Abstract
This study was aimed at generating the full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt) using single-molecule real-time (SMRT) sequencing. Four developmental stages of A. hygrophila, including eggs, larvae, pupae, and adults were harvested for isolating total RNA. The mixed samples were used for SMRT sequencing to generate the full-length transcriptome. Based on the obtained transcriptome data, alternative splicing event, simple sequence repeat (SSR) analysis, coding sequence prediction, transcript functional annotation, and lncRNA prediction were performed. Total 9.45 Gb of clean reads were generated, including 335,045 reads of insert (ROI) and 158,085 full-length non-chimeric (FLNC) reads. Transcript clustering analysis of FLNC reads identified 40,004 consensus isoforms, including 31,015 high-quality ones. After removing redundant reads, 28,982 transcripts were obtained. Total 145 alternative splicing events were predicted. Additionally, 12,753 SSRs and 16,205 coding sequences were identified based on SSR analysis. Furthermore, 24,031 transcripts were annotated in eight functional databases, and 4,198 lncRNAs were predicted. This is the first study to perform SMRT sequencing of the full-length transcriptome of A. hygrophila. The obtained transcriptome may facilitate further exploration of the genetic data of A. hygrophila and uncover the interactions between this insect and the ecosystem.
Similar content being viewed by others
Introduction
Alligator weed Alternanthera philoxeroides (Mart.) (Amaranthaceae) that originated from South America1,2, was introduced into China in the 1930 s. In China, A. philoxeroides is an important invasive species and has resulted in ecological and economic damage3. To control A. philoxeroides infestations, the flea beetle Agasicles hygrophila (Selman and Vogt) (Coleoptera: Chrysomelidae) was introduced as a biological control agent4. The use of A. hygrophila as a biological control of A. philoxeroides is acknowledged to be the world’s first successful example of aquatic weed control5.
Study has reported that the physiological adaptations of biological control agents determine where and when they will be successful6,7. Host specificity and potential host shifting are considered as the most important factors for the evaluation of A. hygrophila in many countries where it is introduced for biological control of A. philoxeroides8. Due to the good performances in host selection and ecological adaptation, A. hygrophila has become a desirable model insect for the investigation of the relationships between insects and plants, as well as insects and ecosystem. Currently, the genome and transcriptome information of A. hygrophila has not been investigated, which hinders the study of the molecular mechanisms underlying the interaction between A. hygrophila and host plant and ecosystem.
Transcriptome could reflect the type and number of intracellular genes and reveal the physiological and biochemical processes at a molecular level9. Several technologies have been applied for transcriptome sequencing. Among these, short-read transcriptome sequencing has become a powerful tool for the description of gene expression levels10,11. However, most of these technologies are incapable of assembling full-length transcripts because of the shortness of sequencing reads, which necessitates efforts for exploring other technologies. Thus, single-molecule real-time (SMRT) sequencing (Pacific Biosciences of California, Inc., CA, USA) is developed12, which overcomes the limitation of short-read sequences by enabling the generation of kilobase-sized sequencing reads13. The full-length transcriptome can be used to analyze the alternative splicing events, and the primary-percusor-mature RNAs structures, which help better understanding the RNA processing.
In this study, SMRT sequencing was performed to generate full-length transcriptome of A. hygrophila. Based on the obtained transcriptome data, we performed alternative splicing analyisis, simple sequence repeat (SSR) analysis, coding sequence prediction, transcript functional annotation, and lncRNA prediction. This study may be a valuable resource for further investigation of A. hygrophila.
Materials and Methods
Insects and host plants
The flea beetles, A. hygrophila, were obtained from South China Agricultural University (Guangdong, China) and were maintained in the insectary of Shanxi Agricultural University (Shanxi, China) under controlled conditions of 25 ± 1°C, with a light: dark photoperiod of 14:10 h and 80 ± 5% relative humidity. The insects were reared for several generations to obtain an experimental population with a consistent genetic background.
The host plant A. philoxeroides was collected from a field greenhouse at Yuhuan County, Zhejiang, China and was planted in the greenhouse of the biosafety and biological control research base at Shanxi Agricultural University.
Sample processing
The eggs laid by A. hygrophila within 24 h, first- to third-instar larvae starved for 24 h, pupae (2–6 days old), and newly emerged adults starved for 24 h were gathered and rinsed three times in precooled normal saline. Finally, 0.3 g of eggs, 0.8 g of larvae, 3.8 g of pupae, and 6.2 g of adults were harvested and frozen in liquid nitrogen for further experiments.
