Abstract
Naturally fermented milk (NFM) products are popular ethnic fermented foods in Arunachal Pradesh and Sikkim states of India. The present study is the first to have documented the bacterial community in 54 samples of NFM products viz. chhurpi, churkam, dahi and gheu/mar by high-throughput Illumina amplicon sequencing. Metagenomic investigation showed that Firmicutes (Streptococcaceae, Lactobacillaceae) and Proteobacteria (Acetobacteraceae) were the two predominant members of the bacterial communities in these products. Lactococcus lactis and Lactobacillus helveticus were the predominant lactic acid bacteria while Acetobacter spp. and Gluconobacter spp. were the predominant acetic acid bacteria present in these products.
Similar content being viewed by others
Introduction
Naturally fermented milk (NFM) products are prepared by one of the oldest processes of milk fermentation in the world using raw or boiled milk to ferment spontaneously or by back-sloping method1. Some naturally fermented milk products are chhu, chhurpi, dahi, lassi, misti dahi, mohi, philu, shoyu, somar and srikhand (cow/buffalo/yak milk) of India, Nepal, Pakistan, Bhutan and Bangladesh2,3,4,5, kurut of China6, aaruul, airag, byasulag, chigee, eezgii, khoormog and tarag of Mongolia7,8,9, ergo of Ethiopia, kad, lben, laban, rayeb, zabady, zeer of Morocco and Northern African and Middle East countries, rob (from camel milk), biruni (cow/camel milk), mish (cow/camel milk) of Sudan, amasi (hodzeko, mukaka wakakora) of Zimbabwe, nunu (from raw cow milk) of Ghana and kule naoto of Kenya10,11, filmjölk and långfil of Sweden12, koumiss or kumis or kumys or kymys of the Caucasian area13. Various cultivation-based studies reported lactic acid bacteria as the predominant microbiota present in the NFM products of the world mostly Lactococcus lactis subsp. cremoris, Lc. lactis subsp. lactis, Lactobacillus casei/Lb. paracasei, Lb. fermentum, Lb. helveticus, Lb. plantarum, Lb. acidophilus, Lb. coryniformis, Lb. curvatus, Lb. kefiranofaciens, Lb. kefiri, Lb. buchneri, Lb. jensenii, Lb. kitasatonis, Enterococcus faecium, E. faecalis and Leuconostoc mesenteroides, Streptococcus thermophilus, and others11,14,15,16,17,18,19. Besides bacteria, yeasts are also present in some NFM products which include Candida lusitaniae, C. parapsilosis, C. rugosa, C. tropicalis, Kluyveromyces marxianus, Saccharomyces cerevisiae, Galactomyces geotrichum, Issatchenkia orientalis, Kazachstania unispora, Pichia mandshurica, P. fermentans, P. kudriavzevii, and others8,11,13,16,20,21,22.
High altitude (upto 4878 m)-naturally fermented milk products of cow (Bos taurus) or yak (Bos grunniens)-milk prepared by back-sloping are common in the Himalayan states of Arunachal Pradesh and Sikkim in India which include chhurpi, churkam, dahi and gheu/mar (Fig. 1a–f) as a protein-rich food supplement and also as a source of livelihood5. Dahi, similar to yogurt, is the first product of milk fermentation by back-sloping, and is consumed as savory non-alcoholic beverage. Gheu/mar (crude butter) is a fat-rich milk product obtained by a process of milk churning in which the casein-rich soft-variety product called chhurpi (cottage cheese-like) is produced, and is consumed as curry/soup in meals; and churkam (hard-variety of chhurpi) is the product of dehydrated chhurpi, which is used as masticatory as chewing gum in high altitudes. Lactic acid bacteria were predominant with the load of 108 cfu/g in the Himalayan fermented milk products17. Lactobacillus bifermentans, Lb. alimentarius, Lb. paracasei subsp. pseudoplantarum, Lactococcus (Lc.) lactis subsp. lactis, Lc. lactis subsp. cremoris; Lb. plantarum, Lb. curvatus, Lb. fermentum, Lb. kefir, Lb. hilgardii, Enterococcus faecium and Leuconostoc mesenteroides were reported from dahi and chhurpi of Sikkim based on phenotypic, biochemical characterization and mol (%) content of G+C of DNA14,17. However, no study has been conducted yet on churkam and gheu/mar.
