplification with RG64 (digested with HaeIII) linked to Pi2. (B) Amplification with P28 linked to Pi9. (C) Amplification with RM444 linked to Gm1. (D) Amplification with RM547 linked to Gm4. (E) Amplification with SUB1BC2 linked to Sub1. (F) Amplification with RM10745 linked to Saltol. Arrows indicates amplicons linked to resistance and susceptible alleles.
Pyramiding of target genes/QTLs
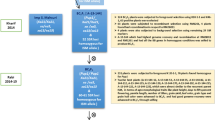
The conventional backcross breeding method was practiced to transfer six new genes/QTLs to complement the four resistance genes present in Improved Tapaswini. In the first cycle, the recurrent parent was crossed with all the six donor parents separately and each of the F1s was backcrossed to the recurrent parent and each population was advanced till BC3F1 generation (Fig. 2). At each generation, marker assisted selection was carried out with the respective markers and plants having resistant alleles were selected. From BC1F1 onwards, stringent selection was also practiced for selecting plants similar to the recurrent parent in morphology along with forward selection for resistant alleles and only plants with desired morphology and alleles were advanced. At the end of the first cycle, six different back cross derivatives each having a desired allele of the target (six) trait(s) were generated. These six recombinants were used to develop three F1 hybrids, using two pyramids in each cross and both forward and background (morphology) selection was employed in the hybrids generated as described earlier. At the end of the second cycle, three recombinants, each with two resistance alleles of target genes/QTLs were developed. In the third cycle, the three recombinants were inter crossed to generate two different F1 hybrids as shown in Fig. 2 and after ensuring their hybrid nature, these two F1s were crossed again to generate a hybrid in the fourth cycle to pool all the genes/QTLs. A large number of hybrid plants were generated and from the population generated, after forward selection, two hundred and seventy plants (having resistance alleles of 4, 5, 6, 7, 8, 9, 10 genes/QTLs) and close similarity to recurrent parent in morphology were selected and carried forward (Fig. 2). From a large BC3F2 population, 250 lines having the resistance alleles in homozygous state were selected. From these lines, 32 lines with different gene combination were selected for different bioassays and characterization for morphological and quality traits in BC3F3 generation. Ten promising lines (Fig. 3) were selected for further experiments. Further refinement of gene pyramids is continuing.
PCR analysis of gene pyramids for the presence target allele(s) (cropped gels displayed). 1. DNA amplification of Gm1 alleles using the primer RM444. 2. DNA amplification of Gm4 alleles using the primer RM547. 3. DNA amplification of Sub1 allele using the primer SUB1BC2. 4. DNA amplification of Saltol alleles using the primer RM10745. 5. DNA amplification of Pi2 alleles using the primer RG64 and digested with HaeIII. 6. DNA amplification of Pi9 alleles using the primer P28. 7. DNA amplification of Xa21 alleles using the primer pTA248. 8. DNA amplification of xa13 alleles using the primer xa13Prom. 9. DNA amplification of xa5 alleles using the primer RG556 and digested with DraI. M = Marker. For Gm1, Gm4, Sub1 and Saltol the M.W. marker was100 bp ladder and for Pi2, Pi9, Xa21, xa13 and xa5 the M.W. marker was 1 kb ladder. S = Recurrent parent (susceptible) R = Resistant (donor) parent. The numbers (1–10) represent different gene pyramids. 1 = ITGP1, 2 = ITGP2, 3 = ITGP4, 4 = ITGP5, 5 = ITGP7, 6 = ITGP9, 7 = ITGP10, 8 = ITGP14, 9 = ITGP18, 10 = ITGP20.
One important observation was the presence of interaction between the BB genes. Of the different combinations possible, Xa4 in all combinations was recovered, while the combinations with xa5 with the other two genes i.e. xa5 + xa13 and xa5 + Xa21 were not recovered in the populations. Since the four gene (Xa4 + xa5 + xa13 + Xa21) combination was observed to confer high levels of resistance, selection was practiced for the combination in all generations and lines homozygous for all the R alleles of all four genes were selected.
Bioassays
Bacterial blight
In the bioassays against bacterial blight, the presence of high levels of resistance was confirmed in Improved Tapaswini, the recurrent parent. Among the tested pyramided lines, the lesion length was in the range of 0.70–2.50 cm while on IRBB 60, the resistant control, the length was ~3 cm (Fig. 4).
Rice gall midge
The bioassays against gall midge were conducted employing Kavya (Gm1) and Abhaya (Gm4), the donors as resistant controls and T(N)1 and Improved Tapaswini as susceptible controls. The donors exhibited high levels of resistance and silver shoot formation was not observed in them. In susceptible controls [T(N)1; Improved Tapaswini], the incidence of galls/silver shoot was 100% suggesting complete susceptibility. Out of the ten gene pyramids (Fig. 3), ITGP7 (IT + Gm1 + Gm4) showed high levels of resistance similar to resistant controls while in four gene pyramids (ITGP1, ITGP2, ITGP4 and ITGP5), the resistance levels are in the range of 66.6–90.9%. In five gene pyramids, the positive plants were below 30% (Fig. 5).
Blast
The screening against blast was carried out with C1O1A51 and WHD-1S-75-1-127 (donors) as resistant controls, and NSN1, HR12 and Improved Tapaswini as susceptible controls. In the bioassays, the resistant controls showed high levels of resistance and exhibited a resistant reaction (R) with a score ‘0’ whereas the recurrent parent exhibited a score of 4 similar to susceptible controls suggesting susceptible nature (S). The gene pyramids ITGP2 and ITGP5 (IT + Pi2 + Pi9) exhibited a high degree of resistant reaction (R) with a score of 0 while ITGP1 and ITGP4 exhibited a moderately resistant reaction (MR) with a score 3. It is interesting to observe that ITGP8, despite having only Pi2 also exhibited a moderately resistant reaction (MR) with a score 3. Five lines that were without any of the blast genes, exhibited a susceptible reaction with a score 4 (Table 2).
Submergence
The screening against submergence was conducted with FR13A, the donor as the resistant control, and IR42 and Improved Tapaswini as susceptible controls. The resistant control FR13A, showed 100% regeneration after 15 days of submergence stress and eight days of desubmergence while both the recurrent parent and IR42 completely died after the submergence stress. All the ten selected pyramid lines having Sub1 QTL showed >83% survival. One gene pyramid, ITGP10, exhibited complete survival (100%) similar to resistant control while six pyramid lines showed >91% survival (Fig. 6).
Salinity
For screening against salinity, FL478 (donor) and SR26B were employed as tolerant controls and IR29 and the recurrent parent as susceptible controls. All the seedling stage gene pyramids with Saltol QTL showed high degree of tolerance to salinity stress with most of them exhibiting >86% survival. The gene pyramid ITGP4, exhibited complete survival with a score 1, a reaction similar to FL478, the resistant control. Two gene pyramids showed less than 80% survival rate, while the susceptible controls, IR29 and Improved Tapaswini, showed only 6 and 8.8% survival respectively (Fig. 7). Our salinity bioassay result implied that the positive gene pyramids showed seedling stage salinity tolerance.
Agronomic performance and grain quality
From the thirty two lines with different gene combinations identified, ten promising lines (Fig. 3), all four gene combinations, were selected and evaluated under field condition for their agronomic performance and produce was used for grain quality studies.