RNA sample preparation
Total RNA samples (at four different developmental stages) were isolated using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA). RNA degradation and contamination were monitored using 1% agarose gels. The purity and concentration of RNA were measured using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE, USA) with a OD260/280 reading. The RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The total RNA samples from four developmental stages were mixed together for the following experiments.
Library preparation and SMRT sequencing
mRNA was purified from 3 µg of mixed total RNA using poly (T) oligo-attached magnetic beads. Fragmentation was conducted using divalent cations under elevated temperatures in the NEBNext First Strand Synthesis Reaction Buffer (5×). The SMART PCR cDNA Synthesis Kit (Clontech, CA, USA) was used for synthesizing full-length cDNA. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3′ ends of the DNA fragments, NEBNext Adaptor with a hairpin loop structure was ligated to prepare for hybridization. BluePippin® (Sage Science, Beverly, MA, USA) was used for size selection of the full-length cDNA and for building libraries of differently sized cDNA. The generated cDNA was then re-amplified using PCR, and the fragment size distribution was quantified using the Qubit fluorometer (Life Technologies, Carlsbad, CA, USA). The quality of the libraries was assessed using the Agilent Bioanalyzer 2100 system. SMRT sequencing was performed using the Pacific Biosciences’ real-time sequencer using C2 sequencing reagents.
Next-generation sequencing
Total RNA (5 μg) was digested by using DNase I (NEB, Frankfurt, Germany). The sample was purified with Agencourt RNAClean XP Beads and fragmented into 130–170 nt. First-strand cDNA was generated by First Strand Master Mix and Super Script II reverse transcription (Invitrogen). Then second-strand cDNA was synthesized using Second Strand Master Mix. After end repairing, adding A and adaptor ligation, several rounds of PCR amplification with PCR Primer Cocktail and PCR Master Mix were performed to enrich the cDNA fragments. The final library is quantitated by using the Agilent 2100 bioanalyzer instrument, and real-time quantitative PCR. The qualified libraries was sequenced pair end on the Illumina HiSeq. 2000 System.
Preprocessing of SMRT reads
Raw SMRT sequencing reads were processed by removing polymerase reads that were <50 bp and had quality of <0.75, obtaining the clean reads. The obtained clean reads were processed into error-corrected reads of inserts (ROIs) with parameters of full passes of ≥0 and quality of >0.75. The ROI reads with both the 5′ and 3′ primer sequences and a poly(A) tail present were considered to be full-length transcripts. During the processes of library preparation, the chimeric sequences formed by the direct linkages of two cDNA template strands due to the low concentrations of adapter or SMRTbell are called artificial chimeric sequences. The non-chimeric sequences in the full-length sequence are called full-length non-chimeric (FLNC) sequences. The FLNC transcripts were determined by searching for the poly(A) tail signal and the 5′tail si cDNA primers in ROIs.
Iterative clustering for error correction (ICE) in SMRT analysis (v2.3.0)12 was used to obtain consensus isoforms by approaching clustering, and the full-length consensus sequences from ICE were refined using Quiver. Full-length transcripts with post-correction accuracy of >99% (high-quality isoforms) were generated for further analysis. Any redundancy in high-quality full-length transcripts was removed by CD-HIT14.
Alternative splicing detection
RNA alternative splicing, occurring after a pre-mRNA transcript, is formed from template DNA, which results in a single gene coding for multiple proteins. During this process, particular exons of a gene may be included within or excluded from the final, processed mRNA produced from that gene15, which results in the proteins translated from alternatively spliced mRNAs containing differences in their amino acid sequence and biological functions. In this study, based on the obtained redundancy removed transcripts, we predicted the alternative splicing events. Briefly, all sequences were aligned to each other with BLAST16. The alignment results according with the following conditions were considered as alternative splicing events17:
-
Both sequences lengths were more than 1000 bp, besides there should be two High-scoring Segment Pairs in the alignment;
-
The alternative splicing Gap was greater than 100 bp, with at least 100 bp distance to the 3′/5′ end;
-
All alternatively spliced transcripts allowed 5 bp overlap.
Simple sequence repeat (SSR) detection
SSR, also known as microsatellite, is a tract of repetitive DNA in which certain DNA motifs (ranging in length from 2–13 base pairs) are repeated, typically 5–50 times18. Transcripts that were >500 bp were selected for SSR analysis using the MIcroSAtellite identification tool (MISA; http://pgrc.ipk-gatersleben.de/misa/http://pgrc.ipk-gatersleben.de/misa/)19. MISA can identify seven SSR types, namely mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, hexanucleotide, and compound SSR, by analyzing transcript sequences.