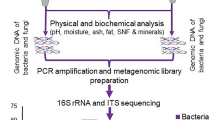
As it is well known that the cultivability of microbiota is still a limiting factor in understanding the natural food fermentation23,24, application of high throughput metagenomic techniques like Illumina amplicon sequencing may serve to give more insight into microbial ecology of natural food fermentation. Metagenomic studies of various fermented milk products like kefir, buttermilk, cheeses etc have shown a realistic view of the microbial community structure involved in the natural milk fermentation21,24,25,26,27,28. In this study we aimed to anlayse the bacterial community structure of fifty-four samples of naturally fermented milk products (chhurpi, churkam, dahi and gheu/mar) of Arunachal Pradesh and Sikkim by Illumina amplicon sequencing. This is the first report on bacterial community in NFM products of the Himalayas using in-depth metagenomic analysis.
Results
Overall microbial community structure
The bacterial composition of the different naturally fermented milk products (chhurpi, churkam, dahi and gheu/mar) was compared at different taxonomic levels (Fig. 2a–c). The bacterial phyla present in four types of NFM products were Firmicutes and Proteobacteria, respectively (data not shown). Phylum Firmicutes was represented by six families belonging to Streptococcaceae (24.2%), Lactobacillaceae (16.8%), Leuconostocaceae (8.0%), Staphylococcaceae (6.8%), Bacillaceae (1.6%), and Clostridiaceae (1.3%); and phylum Proteobacteria included Acetobacteraceae (26.8%), Pseudomonadaceae (3.3%) and Enterobacteriaceae (1.2%) (Fig. 1a). The overall bacterial diversity of these NFM products were predominated by species belonging to the lactic acid bacteria: Lactococcus lactis (19.7%) and Lactobacillus helveticus (9.6%) and Leuconostoc mesenteroides (4.5%) (Fig. 2b,c). Additionally, species belonging to the acetic acid bacteria: Acetobacter lovaniensis (5.8%), Acetobacter pasteurianus (5.7%), Gluconobacter oxydans (5.3%), and Acetobacter syzygii (4.8%) were also observed (Fig. 2b,c). The percentage of Enterobacteriaceae was 1.2% (Fig. 2a), whereas the percentage of genus Enterococcus was below 0.5% (data not shown), hence it was not shown at the genus level (Fig. 2b). Percentage of Streptococcus thermophilus was below 0.1% (data not shown). The percentage of unclassified bacteria at the taxonomical levels was 7.9% (Fig. 2a–c). Presence of uncultured bacterium was shown in all samples (Fig. 2c).
Multivariate analysis
PCA using species-level OTUs data showed significant differences among the NFM products studied (Fig. 3). The NFM products collected from two regions (Arunachal Pradesh and Sikkim) showed significant difference in the bacterial community structure (ANOSIM, p = 0.005, R = 0.16), but however, there was no significant difference between the same products prepared from different sources of milk (cow or yak). This reflects the regional contribution to the bacterial diversity of these products with respect to their location of preparation, but not from the milk source whereby these products are being prepared.