The results revealed that majority of the selections were close to the recurring parent (Table 3) with respect to yield components. Significant variation was not observed for traits like days to 50% flowering (except ITGP2, 20), plant height and number of ear bearing tillers (except ITGP10). The plant height of the Improved Tapaswini gene pyramids was in the range of 95 cm (ITGP4) to 104 cm (ITGP2) while the parental value was 99.0 cm. In case of panicle length, the range observed in the gene pyramids was 23.0 cm (ITGP1) to ITGP20 (29 cm) and the minimum value is closer to the parental value (23.5 cm). All the gene pyramids except ITGP1, had longer panicles than the parent. In general, the values of leaf traits i.e. length, width and the thousand grain weight are on the higher side in the gene pyramids. The line yields of all the gene pyramids were lower than the recurrent parent. The results on the grain quality suggest that some gene pyramids were better than the recurrent parent for important quality traits like kernel length and KLAC and all pyramid lines showed a desirable KALC value, which is identical to Improved Tapaswini. When the grain and quality traits were compared, five pyramids and the parent had similar HRR value. All gene pyramids had amylose content in the range of 25.04 to 22.79% (intermediate range) and the values are close to the value of Improved Tapaswini (24.52%) (Table 4).
Of the 600 SSR markers screened, 44 markers showed polymorphism (Supplementary Table 1). From the 32 lines available at the BC3F3 generation, based on the SSR background selection, 10 gene pyramids having closer similarity to the recurrent parent were selected. In the ten gene pyramids and the recurrent parent, a total of 69 amplicons were amplified with the number of alleles were in the range of 1–3 and the mean PIC value of all markers was 0.20 with a highest value of 0.87 (RM5711) and a lowest value of 0.07 (RM1278) (Supplementary Table 1).
The gene combinations of the selected gene pyramids are: Four gene pyramids (ITGP1, ITGP2, ITGP4, ITGP5) with all ten target genes/QTLs (Xa4 + xa5 + xa13 + Xa21 + Gm1 + Gm4 + Pi2 + Pi9 + Sub1 + Saltol), one (ITGP 7) with 8 genes/QTLs (Xa4 + xa5 + xa13 + Xa21 + Gm1 + Gm4 + Sub1 + Saltol), one (ITGP 8) with 7 genes/QTLs (Xa4 + xa5 + xa13 + Xa21 + Pi2 + Sub1 + Saltol) and four (ITGP9, ITGP1, ITGP14, ITGP20) with 6 genes/QTLs (Xa4 + xa5 + xa13 + Xa21 + Sub1 + Saltol).
Background profiling with morphological and quality data
Using combined values of agronomic and quality traits, genetic distances between the 10 pyramid lines and the parent was calculated between each pair of observations. The average distance of recurring parent to all pyramid lines was 0.43. The result of pair wise comparisons indicated that ITGP 5 and ITGP10 were closer to IT parent with 0.28 distance. ITGP1, ITGP7 and ITGP4 were closer to parent with a distance of 0.34, 0.32 and 0.39 respectively while ITGP2, ITGP8, ITGP9 and ITGP20 were related to the parent with 0.53, 0.48, 0.47 and 0.49 distances respectively.
In the dendrogram generated based on a combination of morphological and quality traits, the eleven entries (10 pyramids + RP) were grouped into three major clusters with cluster I having only one entry (ITGP9). The cluster II consists of ITGP 5 and RP while cluster III was further divided into two sub clusters, Cluster III-A and III-B with Cluster III-A having ITGP 1 while cluster III-B had seven pyramids (Fig. 8). The analysis suggest that ITGP 5 was closer to the recurrent parent while all other pyramids except ITGP 9 are closer to the parent at various levels.
Discussion
In the beginning of our study, clear polymorphism was observed with all the markers linked to the genes/QTLs (Table 1) between the Improved Tapaswini (recurrent parent) and the donors (Fig. 1). The novel feature of the study was the stacking and successful demonstration of the function of ten different genes/QTLs that govern the resistance/tolerance of five different stresses into an elite variety (Fig. 2) a first report in conventional breeding. Of these, two genes having broad spectrum resistance against blast (Pi 9 from O. minuta) and bacterial blight (Pi2 from O. longistaminata) had their origin from wild relatives. The two QTLs that confer tolerance to submergence (Sub1) and salinity (Saltol) are well known for their function. While the desired alleles of genes/QTLs was accomplished through effective foreground selection over ten generations using appropriate co-dominant markers, maximum recovery of the recurrent parental genome was attempted using both conventional (agronomic, grain quality) and using SSR markers. Out of the thirty two lines, ten selected promising lines with the presence and absence of targeted genes/QTLs with different gene combinations (Fig. 3), were evaluated in further studies.
Development of broad-spectrum resistance against bacterial blight in India is a great challenge due to the presence of a number of genetically distinct virulent Xoo strains in different geographical locations13. As probability of effectiveness of a combination of two or more genes is much higher than a single gene to defeat simultaneous pathogen mutations for virulence58, assemblage of several resistance genes into the host plant is a viable and practical strategy.
In the bioassays, all the gene pyramids, having all the BB resistance genes, displayed high levels of resistance against BB (Fig. 4) comparable to resistance shown by Improved Tapaswini, the recurrent parent. Three pyramids i.e. ITGP4, ITGP7 and ITGP8 displayed high levels of resistance with less than 0.9 cm lesion lengths, a result similar to Improved Tapaswini, the recurrent parent (Fig. 4) against BB and the results are in agreement with earlier reports13,14,15,59,60,61.
Though Xa21 gene was reported to provide broad spectrum resistance against BB races that are spread over Africa and Asia62,63, in the study, selection was practiced to stack all the four genes (Xa4, xa5, xa13 and Xa21) in the gene pyramids as the combination of Xa4 + xa 5 + xa 13 + Xa21 was superior to all other gene combinations of 1,2,3 genes13,14,64. In contrast, Tapaswini, the original parent of Improved Tapaswini, was highly susceptible to BB despite the presence of Xa 4 gene suggesting its breakdown. The enhanced level of resistance conferred by more than one gene against a single pathogen has been described as quantitative complementation or synergistic action65.
Breakdown of resistance is well-known against blast disease and pyramiding of multiple resistance genes into a single genetic background is considered as a practical strategy to prevent/delay the breakdown of resistance of pathogens66. Since Pi2 + Pi9 combination was considered ideal, programs were taken up at CRRI to address the problem of blast in upland varieties66,67,68.
In the bioassays conducted at Central Rainfed Rice Research Station, Hazaribagh, Jharkhand, India, a hot spot for blast disease, ITGP2 and ITGP5 showed a highly resistant reaction with the score ‘0’, while ITGP1 and ITGP4, despite having Pi2 + Pi9, showed moderately resistant reaction with the score ‘3’. The other line, ITGP8, having only Pi2 also showed a moderately resistant (MR) reaction with the score ‘3’. The parent and the susceptible genotype showed a score of ‘4’ while both the donors (Pi2 or Pi9) showed a highly resistant reaction. It was evident from the results that the gene combination Pi2 + Pi9 were more effective as they showed resistant reaction (Table 2). In the present study, two lines (ITGP1; ITGP4) showed a moderate reaction even with the two gene combination. This deviation could be a result of either admixtures or errors in sowing as these tests are conducted in screening nurseries at seedling stage and all the entries are surrounded by susceptible checks. This explanation is reasonable in the context as ITGP8, a line having only Pi2 showed moderate reaction. Further studies are on to resolve the deviation exhibited by these two gene pyramids.