Prediction of coding sequences
The coding sequences and corresponding amino acid sequences within the transcript sequences were predicted using TransDecoder (https://github.com/TransDecoder/TransDecoder/releases). TransDecoder could identify candidate protein-coding regions based on nucleotide composition, open reading frame (ORF) length, log-likelihood score, and (optional) Pfam domain content20.
Functional annotation of transcripts
The obtained non-redundant transcript sequences were mapped to eight databases to obtain the annotation information of the transcript. These databases included NR21, Swiss-Prot22, Gene Ontology (GO; http://www.geneontology.org)23, Clusters of Orthologous Groups of proteins (COG; http://www.ncbi.nlm.nih.gov/COG)24, euKaryotic Ortholog Groups (KOG)25, Pfam (http://pfam.janelia.org/)26, evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG; http://eggnog.embl.de), and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.ad.jp/kegg/)27.
lncRNA prediction
The most widely used methods for analyzing coding potential are Coding Potential Calculator (CPC)28, Coding-Non-Coding Index (CNCI)29, Coding Potential Assessment Tool (CPAT)21, and pfam protein structure domain analysis. In this study, lncRNAs were predicted by screening the coding potential of transcripts using these four methods above.
Results
SMRT sequencing data output
Bases on the Pacific Biosciences’ SMRT sequencing technology, 456,994 polymerase reads were generated. After preprocessing, 9.45 Gb of clean reads were obtained (Table 1). On the basis of the conditions of full passes of ≥0 and quality of >0.75, 335,045 ROIs were obtained (Table 2). In addition, 158,085 FLNC sequences were identified (Table 3).
Comparison of results of SMRT sequencing and next-generation sequencing
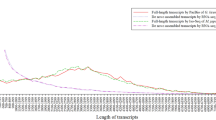
Most of the assembled contigs (70.41%) from next-generation sequencing were with length between 200–300 bp and only 2.31% were more than 2 kb. A total of 11,994 unigenes (25.99%) had length of 200–300 bp and 1,1981 unigenes (25.96%) had length of 300–500 bp. The comparison results between SMRT sequencing transcript and Illumina sequencing contig and unigene are shown in Table 4. Additionally, a total of 28,982 transcripts with total length of 48,811,662 bp were obtained from SMRT sequencing. For Illumina sequencing, 95,700 contigs and 46,151 unigenes (38,506,958 bp) were obtained (Table 5).
Transcript clustering analysis
In total, 40,004 consensus isoforms were obtained, including 31,015 high-quality isoforms and 8,989 low-quality ones. The ICE clustering results are shown in Table 6. Finally, 28,982 transcripts were obtained after removing redundant sequences from the high-quality transcripts.
Alternative splicing analysis
Total 146 alternative splicing events were identified, as shown in Supplementary Table S1. Additionally, since no reference genome is available for SMRT sequencing of transcriptome in A. hygrophila, we could not determine the types of alternative splicing events.
SSR detection
A total of 27,318 sequences (48,121,807 bp) were subjected to SSR analysis, including 12,753 SSRs and 8,535 SSR-containing sequences. The number of sequences containing more than one SSR was 2,733, and the number of SSRs present in compound formation was 966. In addition, the numbers of mono nucleotides, di nucleotides, tri nucleotides, tetra nucleotides, penta nucleotides, and hexa nucleotides were 10,810, 922, 952, 59, 6, and 4, respectively.
Prediction of coding sequences
Using TransDecoder, 24,040 ORFs were identified, which included 16,205 complete ORFs. The distribution of the coding sequence lengths of complete ORFs is shown in Fig. 1.
Functional annotation of transcripts
In total, 8,292 transcripts were annotated in the COG database; 13,197 were annotated in GO; 12,592 in KEGG; 16,955 in KOG; 20,940 in Pfam; 15,025 in Swiss-Prot; 22,887 in eggNOG; and 23,793 in NR. Moreover, 24,031 transcripts were annotated in all of the eight databases.
NR annotation
NR is a non-redundant protein database in NCBI, which contains protein data from the Swiss-Prot, Protein Information Resource, Protein Research Foundation, Protein Data Bank, GenBank, and RefSeq.21 databases. The homologous species of A. hygrophila were predicted by sequence alignment on the basis of the NR database. Approximately 56.21% of sequences were aligned to Tribolium castaneum, followed by Dendroctonus ponderosae (22.3%) (Fig. 2).