Alpha diversities
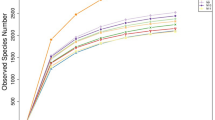
Alpha diversities were compared on the basis of states (Sikkim and Arunachal Pradesh)/places of collection of samples, animal’s milk source (cow/yak) and product types (Table 1). There was no significant difference between the states/regions and animal’s milk source, respectively. However, significance difference (p = 0.0125) was observed in terms of product types i.e., chhurpi and churkam in Chao1 species richness (Fig. 4). Chhurpi and churkam are two final products of milk fermentation where the latter is produced through a process of dehydration of the former and is usually kept for a longer fermentation. Multivariate analysis of species level OTUs showed a significant difference (ANOSIM p = 0.002, R = 0.16) between the two products. However, there is no significant difference among the general fermenting bacteria. Also, we observed a significant difference in Clostridiaceae (p = 0.0004) and Pseudomonadaceae (p = 0.013) between these two food types (Fig. 5).
Discussion
In this study, bacterial diversity was explored by barcoded Illumina MiSeq amplicon sequencing of the 16 S rRNA gene (V4-V5 region). The applied method using high throughput sequencing detected Lactococcus lactis, Lb. helveticus, Acetobacter lovaniensis, A. pasteurianus, A. syzygii, Gluconobacter oxydans and Leuconoctoc mesenteroides (above 1%) in all 4 samples of NFM products. Reads of OTUs in present study could not detect Lb. farciminis, Lb. biofermentans, Lb. hilgardi, Lb. paracasei subsp. pseudoplantarum, Lb. hilgardii, Lb. paracasei subsp. paracasei which were reported earlier in chhurpi and dahi based on limited phenotypic characterization14,17. However, Lb. helveticus (9.6%) was detected in the present culture-independent method which was not reported in culture dependent method earlier. Lb. helveticus is known to be present in dairy products29. A major composition of Lactococcus lactis (Streptococcaceae) and Lb. helveticus (Lactobacillaceae) was found to be the most predominant species along with Leuc. mesenteroides (Leuconostocaceae) in the NFM products of India, which still form what are commonly known as the primary cultures in milk fermentation1. Metagenomics-based studies of other milk products around the world like kefir, cheeses, have also reported to harbour species of Lactobacillus, Lactococcus and Leuconostoc25,26,30,31 as the dominant bacteria in general. Apart from the common known lactic acid bacteria group, a relatively high abundance of Proteobacteria-associated Acetobacteraceae (acetic acid bacteria) was observed in gheu/mar products. Acetobacteraceae members have also been reported in milk-related products19,25,32,33, and their dominance in gheu/mar (churned before heating) products than the subsequent downstream products (chhurpi and churkam) may be due to the effect of heating during the processing steps. Even though the Acetobacteraceae members were still present in chhurpi and churkam, the abundance was generally low. During the fermentation of chhurpi and churkam, we observed an increase in the abundance of Streptococcaceae (Lactococcus) and subsequently a build-up in the Lactobacillaceae (Lactobacillus) population in churkam.
Based on OTUs system, the percentage of Enterobacteriaceae and genus Enterococcus was very low in NFM samples analyzed. Enterococcus faecalis, Ent. faecium along with Lactococcus lactis subsp. lactis were reported from dahi of Bhutan based on 16 S rRNA gene sequencing6. Nunu, African NFM product, is frequently contaminated with pathogenic Enterobacteriaceae, demonstrated by short-read-alignment-based bioinformatics tools which may be used for high-throughput food safety testing34. Staphylococcaceae, Bacillaceae, Clostridiaceae and Pseudomonadaceae were observed at relatively low level in this study probably as contaminants. Pseudomonadaceae (Pseudomonas fluorescens) is usually present in milk and milk products as sources of contaminants35 and Clostridiaceae (Clostridium tyrobutyricum) is another bacterium found in cheese causing late blowing defect36. These contaminants were probably associated with the overall handling process, since samples are naturally fermented milk products, and there is no controlled process involved. Contamination of unwanted or rather non-fermenting bacteria are known to have acquired from various sources of production environment37,38. Presence of uncultured bacterium was shown in all samples analyzed. Uncultured bacterium group at species level were obtained using OTUs method, as the database could not assign them to any of their closest taxa. OTUs system put sequences into bins based on similarity of sequences within a data set to each other39. Moreover, limitations to using OTUs-based method is that the clustering algorithms are computationally intensive, relatively slow, and require significant amounts of memory40.