In the bioassays against gall midge, the gene pyramids, carrying both the gall midge resistance genes Gm1 and Gm4, displayed complete resistance against gall midge. The gene pyramids such as ITGP2, ITGP4 and ITGP5 showed high degree of resistance with 80%, 90.9% and 86.6% positive plants respectively and in ITGP7, high degree of resistance was observed with 100% positive plants while in ITGP1, the survival was 66.6%. The other gene pyramids, without any of the gall midge resistance genes, as expected, showed complete susceptibility similar to recurrent parent and susceptible control T(N)1 (Fig. 5). The current result suggest the effectiveness of this gene combination against gall midge, a devastating insect pest on rice.
Ensuring food security in the context of climate change is the foremost challenge and improvement of rice varieties that can resist adverse conditions such as submergence and salinity is a priority. With a wide range of changes happening with El Nino effect, ensuring stable production and productivity of rice, the crop of millions of poor farmers of Asia, is a major concern as millions of hectares of monsoon based rice crop is severely affected by submergence or salinity or both, year after year. Though several genes and quantitative traits loci (QTL) linked with tolerance to salinity and submergence were reported, the most prominent and effective are Sub 1 for submergence and Saltol for salinity. Till date, only FR13A, a submergence tolerant landrace has been extensively exploited in breeding69. FR13A, follows a quiescence strategy (i.e., the low-oxygen quiescence syndrome70) in which shoot elongation is suppressed to preserve carbohydrates for a long period (10–14 days) under flash-flood conditions and can restart its growth during de submergence by using preserved carbohydrates. The Sub1 QTL positioned on chromosome 9 rice was eminent as a leading gene conferring submergence tolerance in FR13A and with the development of Swarna Sub 1, through transfer of Sub1 from FR13A into Swarna, the efficacy of the Sub1 QTL was well established71. The effectiveness of Sub1 QTL was also evident in the present study, as all the selected gene pyramids having Sub1 QTL, displayed high degree of tolerance to submergence similar to FR13A with high survival rates [83.3% (ITGP1) to 100% (ITGP10)] (Fig. 6). Sub1BC2, an Indel marker, located between Sub1B and Sub1C genes, is closely linked with Sub1A gene, is being widely used as the marker for Sub 1A into elite cultivars72 and the variations observed in the gene pyramids in its expression needs further studies. However, with the use of gene specific markers like GNS2 and AEX, further improvements are possible.
Rice is reported to be sensitive at both vegetative and reproductive stages to salinity73 and Salt tolerance is a complex quantitative genetic character controlled by many genes74,75. Even though several genes are known to contribute salt tolerance, Saltol, a major QTL located on chromosome1, present in Pokkali53, confers tolerance in rice at vegetative stage through control of Na-K absorption ratio. Though Pokkali is the most widely used donor for salt tolerance, FL478, a derivative from IR29/Pokkali cross, has become a novel source of salinity tolerance because it possessed higher salinity tolerance than Pokkali and possessed many desirable traits like plant type, grain quality76.
All the seedling stage gene pyramids having Saltol QTL survived the saline stress showing varying degrees of tolerance. The gene pyramid ITGP4 showed complete survival after stress, a result similar to tolerant control (FL478) while other gene pyramids showed >86% (87–95%) survival except ITGP10 and ITGP20 which recorded 77% survival (Fig. 7). The varying degrees of tolerance expressed by different gene pyramids could be due the combination of different genes/QTLs (other than Saltol) that govern tolerance to salinity present in different pyramids. Further refinement of recombinants is in progress.
One of the observations in the study was the interaction between the stacked genes. Of the gene combinations possible with the four genes of BB, two combinations i.e. Xa4 + xa5 + xa13 and Xa4 + xa5 + Xa21 were not recovered and both the combinations involve xa5. Similar observation was made with the same genotype in our earlier study that is related to the development of Improved Tapaswini13. In our studies with IR 64, structural changes were observed in case of Xa21 gene in gene pyramids (Das, Personal communication) while such structural changes were not recorded in the present study though large populations were screened in each generation. Reports of interaction between introgressed gene and reporter sequences having homology are available in transgenic rice77. The non-recovery of some combinations appears to be genotype specific as we could recover all the gene combinations with the same set of genes14,78 in our studies with other genotypes while non-recovery of some combinations with BB genes in a MAS based study was reported79 in addition to our reports with Tapaswini. Further studies are needed to draw definite conclusions on the gene-gene interactions. We also could not find any interaction between genes associated with resistance genes of blast and gall midge. Though Pi2 and Pi9 are located at the same locus on chromosome 6, both genes remained distinct with demonstrated independent function and this might be the reason for not observing any interaction between them. The probable reason for non-interaction between the GM genes which are located on different chromosomes, might be some structural dissimilarity. Of the two QTLs employed in the study, Sub1 is known to be an ethylene response factor while Saltol is associated with Na-K absorption and due their differences in function, no interaction was observed.
The pyramiding of genes was accomplished using a systematic approach and at the end of the first cycle itself, six populations each having a different gene/QTL but similar to the recurrent parent in morphology and grain shape were generated. Practice of both foreground and background selection has resulted in identification of lines closer to the parent in morphology but with different combination of genes. The use of molecular markers (SSR) that were unlinked to the assembled genes/QTLs for background selection, has enhanced the proportion of recovery of the recipient genome. The high variance present at BC1F1 than those in subsequent generations, indicate a wider frequency distribution in the BC1F1. The desired genotype with recombination between the target gene locus and either one of the flanking markers is expected to occur at a much higher frequency in BC1F1 than BC2F1 suggesting the feasibility of practicing background selection in the BC1F1 generation. Thus, in addition to the background selection in the BC3F1, adding one round of background selection in BC1F1 to the MAS scheme may greatly increase the efficiency of the program and maximum recovery of recurrent parent genome was reported using SSR based background selection in the BC1F1 generation80,81,82. In the present study, morphology and grain quality based selection at each stage has greatly helped in recovery of the recurrent parent genome to the maximum extent despite the use of seven different parents and the back ground selection with SSR markers led to the selection of pyramids having closer similarity to the recurring parent (Improved Tapaswini), a result which is conformity with earlier reports which used varied markers like RFLP79, AFLP81 and SSR13,14,15,30,36,83 to achieve the same.
Furthermore in the present study, the Improved Tapaswini gene pyramids were evaluated for their closeness according to their agro-morphological and grain quality traits with the recurrent parent Improved Tapaswini. Combining both morphological and grain qualities the dendrogram showed that the selected ten gene pyramids were closest to the recurrent parent with very less distance. The average genetic distance of recurring parent to ten pyramid lines was less than 0.50 distance using combined values of agronomic and quality traits (Fig. 8). However, selection studies with more number of polymorphic SSR markers can clearly assess the proportion of the recurrent parent genome in these gene pyramids.