GO annotation
The GO database is produced by the Gene Ontology Consortium and features a structured, precisely defined, common, controlled vocabulary for describing the roles of genes and gene products in any organism. GO annotation system is a directed acyclic graph, including three categories: biological process (BP), molecular function (MF), and cellular component (CC). In this study, GO analysis revealed that the transcripts were enriched in several BP, MF, and CC associated terms (Fig. 3).
Gene Ontology (GO) functional annotation of Agasicles hygrophila transcripts. Green represents biological process; blue represents molecular function; and red represents cellular component. The x-axis represents GO categories; the y-axis (right) represents the number of transcripts; and the y-axis (left) represents the percentage of transcripts.
COG annotation
The COG database is an attempt at phylogenetically classifying proteins encoded in 21 complete genomes of bacteria, archaea, and eukaryotes. The database can be used for the functional and phylogenetic annotation of newly sequenced genomes. This study also found that the number of transcripts that were enriched in function R was the most, followed by function J and function O (Fig. 4).
lncRNA prediction
The number of lncRNA transcripts, as predicted by CPC, CNCI, pfam protein structure domain analysis, and CPAT is shown in Fig. 5. In total, 4,198 lncRNA transcripts were predicted by all four methods.
Discussion
The methodological strengths of SMRT sequencing have been comprehensively investigated in human13, which is superior to methods of short read sequencing due to the advantage of obtaining full-length transcripts. Besides, it could be used for the analysis of alternative splicing events, and the primary-percusor-mature RNAs structures to help better understand the RNA processing. In this study, 9.45 Gb of clean data were generated after SMRT sequencing, including 335,045 ROI and 158,085 FLNC reads. Total 40,004 consensus isoforms were identified through transcript clustering analysis of FLNC reads, which included 31,015 high-quality isoforms. After removing redundant sequences, 28,982 transcripts were obtained, and 145 alternative splicing events were predicted. SSR analysis revealed that 12,753 SSRs and 16,205 coding sequences were identified. Furthermore, 24,031 transcripts were annotated in eight functional databases. A total of 4,198 lncRNAs were predicted.
Based on 28,982 high-quality transcripts, a series of annotation analyses were performed. NR annotation revealed that 56.21% sequences were aligned to T. castaneum, followed by D. ponderosae (22.3%). T. castaneum is a member of the most species-rich eukaryotic order, an important model organism for studying generalized insect development30. Both T. castaneum and D. ponderosae belong to Coleoptera.
Genomic sequencing clearly revealed that the great majority of genes specifying the core biological functions are shared by all eukaryotes31. The rational classification of proteins encoded in sequenced genomes is critical for maximizing the use of genome sequences for functional and evolutionary studies24. In this study, these transcripts were enriched in various subcategories such as metabolic process, cellular process, cell, cell part, binding, and catalytic activity in the three main categories BP, MF, and CC according to the GO annotation analysis. The results of COG annotation showed that the largest number of transcripts were enriched in the function of general function prediction only. The results suggested that the transcripts of A. hygrophila were associated with the abovementioned functions.
LncRNAs, a novel class of nonprotein coding transcripts longer than 200 nt, are key regulatory molecules that can regulate gene expression at many different levels. Recently, increasing number of research has focused on the functions of lncRNAs in entomology, such as in Drosophila melanogaster, Plutella xylostella, and Nilaparvata lugens32, which provides a foundation for exploring the functions of lncRNA in insect development. This study identified 4,198 lncRNA transcripts with four analytical methods. However, their functions in A. hygrophila require further investigations.
In conclusion, our study, for the first time, completes SMRT sequencing of the full-length transcriptome of A. hygrophila. The obtained transcriptome may facilitate further studies on the genetic data of A. hygrophila and may help clarify the interactions between A. hygrophila and the ecosystem.
References
Maddox, D. Bionomics of an Alligatorweed Flea Beetle, Agasicles sp. 1 in Argentina. Annals of the Entomological Society of America 61, 1299–1305 (1968).
Vogt, G. B., McGuire, J. U. & Cushman, A. Probable evolution and morphological variation in South American Disonychine flea beetles (Coleoptera: Chrysomelidae) and their Amaranthaceous hosts. (Department of Agriculture, Science and Education Administration, 1979).
Wang, R., Wang, Y., Zhang, G. & Li, J. Host specificity tests for Agasicles hygrophila (Coleoptera: Chrysomelidae) a biological control agent of alligatorweed. Chinese Journal of Biological Control 4, 14–17 (1988).