However, the predominance of few species were observed in a particular product showing the remarkable diversity of microbiota among 4 analyzed samples of NFM products and subsequently a build-up in the Lactobacillaceae (Lactobacillus) population in churkam. Lactococcus lactis was predominant in chhurpi, dahi and churkam, whereas in gheu/mar samples, it was relatively less. Lb. helveticus was dominant in churkam comparable to other 3 NFM products. However, Leuc. mesenteroides was predominant in dahi samples. Though we observed a fairly equal distribution between Lactococcus and Acetobacter species in 4 NFM products, however, at species level Lactococcus was represented only by Lc. lactis whereas Acetobacter was represented by A. lovaniensis, A. pasteurianus, A. syzygii and Gluconobacter oxydans. Diversity in bacterial species among the 4 NFM products was observed based on alpha diversity analysis. However, significance difference was observed only in between chhurpi and dahi (p = 0.0152) and chhurpi and churkam (p = 0.0125), respectively.
Conclusion
Earlier reports on chhurpi and dahi of North East India was based on limited culture-dependent analysis with some species of lactic acid bacteria. However, in the present study the NGS data of chhurpi, churkam, dahi and gheu showed the abundance of Lactococcus lactis (Streptococcaceae), Lb. helveticus (Lactobacillaceae) with Leuc. mesenteroides (Leuconostocaceae) as one the main bacterial species which may be the reliable information on microbial profile of NFM products. The application of NGS culture-independent methods to study the microbial ecology of fermented foods is of great significance in understanding the products, where Illumina sequencing has been shown to be one of the reliable tools in this study. Further studies on selective culturing of dominant bacteria, development of probiotic starter cultures and standardisation of processing methods may lead to industrialisation of ethnic food products.
Materials and Methods
Sampling
Fifty-four samples of naturally fermented milk products (chhurpi, churkam dahi and gheu/mar) were collected from high altitude mountains (1650–2587 meter) in Arunachal Pradesh (n = 35) and hills and mountains (381–4878 meter) in Sikkim (n = 19) of India (Table 2). The products were aseptically collected from the traditional production centres, transported in an ice-box and stored in the laboratory at −20 °C.
Metagenomic DNA extraction
Metagenomic DNA was extracted by two different methods based on the nature of the samples i.e., lipid-rich sample (gheu/mar) and casein-based samples (dahi, chhurpi and churkam). For the gheu/mar (lipid-rich) samples, extraction of DNA was performed as per method I as described in48 with some modifications. This method was chosen on the basis of the product being rich in its fatty content. The usage of a combination of petroleum ether:hexane (1:1) serves the purpose of dissolving the fat content resolving the product into two phases after rigorous vortexing. Briefly, 2 mL of the sample melted in low temperature was homogenized with 2 ml citrate buffer (2%). To this, 4 ml of petroleum ether: hexane (1:1) was added followed by vortexing and 10 min incubation at room temperature. 2 mL of the lower part of the homogenate was transferred to a sterile 2 ml screw-cap tube containing 0.5 g of zirconia/silica beads (0.1 mm) and 4 glass beads (2 mm). The tubes were centrifuged and the pellet resuspended in 150 µl proteinase-K buffer [50 mM Tris-Cl, 10 mM EDTA (pH 8), 0.5% (w/v) SDS]. After overnight incubation at 65 °C with 25 µl proteinase K (25 mg/ml), it was treated with 150 µl of 2X breaking buffer [4% Triton X-100 (v/v), 2% (w/v) SDS, 200 mM NaCl, 20 mM Tris (pH 8), 2 mM EDTA (pH 8)]. After addition of phenol (pH 8.0), the samples were treated in a bead beater three times (30 sec beating, 10 sec in ice) and further purified with chloroform: isoamyl alcohol mixture (24:1). Lastly, DNA was precipitated with ethanol and the pellet is dissolved in 50 µl of TE buffer (10 mM Tris, 1 mM EDTA).