From the earlier study, with the mapping and identification of microsatellite markers like RM444 linked to Gm1, all the major gall midge resistance genes have tags, flooring the route for gene pyramiding of two or more genes into elite rice cultivars for durable resistance. Gm1 was also introgressed into popular high yielding, rice cultivars Swarna and Samba Mahsuri, using the closest marker RM444 in a backcross breeding programme. It was also previously reported that, for disease resistance in Gm4 region, RPP13- like protein were identified.The marker RM547 flank the Gm4 gene at a distance of 1.9 cM. can be successfully used in MAB for development of gall midge resistant varieties84,85,86. As the same as previously reported we successfully transferred two gallmidge resistance genes Gm1 and Gm4 in Improved Tapaswini background using the flanking markers RM444 and RM547 respectively. Through map-based cloning approach the broad-spectrum rice blast resistance gene Pi9 was cloned,which led to the identification of tandemly arranged six resistance-like genes with a NBS (nucleotide-binding site) and LRRs (leucine-rich repeats) in the Pi9 locus. Both the rice Pi2 and Pi9 locus harbors multiple resistance (R) genes each controlling broad-spectrum resistance against diverse isolates of fungal pathogen, Magnaporthe oryzae causing devastating blast disease to rice. As per the previous report we succesfully transferred these two broad-spectrum rice blast resistance genes Pi2 and Pi9 in Improved Tapaswini background using the closest markers RG64 and P28 respectively87,88.
The molecular and physiological mechanisms involved in the submergence responses in plants have largely been characterized in two tolerant species, rice (Oryza sativa) and Rumex palustris. Rice SUB1A is an ethylene responsive factor (ERF) like transcriptional regulator, a member of the ERF gene family, dramatically improves submergence tolerance in controlled and field conditions. From the earlier report it was confirmed that SUB1A was highly expressed in FR13A and almost not detected in IR42. With reference to previous reports we have succesfully transferred the highly tolerant rice submergence QTL Sub1 using FR13A, in Improved Tapaswini background using the closest markers SUB1BC289,90,91. In a genome scale gene expression analysis it was found that between the two genotypes salinity suceptable IR29 and tolerant FL478 both are strikingly different at their transcriptional perspective, under salinity stress. Expression of IR29 was of relatively large number of genes as compared to the tolerant FL478. FL478, was responsible for maintaining low Na+, high K+, and Na+/K+ homeostasis in shoots of rice. Several salt tolerant rice lines has been developed by incorporating Saltol QTL into modern high yielding and salt-sensitive rice varieties through a targeted marker assisted backcrossing and marker assisted selection approach. In the previous study it was also further verified that, RM10745 is beneficial for marker-assisted selection of Saltol QTL92,93. Same as earlier study we have succesfully transferred the highly tolerant rice salnity QTL Saltol using FL478, in Improved Tapaswini background.
Four gene pyramids (ITGP1, ITGP2, ITGP4 and ITGP5) that have all the ten genes hold huge potential in rice breeding as they possess high levels of resistance/tolerance against multiple stresses. Though the integration and expression of ten or more genes using transgenic approach was demonstrated in rice94. The release of transgenic rice for general cultivation remains uncertain unless the issues related to human biosafety and transgenics are resolved. After their release, the MAS products can address both the biotic and abiotic stress simultaneously and having morphology and grain quality similar to the parent, the farmers with their popular variety can realize significant yield grains thereby enhancing their farm income and the variety with its stable yields, can lead to enhanced rice production in eastern India.
The rising number of pathogenic variants of different diseases and appearance of new and active insect biotypes, necessitates immediate attention. Through conventional breeding, pooling of ten different genes/QTLs will be a herculean task and the advances in marker technology has provided us a great opportunity to achieve the precise transfer and stacking of ten genes/QTLs with significant savings in time, space, labour and money. In addition, the end products can reach farmers directly as they need not pass thorough any regulatory approvals. Though the yield levels of the gene pyramids is lower than the recurrent parent, large scale field trials are needed to confirm the observation and if any yield penalty exists, detailed investigations can dissect the influence of individual gene(s)/QTL(s), gene combinations/interactions on yield and yield attributes using the large number of gene pyramids developed in the study. This novel demonstration of pyramiding ten (six + existed four) different genes/QTLs and their expression at desired levels in a new genetic background can herald a new revolution in molecular plant breeding. This achievement is a clear demonstration of the immense potential of modern plant breeding to confront several biotic and abiotic stresses at the same time and can greatly help in increasing both production and productivity of rice in the years to come.
Materials and Methods
Plant material and breeding scheme
The recurrent parent employed in the study was Improved Tapaswini, a gene pyramid of Tapaswini, developed at Central Rice Research Institute, Cuttack68. Tapaswini (CR 333-6-4), is a derivative of Jagannath/Mahsuri cross combination, is a highly popular indica cultivar with the rice farmers of Orissa in 1996 because of its high yield, medium slender grains and good cooking quality and can be grown in both the seasons. The variety was initially released for cultivation in Orissa, an Eastern state in India, and now its cultivation has now spread to five more eastern states.
Though high in popularity, it is susceptible to bacterial blight and several other stresses. To address the problem, in an earlier study, four bacterial blight resistance genes (Xa4, xa5, xa13 and Xa21) were introduced into Tapaswini using MAS approach and NILs having desired levels of resistance to BB were developed and one of the NILs [CRMAS 2622-7-6 (IET 21070)] was released as Improved Tapaswini for cultivation in BB endemic areas. The Improved Tapaswini did show higher levels of resistance against BB and did not record any yield penalty in the two year mandatory multi-location trials (10 and 18 locations) conducted by AICRIP, the nodal agency in India95. The new cultivar has been accepted by the farmers as it closely resembles the parent in both morphology, yield and grain and cooking quality but with enhanced resistance to BB. But, the variety despite its popularity is susceptible to pathogens like blast and gall midge and environmental stresses like submergence and salinity.
The donors used in the study include two breeding lines WHD-1S-75-1-127 (O. minuta derivative-Pi9)66 and C1O1A51(Pi2)96, for blast resistance, two released varieties Kavya (Gm1)97, a derivative of the cross WGL 27120/WGL 7672//Mahsuri/Surekha, and Abhaya(Gm4)34 are resistant to different gall midge biotypes, the popular FR13A98 for submergence tolerance QTL (Sub1) and a breeding line FL478, for the salinity tolerance QTL (Saltol)53. FL478, a breeding line from IRRI, Philippines, contains a 1 Mb DNA fragment (10.6–11.5 Mb on chromosome1) that contains the entire Saltol locus of Pokkali, a well-known donor for salinity tolerance, and the fragment was flanked by IR29 alleles54 (Kim et al. 2009). The primers employed for all the ten target genes/QTLs were all from published reports (Table 1). Due to its popularity and farmers preference, the Institute (CRRI) has initiated a program to enhance the defence capabilities of improved Tapaswini further through transfer of most effective gene (s)/QTL(s) to address submergence (Sub1) and salinity (Saltol) or gene combinations to address blast (Pi2 + Pi9), gall midge (Gm1 + Gm4).
Parental polymorphism survey
For the parental polymorphism survey, all the parents involved were included i.e. Improved Tapaswini, the recipient parent and donors: Kavya (Gm1-gall midge), Abhaya (Gm4-gall midge), C1O1A51 (Pi2-blast), WHD-1S-75-1-127 (Pi9-blast), FR13A (Sub1 QTL-submergence) and FL478 (Saltol QTL-salinity). For the survey, markers linked to different genes/QTLs like RM444 (Gm1), RM547 (Gm4), RG64 (Pi2), P28 (Pi9), SUB1BC2 (Sub1) and RM10745 (Saltol) were employed to examine the relation between the markers and the resistance/tolerance and susceptible nature of the parents for each trait under study.