Lu, J. J. et al. Performance of the biological control agent flea beetle Agasicles hygrophila (Coleoptera: Chrysomelidae), on two plant species Alternanthera philoxeroides (alligatorweed) and A. sessilis (joyweed). Biological Control 54, 9–13 (2010).
Buckingham, G. R. Biological control of alligatorweed, Alternanthera philoxeroides, the world’s first aquatic weed success story. Castanea, 232–243 (1996).
Zhao, L. et al. Cold hardiness of the biological control agent, Agasicles hygrophila, and implications for its potential distribution. Biological Control 87, 1–5 (2015).
Lu, X. et al. Climate warming increases biological control agent impact on a non‐target species. Ecology Letters 18, 48–56 (2015).
Li, N. et al. Manipulating two olfactory cues causes a biological control beetle to shift to non-target plant species. Journal of Ecology 105, 1534–1546 (2017).
Yang, F., Huang, L. & Zhang, A. High-throughput transcriptome sequencing technology and its applications in Lepidoptera. Acta Entomologica Sinica 57, 991–1000 (2014).
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101 (2012).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. science 323, 133–138 (2009).
Sharon, D., Tilgner, H., Grubert, F. & Snyder, M. A single-molecule long-read survey of the human transcriptome. Nature Biotechnology 31, 1009–1014 (2013).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry 72, 291 (2003).
Henzinger, T. A., Jhala, R. & Majumdar, R. In International Conference on Model Checking Software. 25–26 (2007).
Liu, X., Mei, W., Soltis, P. S., Soltis, D. E. & Barbazuk, W. B. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Molecular Ecology Resources 17, 1243–1256 (2017).
Gulcher, J. Microsatellite markers for linkage and association studies. Cold Spring Harbor Protocols 4, 425 (2012).
Thiel, T. MISA—Microsatellite identification tool.(2003) Available at: http://pgrc.ipk-gatersleben.de/misa/. (accessed 17 June 2016)
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nature Protocols 8 (2013).
Deng, Y. et al. Integrated nr database in protein annotation system and its localization. Computer Engineering 32, 71–74 (2006).
Apweiler, R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Research 32, 115–119 (2004).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nature Genetics 25, 25 (2000).
Tatusov, R. L., Galperin, M. Y., Natale, D. A. & Koonin, E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research 28, 33–36 (2000).
Koonin, E. V. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biology 5, R7 (2004).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Research 42, D222–D230 (2013).
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Research 32, D277–D280 (2004).
Li, A., Zhang, J. & Zhou, Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 15, 311 (2014).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402 (1997).
Richards, S. et al. The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 (2008).
Botstein, D. et al. Gene Ontology: tool for the unification of biology. Nature Genetics 25, 25–29 (2000).
Zhu, B., Liang, P. & Gao, X. Long noncoding RNAs (lncRNAs) and their research advances in entomology. Acta Entomologica Sinica 59, 1272–1281 (2016).
Acknowledgements
This work was supported by National Natural Science Foundation of China (Nos 31500304 and 31570436), Applied Basic Research Program of Shanxi Province (No. 201601D021096) and Doctor of Shanxi agricultural university introduced talent support program (No. 2015ZZ12).
Author information
Authors and Affiliations
Contributions
Conception and design of the research: Dong Jia; Acquisition of data: Yuanxin Wang and Yanhong Liu; Analysis and interpretation of data: Jun Hu, Yanqiong Guo and Lingling Gao; Drafting the manuscript: Dong Jia; Revision of manuscript for important intellectual content: Ruiyan Ma.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jia, D., Wang, Y., Liu, Y. et al. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt). Sci Rep 8, 2197 (2018). https://doi.org/10.1038/s41598-018-20181-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20181-y
This article is cited by
-
Characterization and analysis of multi-organ full-length transcriptomes in Sphaeropteris brunoniana and Alsophila latebrosa highlight secondary metabolism and chloroplast RNA editing pattern of tree ferns
BMC Plant Biology (2024)
-
PacBio single-molecule long-read sequencing provides new insights into the complexity of full-length transcripts in oriental river prawn, macrobrachium nipponense
BMC Genomics (2023)
-
Tolerance to dietary linalool primarily involves co-expression of cytochrome P450s and cuticular proteins in Pagiophloeus tsushimanus (Coleoptera: Curculionidae) larvae using SMRT sequencing and RNA-seq
BMC Genomics (2023)
-
Genomics and transcriptomics of the Chinese mitten crabs (Eriocheir sinensis)
Scientific Data (2023)
-
SSR Marker Acquisition and Application from Transcriptome of Captive Chinese Forest Musk Deer (Moschus berezovskii)
Biochemical Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.