For the casein-based samples (dahi, chhurpi and churkam), metagenomic DNA was extracted using the method of Keisam et al.41. This method was shown to recover maximum DNA yield from fermented milks41, hence it was also applied in this study. Briefly, 10 g or 10 ml of the samples were mixed with 90 mL 2% sodium citrate buffer and homogenized in a stomacher at 200 rpm for 2 min. Churkam (hard-cheese) samples were first grinded into powder before the homogenization. 1.5 mL of the homogenate was transferred to a sterile centrifuge tube and centrifuge for 10 min at 18000 × g. To the pellet, 400 µl TES buffer [50 mM Tris, 1 mM EDTA, 8.7% sucrose] 50 KU lysozyme, 25 U mutanolysin and 20 U lyticase were added and incubated at 37 °C for 1 h. After incubation, proteinase-K (25 mg/mL) was added to the mixture and further incubated at 65 °C for 1 h, followed by addition of GES reagent (5 M guanidine thiocyanate, 100 mM EDTA, and 0.5% sarkosyl). The sample was treated with 7.5 M ammonium acetate followed by purification with choloroform: isoamyl alcohol (24:1). Finally, DNA was precipitated with ethanol and the pellet dissolved in 50 µl of TE buffer (10 mM Tris, 1 mM EDTA). In all cases, absence of contaminating DNA in the laboratory prepared reagents was confirmed by extracting DNA from sterile water and observing negative PCR amplification with universal bacterial primers. The quality (A260/280) and quantity of the extracted DNA was checked using a spectrophotometer (NanoDrop ND-1000, USA). DNA was stored at −20 °C until required.
Barcoded Illumina MiSeq Sequencing
For in-depth bacterial community analysis, barcoded Illumina MiSeq amplicon sequencing targeting the V4-V5 region of the 16 S rRNA gene was conducted as described earlier49. The forward primer F563–577 (5′-AYTGGGYDTAAAGNG-3′) and barcoded reverse primers R924–907 (5′-CCGTCAATTCMTTTRAGT-3′) with an 8 bp barcode in its 5′-end was used for sample multiplexing42. Each PCR reaction was performed in a total volume of 25 µl with a template-free reaction that acts as a control. The following PCR conditions were used for amplification- initial denaturation (98 °C for 5 min); denaturation (98 °C for 15 sec), annealing (55 °C for 30 sec) and elongation (72 °C for 30 sec). The PCR reaction was run for 28 cycles with a final extension process of 72 °C for 5 min. The 430 bp sized products were separated in a 1.5% agarose gel (w/v) and the target bands were carefully excised from the gel with a sterile scalpel blade and then purified using QIAquick gel extraction kit (Qiagen, New Delhi, India) as per the manufacturer’s instructions. The purified DNA was quantified with Qubit dsDNA BR Assay Kit (Invitrogen) in a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA) and the individual were samples pooled in equimolar proportions. The final DNA pool was sent to the NGS facility in Xcelris Genomics (Ahmedabad, India) for paired-end MiSeq sequencing (2 × 300 bp). The raw sequence reads obtained was analysed using the default settings in MG-RAST43 and an open-source bioinformatics pipeline QIIME v1.8.044. A total of 7,614,683 post-quality filtered sequences originating from 54 samples belonging to 4 food types of NFM samples were uploaded to MG-RAST server with the MG-RAST ID number 4732361 to 4732414. The reads were subjected to secondary quality filtering to remove non-rRNA sequences before clustering into operational taxonomic units (OTUs) and subsequent generation of OTU tables at four different taxonomic levels (phylum, family, genus and species) using the SILVA SSU database in MG-RAST. Eukaryota-specific and unassigned OTUs were removed before performing further analysis.