DNA isolation and PCR analysis
Mini scale DNA isolation for PCR analysis was carried out as per Dellaporta et al.99. As stated earlier, the primers employed for the target genes/QTLs were all from published reports53,84,100,101,102,103,104,105,106. The reaction mixture for PCR analysis contained 50 ng of DNA template, 5 pico M of each of the primers forward and reverse, 200 μM dNTPs, 1× PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM of MgCl2 and 0.01 mg/ml gelatin) and 0.5 U of Taq DNA polymerase (DreamTaq DNA Polymerase, Thermo Scientific) in a quantity of 20 µl. All the PCR assays were performed as described earlier for each marker and the separated PCR products were visualized under UV light and photographed using an alpha imager (Alpha Innotech, USA). The PCR products of RG64 (Pi2) and RG556 (xa5) were digested with restriction enzymes Hae III and DraI respectively as per manufacturer’s instructions. The products amplified by RG 64 and RG 556 primers were monomorphic, hence restriction digestion was carried out and the digested products did show polymorphism associated with resistance and susceptibility.
Bioassays against bacterial blight
For screening of Tapaswini gene pyramids against the bacterial blight pathogen (BB), the inoculum of BB isolates recommended at CRRI was used. The inoculum was prepared by suspending the bacterial mass in sterile water to concentration of approximately 109 cells/ml. The upper three leaves of the plants were cut and clip-inoculated with the bacterial suspension at the maximum tillering stage as per107. The observations were recorded based on visual scoring and lesion length (LL) measurement 20 days later. The threshold level employed for distinguishing resistant and susceptible reaction based on lesion length was <5 cm (resistant) and >5 cm (susceptible).
Bioassays against blast
The screening was carried in the Uniform Blast Nursery (UBN) pattern, where each test entry (gene pyramids, parent, donor, susceptible controls) was sown as a single line with a 10 cm distance between consecutive rows and the susceptible control (B 40) are placed at the end of 20 rows of test entries. All the rows in the nursery were surrounded all around by two rows of HR12, a well-known blast susceptible line to spread the disease. High dosage (120 kg N/ha) of nitrogen was applied to the nursery for promotion of infection. Supplementary inoculums, in the form of chopped diseased leaves were scattered over the nursery and transplanting of infected seedlings in the midst of border rows. The scoring on SES scale108 was recorded at 10 days intervals starting from 25 to 30 days, depending to the severity of leaf blast and at least two readings of blast score were recorded. The bioassay was carried out with three replications with 18 plants constituting each replication.
Bioassays against gall midge
Ten days old seedlings of selected pyramids, donor parents (Kavya, Abhaya) and a susceptible control [T(N)1] were planted in plastic trays and the bioassays were conducted as per standard protocol109. The trays were kept in cages and gall midge larvae were released on the seedlings inside the cages and the reaction scores like seedling damage (in %), emergence and frequency of silver shoot (galls) were recorded after twenty days. The classification of lines into resistant and susceptible classes was recorded when the susceptible plants showed 90–100% plant damage with the emergence of galls. The plants free of galls were cut to observe the dead maggots and tissue necrosis to confirm the resistance status. The threshold level employed for distinguishing resistant and susceptible reaction based on injury (%) was 0–20 percent (resistant) and >20 percent (susceptible). The bioassay was carried out with three replications with 18 plants constituting each replication.
Screening for submergence tolerance
Three week old seedlings of gene pyramids, Improved Tapaswini (recipient parent) and controls FR13A (resistant) and IR42 (susceptible), raised in plastic trays, were subjected to submergence stress by placing the trays in submergence tanks of CRRI. The tanks were filled with water slowly and the plants were subjected to stress of total submergence by keeping 1.8 m standing water in the tanks. At the end of 15 days, the tanks were drained and the survival rates were recorded. The trays with the stressed plants were kept in open for desubmergence and the survival rates were recorded again after 8 days of desubmergence110. The screening was carried out with three replications with 18 plants constituting each replication.
Screening for salinity tolerance
Three week old seedlings of gene pyramids were transplanted along with Improved Tapaswini (recipient parent) and controls i.e. FL478 (resistant), SR26B (resistant) and IR29 (susceptible) and the screening was conducted as per Fageria111. After two days, the experimental tanks were filled with salt water and a stress of 8–10 dS m−1 EC was maintained for ten days and the stress was increased to 16 dS m−1 and at the end of 60 days (when the susceptible controls are completely dead), the survival rates and data on other parameters was recorded. The experiment continued with the surviving lines till maturity and spikelet fertility (%) was recorded. The screening was carried out with three replications with 18 plants constituting each replication.
Characterization for morphological and quality traits
Thirty-day-old seedlings of the recurrent parent and selected gene pyramids were transplanted with 15 × 20 cm spacing in a randomized complete block design with three replications at the experimental farm of Central Rice Research Institute (CRRI) Cuttack. Data was recorded on five plants from each line for agronomic traits, namely DFF (days to 50% flowering), plant height, tillers/plant, panicle length, number of filled grains/panicle and 1000-grain weight and line yields were calculated and extrapolated to kg/ha.
Characterization of grain and cooking quality traits was performed on the grains harvested from BC3F3 generation. For physical characters, data was collected from ten fully developed milled rice kernels. The kernel elongation ratio (KER) was expressed as the ratio of the average length of the cooked kernels to that of the uncooked kernels. The average ratio of the length of cooked rice to kernel length of milled rice of ten kernels was elongation ratio (VER). For calculating volume expansion, the standard method112 was followed. By the alkali digestion test113, alkali spreading value (ASV), gelatinization temperature was estimated indirectly. Following the method described by Juliano114, amylose content was determined.
Statistical analyses
Amplified products were scored for presence (1) or absence (0) for each primer genotype combination and the PIC value of a marker was calculated according to94. Dice similarity co-efficient was computed and used for generating a dendrogram following UPGMA. The computer package NTSYS-PC-2.02f115 was used for cluster analysis.