Statistical Analysis
Normalisation of the OTUs relative abundance data was performed by log transformation log10 (xi + 1). To understand the variation in the microbial community structure of different food types, PCA was plotted using Canoco software v4.52 (Wageningen University, The Netherlands). Significant difference in the bacterial community structure amongst the four food type was evaluated by ANOSIM with 10,000 permutations using Bray-Curtis similarity index in PAST v2.17. Any significant difference in the abundance of individual taxa at four different taxonomic levels between the four food types was tested by p-value calculation using Student’s twov-tailed paired t-test and ANOVA. p-value < 0.05 was considered statistically significant and the differences in taxon abundance were represented as boxplots using BoxPlotR45,46. Species level-OTUs table was rarefied at a depth of 50 to 6482 sequences using the multiple_rarefactions.py script in QIIME for generation of alpha diversities rarefaction curves. Rarefaction plots were generated for Chao1 richness, diversity indices (Fisher alpha, Shannon), Shannon’s equitability and Good’s coverage using the make_rarefaction_plots.py script44. Significant differences in the alpha indices amongst the food types were calculated using the script compare_alpha_diversity.py in QIIME.
Data availability
Sequence data associated with this present work have been uploaded to MG-RAST server with the MG-RAST ID number 4732361 to 4732414.
References
Tamang, J. P., Watanabe, K. & Holzapfel, W. H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7, 377, https://doi.org/10.3389/fmicb.2016.00377 (2016).
Harun-ur-Rashid, M., Togo, K., Useda, M. & Miyamoto, T. Probiotic characteristics of lactic acid bacteria isolated from traditional fermented milk “Dahi” in Bangladesh. Pakistan J. Nutr. 6, 647–652 (2007).
Sarkar, S. Innovations in Indian fermented milk products-a review. Food Biotechnol. 22, 78–97 (2008).
Tamang, J. P. Himalayan fermented foods: microbiology, nutrition and ethnic value. CRC Press. (Taylor and Francis Group, New York, 2010).
Rai, R., Shangpliang, H. N. J. & Tamang, J. P. Naturally fermented milk products of the Eastern Himalayas. J. Ethn. Foods. 3, 270–5 (2016).
Shangpliang, H. N. J., Sharma, S., Rai, R., Tamang J. P. Some technological properties of lactic acid bacteria isolated from Dahi and Datshi, naturally fermented milk products of Bhutan. Front. Microbiol. 8, https://doi.org/10.3389/fmicb.2017.00116 (2017).
Sun, Z. et al. Identification and characterization of the dominant lactic acid bacteria from kurut: the naturally fermented yak milk in Qinghai, China. J. Gen. Appl. Microbiol. 56, 1–10 (2010).
Watanabe, K. et al. Diversity of lactic acid bacteria and yeasts in airag and tarag, traditional fermented milk products from Mongolia. World J. Microbiol. Biotechnol. 24, 1313–1325 (2008).
Takeda, S. et al. The investigation of probiotic potential of lactic acid bacteria isolated from traditional Mongolian dairy products. Animal Sci. J. 82, 571–579 (2011).
Mathara, J. M. et al. Isolation, identification and characterization of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int. J. Food Microbiol. 94, 269–278 (2004).
Akabanda, F. et al. Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 34, 277–283 (2013).
Mayo, B., Ammor, M. S., Delgado, S., Alegría, A. Fermented milk products. In: Tamang, J. P. & Kailasapathy, K. (Eds) Fermented foods and beverages of the World, CRC Press, Taylor & Francis Group, New York. pp. 263–288 (2010).
Wu, R. et al. Isolation and preliminary probiotic selection of lactobacilli from Koumiss in Inner Mongolia. J. Basic Microbiol. 49, 318–326 (2009).
Tamang, J. P. et al. Identification and enzymatic profiles of the predominant lactic acid bacteria isolated from soft-variety Chhurpi, a traditional cheese typical of the Sikkim Himalayas. Food Biotechnol. 14, 99–112 (2000).
Mathara, J. M. et al. Functional characteristics of Lactobacillus spp. from traditional Maasai fermented milk products in Kenya. Int. J. Food Microbiol. 126, 57–64 (2008).
Dewan, S. & Tamang, J. P. Microbial and analytical characterization of Chhu - A traditional fermented milk product of the Sikkim Himalayas. J. Sci. Ind. Res. 65, 747–52 (2006).
Dewan, S. & Tamang, J. P. Dominant lactic acid bacteria and their technological properties isolated from the Himalayan ethnic fermented milk products. Antonie Van Leeuwenhoek. 92, 343–52 (2007).
Yu, J. et al. Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia. J. Dairy Sci. 94, 3229–3241 (2011).
Oki, K., Dugersuren, J., Demberel, S. & Watanabe, K. Pyrosequencing analysis on the microbial diversity in Airag, Khoormog and Tarag, traditional fermented dairy products of Mongolia. Bio. Microbiota Food Health 33, 53–64 (2014).
Gadaga, T. H., Mutukumira, A. N. & Narvhus, J. A. Enumeration and identification of yeast isolates from Zimbabwean traditional fermented milk. Int. Dairy J. 10, 459–466 (2000).
Liu, W. et al. High-throughput sequencing for the detection of the bacterial and fungal diversity in Mongolian naturally fermented cow’s milk in Russia. BMC Microbiol. 15, 45 (2015).
Qviris, L. A. et al. Isolation, identification and characterization of yeasts from fermented goat milk of the Yaghnob valley in Tajikistan. Front. Microbiol. 7, https://doi.org/10.3389/fmicb.2016.01690 (2016).
Oliver, J. D. The viable but nonculturable state in bacteria. J Microbiol 43(1), 93–100 (2005).
Delbès, C., Ali-Mandjee, L. & Montel, M. C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 73, 1882–91 (2007).
Dobson, A., O’Sullivan, O., Cotter, P. D., Ross, P. & Hill, C. High-throughput sequence-based analysis of the bacterial composition of kefir and an associated kefir grain. FEMS Microbiol. Lett. 320, 56–62 (2011).
Alegría, Á., Szczesny, P., Mayo, B., Bardowski, J. & Kowalczyk, M. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and -independent approaches. Appl. Environ. Microbiol. 78, 1890–8 (2012).
Quigley, L. et al. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 78, 5717–23 (2012).
Jayashree, S., Pushpanathan, M., Rajendhran, J. & Gunasekaran, P. Microbial diversity and phylogeny analysis of buttermilk, a fermented milk product, employing 16S rRNA-based pyrosequencing. Food Biotechnol. 27, 213–21 (2013).
Moser, A., Berthoud, H., Eugster, E., Meile, L. & Irmler, S. Detection and enumeration of Lactobacillus helveticus in dairy products. Int. Dairy J. 68, 52–9 (2016).
Marsh, A. J., O’Sullivan, O., Hill, C., Ross, R. P. & Cotter, P. D. Sequencing-based analysis of the bacterial and fungal composition of kefir grains and milks from multiple sources. PLoS One. 8, 7 (2013).
Planý, M. et al. Metagenomic analysis of Slocak Bryndza cheese using next-generation 16S rDNA amplicon sequencing. Nova Biotechnol. Chimica. 15(1), 23–34 (2016).
Liu, W. J. et al. A survey of the bacterial composition of kurut from Tibet using a culture-independent approach. J. Dairy Sci. 95, 1064–72 (2012).
Leite, A. M. O. et al. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiol. 31, 215–21 (2012).
Walsh, A. M. et al. Strain-level metagenomic analysis of the fermented dairy beverage nunu highlights potential food safety risks. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.01144-17 (2017).
Wiedmann, M., Weilmeier, D., Dineen, S. S., Ralyea, R. & Boor, K. J. Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66(5), 2085–2095 (2000).