Conclusion
Improvement of high degree of resistance against diseases (BB and blast) and insects (gall midge) in the Indian subcontinent is a leading challenge because of high diversity of agro-climatic zones that conferring tolerance to submergence and salinity. The two major abiotic stresses is critical in the climate change scenario. The current study, presents the gene pyramiding of six tolerance /resistance, genes/QTLs such as submergence (Sub1), salinity (Saltol), and resistance to blast (Pi2, Pi9), gall midge (Gm1, Gm4) into an elite cultivar, Improved Tapaswini, which already have four BB resistance genes (Xa4, xa5, xa13, Xa21). Out of a large BC3F2 population we obtained 250 lines, with resistance alleles in homozygous state. Afterward in BC3F3 generation we carefully selected 32 lines with diverse gene combinations. Finally we selected 10 promising lines having closer similarity to the recurrent parent, based on the SSR background selection with diverse gene combinations, for further refinement of these gene pyramids. The recurrent parent along with the pyramided lines showed high levels of resistance against the bacterial blight bioassay. Against gall midge, blast, submergence and salinity (seedling stage) stresses the recurrent parent showed complete susceptibility while the pyramided lines showed different percentages of resistance/tolorence. Among the 4 pyramided lines with ten genes, the ITGP2 and ITGP5 showed high degree of resistance to all the biotic stresses and tolerance to the abiotic stresses, while ITGP4 showed highest degree of resistance to all targeted stresses and moderate resistance to blast. The line ITGP1 showed resistance to all targeted stresses, but moderate resistance to blast. The other six gene pyramids in different gene combinations also showed high degree of tolerance/resistance to each specific stress. In case of the background selection using morphological and grain quality traits we obtained all the ten gene pyramids with more than 90% recovery of recurrent parental genome. Through utilization of phenotypic selection tied with marker-assisted selection, out of 10 gene pyramids first time we obtained 4 gene pyramids with desired 6 abiotic and biotic tolerance/resistance (Sub1, Saltol, Pi2, Pi9, Gm1 and Gm4) genes/QTLs along with 4 bacterial blight resistance genes (Xa4, xa5, xa13, Xa21) in Improved Tapaswini background. Among the 4 pyramided lines with ten genes, ITGP2 and ITGP5 were obtained as best lines with high degree of resistance/tolerance to all the targeted biotic and abiotic stresses with maximum recovery of recurrent parental genome. The success of the study clearly demonstrates the efficacy of the marker technology in stacking of several agronomically important genes/QTLs into an elite genetic background and the tests on gene pyramids suggest that incorporation of strong and wide-ranging resistance/tolerance against many stresses at the same time is feasible. The other selected gene pyramids (having combinations of fewer genes <10), showing strong resistance/tolerance against each individual stress can be employed to address specific stress(s) present in different regions of India. The future prospect of the study intended on these gene pyramids, comprises a further background selection with additional numbers of high density markers and a multi-location trial against each targeted trait(s) to confirm the expression of the stacked genes/QTLs before their deployment. The achievement may possibly also encourage, several such studies to understand the prospective of molecular plant breeding as the basis for crop enhancement.
References
Finatto, T. et al. Abiotic stress and genome dynamics: specific genes and transposable elements response to iron excess in rice. Rice. 8, 1–18 (2015).
Mew, T. W. Current status of future prospects of research on bacterial blight of rice. Annu. Rev. Phtopathol. 25, 359–382 (1987).
Zhai, W. X. & Zhu, L. H. Rice bacterial blight resistance genes and their utilization in molecular breeding. Adv. Biotech. 19, 9–15 (1999).
Khush, G. S., Mackill, D. J. & Sidhu, G. S. Breeding rice for resistance to bacterial leaf blight. In: IRRI (ed) Bacterial blight of rice. IRRI, Manila, Philippines, 207–217 (1989).
Wang, C. et al. High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genomics. 289, 745–753 (2014a).
Zhang, F. et al. 2014. Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 64, 568–575.
Tian, D. et al. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 26, 497–515 (2014).
Wang, C. et al. Xa23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant. 8, 290–302 (2014b).
Kim, S. M. et al. Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theor. Appl. Genet. 1238, 1933–1943 (2015).
Mew, T. W., Cruz, V. C. M. & Medalla, E. S. Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis. 76, 1029–1032 (1992).
Nelson, R. J. et al. Relationship between phylogeny and pathotype for the bacterial blight pathogen of rice. Appl. Environ. Microbiol. 60, 3275–3283 (1994).
George, M. L. C., Bustamam, M., Cruz, W. T., Leach, J. E. & Nelson, R. J. Movement of Xanthomonas oryzae pv. oryzae in Southeast Asia detected using PCR-based DNA fingerprinting. Phytopathol. 87, 302–309 (1997).
Dokku, P., Das, K. M. & Rao, G. J. N. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker assisted selection. Euphytica. 192, 87–96 (2013a).
Dokku, P., Das, K. M. & Rao, G. J. N. Genetic enhancement of host plant-resistance of the Lalat cultivar of rice against bacterial blight employing marker-assisted selection. Biotechnol. Lett. 35, 1339–1348 (2013b).
Das, G. & Rao, G. J. N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6, 1–18 (2015).
Shafaullah, M. A. K., Khan, N. A. & Mahmood, Y. Effect of epidemiological factors on the incidence of paddy blast (Pyricularia oryzae) disease. Pak. J. Phytopathol. 23, 108–111 (2011).
Bevitori, R. & Ghini, R. Rice Blast Disease in Climate ChangeTimes. J. Rice Res. 3, 1–2 (2014).
Motlagh, M. R. S., Hbibi, F. & Ebadi, A. A. Genetic diversity of pyricularia grisea, the causal agent of rice blast by SSR. Acta Sci. Pol. Hortorum Cultus. 14, 15–28 (2015).
Ribot, C. et al. Susceptibility of rice to the blast fungus. Magnaporthe grisea. J. Plant Physiol. 165, 114–124 (2008).
Challagulla, V., Bhattarai, S. & Midmore, D. J. In-vitro vs in-vivo inoculation: screening for resistance of Australian rice genotypes against blast fungus. Rice Sci. 22, 132–137 (2015).
Sukanya, S. L., Yamini, D. & Fathima, S. K. Eco-friendly management of Pyricularia oryzae: The causal agent of blast of paddy. Curr. Bot. 2, 46–49 (2011).
Khanna, A. et al. Marker assisted pyramiding of major blast resistance genes Pi9 and Pita in the genetic background of an elite Basmati rice variety, Pusa Basmati 1. Indian J. Genet. 75, 417–425 (2015).
Hayashi, K., Yasuda, N., Fujita, Y., Koizumi, S. & Yoshida, H. Identification of the blast resistance gene Pit in rice cultivars using functional markers. Theor. Appl. Genet. 121, 1357–1367 (2010).
Huang, H. et al. Molecular mapping of the new blast resistance genes Pi47 and Pi48 in the durably resistant local rice cultivar Xiangzi 3150. Phytopathol. 101, 620–626 (2011).
Das, A. et al. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genomics. 12, 215–228 (2012).
Hua, L. et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 125, 1047–1055 (2012).
Liu, Y. et al. Fine-mapping and molecular marker development for Pi56 (t), a NBS-LRR gene conferring broad spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 126, 985–998 (2013).
Sharma, T. R. et al. Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52 (2012).
Fukuoka, S. Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci. Rep. 4, 1–7 (2014).
Hua, L. X. et al. Development of a marker specific for the rice blast resistance gene Pi39 in the Chinese cultivar Q15 and its use in genetic improvement. Biotech. Biotechnol. Equip. 29, 448–456 (2015).
Fukuoka, S. et al. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 5, 1–7 (2015).
Bentur, J. S., Amudhan, S., Pasalu, I. C., Sarma, N. P. & Prasad, R. U. New approaches to gall midge resistance in rice, International Rice Research Institute; Los Banos, Philippines: Insect-host plant interactions in gall Midge. Rice. 23–29 (2004).
Kumar, A. et al. Genetic analysis of resistance genes for the rice gall midge in two rice genotypes. Crop Sci. 45, 1631–1635 (2005).
Himabindu, K., Suneetha, K., Sama, V. S. A. K. & Bentur, J. S. A new rice gall midge resistance gene in the breeding line CR57-MR1523, mapping with flanking markers and development of NILs. Euphytica. 174, 179–187 (2010).
Jain, A. et al. Tagging and mapping of a rice gall midge resistance gene, Gm8, and development of SCARs for use in marker-aided selection and gene pyramiding. Thero. Appl. Genet. 109, 1377–1384 (2004).
Iftekharuddaula, K. M. et al. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 178, 83–97 (2012).