Panelli, S., Brambati, E., Bonacina, C., Feligini, M. Detection of Clostridium tyrobutyricum in milk to prevent late blowing in cheese by automated ribosomal intergenic spacer analysis. J. Food Sci. 78 (2013).
Doyle, C. J., Gleeson, D., O’Toole, P. W., Cotter, P. D. High-throughput sequencing highlights the significant influence of seasonal housing and teat preparation on the raw milk microbiota. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.02694-16 (2016).
Ssajjakambwe, P. et al. Milk hygiene in rural southwestern Uganda: prevalence of mastitis and antimicrobial resistance profiles of bacterial contaminants of milk and milk products. Vet. Med. Int. 6, https://doi.org/10.1155/2017/8710758 (2017).
Schloss, P. D. & Westcott, S. L. Assessing and improving methods used in Operational Taxonomic Unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77(10), 3219–3226 (2011).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Keisam, S., Romi, W., Ahmed, G. & Jeyaram, K. Quantifying the biases in metagenome mining for realistic assessment of microbial ecology of naturally fermented foods. Sci. Rep. 6, 34155 (2016).
Romi, W., Ahmed, G. & Jeyaram, K. Three-phase succession of autochthonous lactic acid bacteria to reach a stable ecosystem within 7 days of natural bamboo shoot fermentation as revealed by different molecular approaches. Mol. Ecol. 24, 3372–89 (2015).
Meyer, F. et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 9, 386 (2008).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–6 (2010).
Spitzer, M., Wildenhain, J., Rappsilber, J. & Tyers, M. BoxPlotR: a web tool for generation of box plots. Nat. Methods. 11, 121–2 (2014).
Ruggirello, M., Dolci, P. & Cocolin, L. Detection and viability of Lactococcus lactis throughout cheese ripening. PLoS One. 9, 1–14 (2014).
Acknowledgements
Authors are grateful to Science and Engineering Research Board-Department of Science and Technology, New Delhi for research project (Sanction order: SB/EMEQ-367/2014, dated: 21.07.2014), DAILAB (DBT-AIST International Laboratory for Advanced Biomedicine), Bioinformatics Centre of DBT and Microbial Resources Division, Institute of Bioresources and Sustainable Development (IBSD), Imphal, Manipur.
Author information
Authors and Affiliations
Contributions
H.N.J.S. and R.R. contributed to this present work equally as first co-authors which is a part of their research work. S.K. helps and assists in all the molecular work and N.G.S. (Bioinformatics and statistical) analyses. K.J. and J.P.T. have framed this research paper along with all the authors involved. All authors critically revised, read and approved the final manuscript with final check by J.P.T.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shangpliang, H.N.J., Rai, R., Keisam, S. et al. Bacterial community in naturally fermented milk products of Arunachal Pradesh and Sikkim of India analysed by high-throughput amplicon sequencing. Sci Rep 8, 1532 (2018). https://doi.org/10.1038/s41598-018-19524-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-19524-6
This article is cited by
-
16S metagenomics and metabolomics unveil the microbial compositions and metabolite profiles in Dahi, a traditional Indian fermented milk product prepared by the backslopping method
Journal of Food Science and Technology (2024)
-
Nutritional quality and microbial diversity of Chhurpe from different milk sources: an ethnic fermented food of high-altitude regions of the Western Himalayas
Discover Food (2024)
-
Coexistence of specialist and generalist species within mixed plastic derivative-utilizing microbial communities
Microbiome (2023)
-
Genome Analysis of Potential Probiotic Levilactobacillus brevis AcCh91 Isolated from Indian Home-Made Fermented Milk Product (Chhurpi)
Probiotics and Antimicrobial Proteins (2023)
-
Microbial Diversity and Functional Potential of Keem: A Traditional Starter Culture for Alcoholic Beverage—Application of Next-Generation Amplicon and Shotgun Metagenome Sequences
Molecular Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.