Iftekharuddaula, K. M. et al. Development of new submergence tolerant rice variety for Bangladesh using marker-assisted backcrossing. Rice Sci. 22, 16–26 (2015).
Voesenek, L. A. C. J. & Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 206, 57–73 (2015).
Tamang, B. G. & Fukao, T. Plant adaptation to multiple stresses during submergence and following desubmergence. Int. J. Mol. Sci. 16, 30164–30180 (2015).
Bailey-Serres, J. & Colmer, T. D. Plant tolerance of flooding stress-recent advances. Plant Cell Environ. 37, 2211–2215 (2014).
Pedersen, O. Long-distance water transport in aquatic plants. Plant Physiol. 103, 1369–1375 (1993).
Ali, A. J. et al. Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Res. 97, 66–76 (2006).
Dey, M. M. & Upadhyaya, H. K. Yield loss due to drought, cold and submergence inAsia. In: Evenson, R. E., Herdt, R. W. & Hossain, M. Rice Research in Asia: Progress and Priorities. UK, CAB International Wallingford: 291–303 (1996).
Xu, K. & Mackill, D. J. A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 2, 219–224 (1996).
Manivong, P. et al. Marker-assisted selection to improve submergence tolerance, blast resistance and strong fragrance in glutinous rice. Thai J. Genet. 7, 110–122 (2014).
Xu, K., Xu, X., Ronald, P. C. & Mackill, D. J. A high-resolution linkage map in the vicinity of the rice submergence tolerance locus. Sub1. Mol. Gen. Genet. 263, 681–689 (2000).
Chen, M. et al. An integrated physical and genetic map of the rice genome. Plant Cell. 14, 537–545 (2002).
Septiningsih, E. M. et al. Applying genomics tools for breeding submergence tolerance in Rice. Translational Genomics for Crop Breeding, Volume II: AbioticStress, Yield and Quality. Edited by Rajeev K. Varshney and Roberto Tuberosa, John Wiley & Sons, Inc. Published by John Wiley & Sons, Inc. (2013).
Dissanayake, P. K. & Wijeratne, A. W. Development of a varietial screening procedure for salt tolerance of Rice (Oryza Sativa L.) varieties at germination stage. J. Agricult. Sci. 2, 63–72 (2006).
Todaka, D., Nakashima, K., Shinozaki, K. & Yamaguchi-Shinozaki, K. Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice. 5, 1–9 (2012).
Lafitte, H. R., Price, A. H. & Courtois, B. Yield response to water deficit in an upland rice mapping population: associations among traits and genetic markers. Theor. Appl. Genet. 109, 1237–46 (2004).
Yeo, A. R., Yeo, M. E., Flowers, S. A. & Flowers, T. J. Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 79, 377–384 (1990).
Bonilla, P. S., Dvorak, J., Mackill, D., Deal, K. & Gregorio, G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp. Agric. Sci. 85, 64–74 (2002).
Kim, D. M., Ju, H. G., Kwon, T. R., Oh, C. S. & Ahn, S. N. Mapping QTLsfor salt tolerance in an introgression line population betweenjaponica cultivars in rice. J. Crop Sci. Biotech. 12, 121–128 (2009).
Tanksley, S. D., Young, N. D., Paterson, A. H. & Bonierbale, M. W. RFLP mapping in plant breeding-new tools for an old science. Biotech. 7, 257–264 (1989).
Young, N. D. & Tanksley, S. D. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet. 77, 95–101 (1989).
Ma, B. J. et al. Studies of PCR marker for the rice bacterial blight resistance gene Xa-4. Hereditas. 21, 9–12 (1999).
Mundt, C. C. Probability of mutation to multiple virulence and durability of resistance gene pyramids. Phytophathol. 80, 221–223 (1990).
Pradhan, S. K. et al. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deep water rice variety, Jalmagna. Rice. 8, 1–14 (2015).
Suh, J. P. et al. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. 6, 1–11 (2013).
Rajpurohit, D. et al. Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 178, 111–126 (2011).
Zhai, W., Wang, W., Zhou, Y., Li, X. & Zheng, X. Breeding bacterial blight resistant hybrid rice with the cloned bacterial blight resistance gene. Xa21. Mol. Breed. 8, 285–293 (2001).
Park, C. et al. Elucidation of Xa21-mediated innate immunity. Cell Microbiol. 12, 1017–1025 (2010).
Dash, A. K. et al. Phenotypic and Marker-Assisted Genetic Enhancement of Parental Lines of Rajalaxmi, an Elite Rice Hybrid. Phenotypic and Marker-Assisted Genetic Enhancement of Parental Lines of Rajalaxmi, an Elite Rice Hybrid. Front. Plant Sci. 7, 1–11 (2016).
Sanchez, A. C., Brar, D. S., Huang, N., Li, Z. & Khush, G. S. Sequence tagged site marker-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 40, 792–797 (2000).
Hittalmani, S., Parco, A., Mew, T. V., Zeigler, R. S. & Huang, N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 100, 1121–1128 (2000).
Qu, S. et al. The broad- spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multi gene family in rice. Genetics 172, 1901–1914 (2006).
Annual Report 2011–12 Central Rice Research Institute Cuttack (2012).
Sarkar, R. K. & Bhattacharjee, B. Rice genotypes with sub 1 QTL differ in submergence tolerance, elongation ability during submergence and regeneration growth at reemergence. Rice. 5, 1–11 (2011).
Colmer, T. D. & Voesenek, L. A. C. J. Flooding tolerance: suites of plant traits in variable environments. Funct. Plant Biol. 36, 665–681 (2009).
Collard, B. C. Y. et al. Developing new flood-tolerant varieties at the international rice research institute (IRRI). SABRAO J. Breed. Genet. 45, 42–56 (2013).
Renu, S. et al. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Science, https://doi.org/10.1016/j.plantsci.2015.08.008.
Shahbaz, M. & Zia, B. Does exogenous application of glycinebetaine through rooting medium alter rice (Oryza sativa L.) mineral nutrient status under saline conditions? J. Appl. Bot. Food Qual. 84, 54–60 (2011).
Shannon, M. Principles and strategies in breeding for higher salt tolerance. Plant Soil. 89, 227–241 (1985).
Yeo, A. R. & Flowers, T. J. Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. In: Effect of drought on plant growth. Salts in soils. Turner, N. C. & Passioura, J. B. (Eds). CSIRO. Melbourne. Australia, 161–173 (1986).
Islam, M. R. et al. Validation of Saltol linked markers and haplotype diversity on chromosome 1 of rice. Mol. Plant Breed. 3, 103–114 (2012).
Rao, V. R. & Riley, K. W. The use of biotechnology for conservation and utilization of plant genetic resources. Plant Genet. Newslett. 97, 3–19 (1994).
A. K. Dash et al. Genetic enhancement of bacterial blight resistance in parental lines of an elite rice hybrid Rajalaxmi, through marker-assisted gene pyramiding approach. Frontiers in Plant Science 7(1005), https://doi.org/10.3389/fpls.2016.01005 (2016).
Sanchez, A. C., Brar, D. S., Huang, N., Li, Z. & Khush, G. S. Sequence Tagged Site marker-assisted selection for three bacterial blight resistance genes in rice. Crop. Sci. 40, 792–797 (2000).
Chen, S., Lin, X. H., Xu, C. G. & Zhang, Q. Improvement of bacterial blight of ‘Minghui-63’ an elite restorer line of hybrid rice by molecular marker assisted selection. Crop. Sci. 40, 239–244 (2000).
Basavaraj, S. H. et al. Marker-assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid. Mol. Breed. 26, 293–305 (2010).
Chen, S., Xu, C. G., Lin, X. H. & Zhang, Q. Improving bacterial blight resistance of ‘6078’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Plant Breed. 120, 133–137 (2001).
Vu, H. T. T., Le, D. D., Ismail, A. M. & Le, H. H. Marker-assisted backcrossing (MABC) for improved salinity tolerance in rice (Oryza sativa L.) to cope with climate change in Vietnam. AJCS. 6, 1649–1654 (2012).
Biradar, S. K. et al. Identification of flanking SSR markers for a major rice gall midge resistance gene Gm1 and their validation. Theor. Appl. Genet. 109, 1468–1473 (2004).
Yasala, A. K. et al. In silico analysis for gene content in rice genomic regions mapped for the gall midge resistance genes. P.O.J. 5, 405–413 (2012).
Nanda, A. et al. Flanking microsatellite markers for breedingvarieties against asian rice gall midge. Trop. Plant Biol. 3, 219–226 (2010).
Qu, S. H. et al. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genet. 172, 1901–1914 (2006).
Xiao, N. et al. Improving of rice blast resistances in Japonica by pyramiding major R genes. Front. Plant Sci. 7, 1–10 (2017).
Campbell, M. T. et al. Genetic and molecular characterization of submergence response identifies subtol6 as a major submergence tolerance locus in maize. PLoS ONE. 10, 1–25 (2015).
Iftekharuddaula, K. M. et al. Development of early maturing submergence-tolerant rice varieties for Bangladesh. Field Crop Res. 190, 44–53 (2016).
Goswami, S., Karb, R. K., Paulc, A. & Dey, N. Genetic potentiality of indigenous rice genotypes from Eastern India with reference to submergence tolerance and deepwater traits ☆. Curr. Plant Biol., https://doi.org/10.1016/j.cpb.2017.10.002 (2017).
Mohammadi-Nejad, G., Arzani, A., Rezai, A. M., Singh, R. K. & Gregorio, G. B. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the Saltol QTL. Afr. J. Biotechnol. 7, 730–736 (2008).
Waziri, A., Kumar, P. & Purty, R. S. Saltol QTL and their role in salinity tolerance in rice. Austin J. Biotechnol. Bioeng. 3, 1–5 (2016).
Anderson, J. A., J. E. Churchill, S. D., Autrique, S. T. & Sorrells, M. E. Optimizing parental selection for genetic linkage maps. Genome. 36, 181–188 (1993).
Deng, Y., Zhu, X., Shen, Y. & He, Z. Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 113, 705–713 (2006).
Kumaravadivel, N., Uma, M. D., Saravanan, P. A. & Suresh, H. Molecular marker-assisted selection and pyramiding genes for gall midge resistance in rice suitable for Tamil Nadu Region. In: ABSTRACTS- 2nd International Rice Congress. pp 257 (2006).
Septiningsih, E. M. et al. Development of submergence tolerant rice cultivars: the Sub1 locus and beyond. Ann. Bot. 103, 151–160 (2009).
Dellaporta, S. L., Wood, J. & Hicks, J. B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21 (1983).
Ma, B. J. et al. Study on the PCR marker for the rice bacterial blight resistance gene Xa4. Hereditas (Beijing). 2, 9 (1999).
Yoshimura, S. A. et al. Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol. Breed. 1, 375–387 (1995).
Singh, A. K. et al. Marker assisted selection: a paradigm shift in Basmati breeding. Indian J. Genet. 71, 120–128 (2011).
Ronald, P. C. et al. Genetic and physical analysis of the rice bacterial blight disease resistance locus Xa 21. Mol. Gen. Genet. 236, 113–120 (1992).
Hittalmani, S., Foolad, M. R., Mew, T., Rodriguez, R. L. & Huang, N. Development of a PCR-based marker to identify rice blast resistance gene, Pi2 (t), in a segregating population. Theor. Appl. Genet. 91, 9–14 (1995).
Nair, S., Kumar, A., Srivastava, M. N. & Mohan, M. PCR based DNA markers linked to a gall midge resistance gene, Gm4t has potential for marker aided selection in rice. Theor. Appl. Genet. 92, 660–665 (1996).
Xu, K. et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 442, 705–708 (2006).
Kauffman, H. E., Reddy, A. P. K., Hsien, S. P. Y. & Merca, S. D. An improved technique for valuating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541 (1973).
IRRI. Standard Evaluation System for Rice (SES). (4 ed., pp. 15–16). Los Banos. Philippines: International Rice Research Institute (IRRI) (2002).
Vijaya, L. P., Amudhan, S., Himabindu, K., Cheralu, C. & Bentur, J. S. A new biotype of the Asian rice gall midge Orseolia oryzae (Diptera: Cecidomyiidae) characterized from the Warangal population in Andhra Pradesh, India. Int. J. Insect. Sci. 26, 207–211 (2006).
Jantaboon, J. et al. Ideotype breeding for submergence tolerance and cooking quality by marker-assisted selection in rice. Field Crop. Res. 123, 206–213 (2011).
Fageria, N. K. Salt tolerance of rice cultivars. Plant Soil. 88, 237–243 (1985).
Verghese, E. J. A standard process for cooking rice for experimental purpose. Mad. Agric. J. 37, 217–221 (1950).
Little, R. R., Hilder, G. B. & Dowsan, E. H. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chem. 35, 111–126 (1958).
Juliano, B. O. A simplified assay for milled rice amylose. Cereal Food World. 35(245), 253 (1971).
Rohlf, F.J. NTSYS-PC Numerical taxonomy and multivariate analysis system, Version 1.60. Applied Biostatistics, New York (1990).
Acknowledgements
The authors are highly grateful to the Director, ICAR-National Rice Research Institute, Cuttack, Odisha, India for providing all the necessary facilities. We also thankful to Dr. JK. Patra (Assistant Professor) Dongguk University, Republic of Korea, for his kind help and support during writing of this manuscript.
Author information
Authors and Affiliations
Contributions
G. Das contributed in performing the whole experiment, analysis of data and writing the manuscript. G.J.N. Rao contributed in planning and designing the study. M. Varier helped in bioassay of blast. A. Prakash helped in bioassay of gallmidge. D. Prasad helped in statistical analysis. G. Das and G.J.N. Rao participated in drafting the manuscript. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Das, G., Rao, G.J.N., Varier, M. et al. Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci Rep 8, 2413 (2018). https://doi.org/10.1038/s41598-018-20495-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20495-x
This article is cited by
-
Pyramiding of blast and bacterial blight resistance genes in premium quality rice variety, BRRI dhan63 through marker-assisted breeding approach
Euphytica (2024)
-
Development of a Temperate Climate-Adapted indica Multi-stress Tolerant Rice Variety by Pyramiding Quantitative Trait Loci
Rice (2022)
-
Genetic diversity and population structure of blast resistance genes in Thai upland rice germplasm
European Journal of Plant Pathology (2022)
-
Marker-assisted introgression of bacterial blight resistance gene xa13 into improved CO43
Euphytica (2022)
-
Development of Basmati lines by the introgression of three bacterial blight resistant genes through marker-assisted breeding
Euphytica (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.