Abstract
Recently, we discovered a novel cDNA encoding the precursor of a small secretory protein, neurosecretory protein GL (NPGL), in the hypothalamic infundibulum of chickens. NPGL plays an important role in the regulation of growth and feeding. A database search indicated that the NPGL gene has a paralogous gene: neurosecretory protein GM (NPGM), also in chickens. We identified cDNA encoding the NPGM precursor in chickens. Morphological analysis showed that NPGM-containing cells are specifically localized in the medial mammillary nucleus (MM) and infundibular nucleus (IN) in the hypothalamus. In addition, we found that NPGM and NPGL are co-localized, especially in the MM. The expression levels of NPGM mRNA gradually decreased during post-hatch development, in contrast to those of NPGL mRNA. Moreover, we investigated the relationship between NPGM and other known factors. NPGM was found to be produced in histaminergic neurons in the MM. NPGM and histidine decarboxylase, a histamine-producing enzyme, displayed similar expression patterns during post-hatch development. Acute intracerebroventricular injection of NPGM decreased food intake, similar to the effect of histamine. To our knowledge, this is the first report of the localization and function of NPGM in the brain of vertebrates. These results will further advance the understanding mechanisms underlying energy homeostasis.
Similar content being viewed by others
Introduction
Energy homeostasis is regulated by hormones and neuropeptides released from a variety of sources, such as the gut, liver, adipose tissue, and brain1. The hypothalamus integrates peripheral and central signals to monitor the energetic state of the organism. Feeding behavior is a critical component of energy homeostasis and poultry, such as chickens, are often utilized in studies on appetite control. These have improved commercial productivity and have helped us understand common mechanisms underlying energy homeostasis in vertebrates2. In avian species, the infundibular nucleus (IN) in the hypothalamic infundibulum is known to possess first-order neurons for feeding regulation3. The IN contains orexigenic neuropeptide Y (NPY) and agouti-related protein (AgRP) neurons, which have complementary functions4,5. In contrast, the IN also contains anorexigenic pro-opiomelanocortin (POMC) neurons6. Second-order neurons for feeding regulation are included in the paraventricular nucleus (PVN), expressing corticotropin-releasing hormone (CRH) and thyrotropin-releasing hormone (TRH)3,7,8. In addition, several bioactive factors, including ghrelin and growth hormone-releasing hormone (GHRH), are also involved in ingestive behavior9. Avian and mammalian species share many common neuropeptides which maintain energy homeostasis, such as NPY, POMC, CRH, melanin-concentrating hormone (MCH), and ghrelin10. However, these peptides do not always play similar roles in birds and mammals10. In mammals, MCH, ghrelin and GHRH have orexigenic effects11,12,13. On the other hand, in birds, MCH does not affect food intake, and ghrelin and GHRH have anorexigenic effects9,10. The mechanism of appetite control has not been fully understood in vertebrates, partly because of such differences in feeding regulation between birds and mammals10.
To further understand the mechanisms regulating energy homeostasis in vertebrates, we sought to identify novel factors involved in energy intake and metabolism. Recently, we identified a novel cDNA encoding the precursor of a small neurosecretory protein in the hypothalamus of chickens, mice, and rats14,15,16. The precursor protein contained a signal peptide sequence, a mature protein sequence, a glycine amidation signal, and a dibasic amino acid cleavage site. Because the predicted C-terminal amino acids of the small protein were Gly-Leu-NH2, the small protein was named neurosecretory protein GL (NPGL)14. In situ hybridization indicated that NPGL mRNA was produced in the medial mammillary nucleus (MM) and the IN within the hypothalamic infundibulum of chicken14. In addition, NPGL mRNA levels were found to have increased during post-hatching development14. Chronic subcutaneous and intracerebroventricular (i.c.v.) infusion of NPGL both increased body mass gain in chicks; the latter also increased food intake14,17. These findings suggest that NPGL participates in the growth of chicks. Recently, we also found that NPGL stimulates feeding behavior in mice and rats15,16.
A genome database search suggested the presence of a paralogous gene, named neurosecretory protein GM (NPGM)14, and analysis suggested that NPGM and NPGL are conserved in vertebrates, including chickens, rats, and humans14. Therefore, we expected that NPGM would have significant biological functions in vertebrates. However, prior to the present study, it was unclear whether NPGM is even expressed in the brain. To investigate the physiological function(s) of NPGM and the relationship between NPGM and NPGL, we therefore performed cDNA cloning of the NPGM precursor and analyzed its localization and the effects of i.c.v. injection of mature NPGM in chicks.
Results
Identification of cDNA encoding NPGM
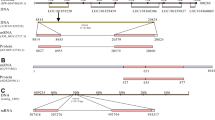
We performed a BLAST search on a genome database (Ensembl Genome Browser; www.ensembl.org) using the amino acid sequence of the NPGL precursor in chicken. We discovered a paralogous gene for the chicken protein. Using PCR primers based on the database sequences in chicken, we cloned a cDNA spanning the entire coding region of the NPGM gene and found it to be identical to that provided in the cDNA database (XM_429770.2). The NPGM cDNA is 408 bp long and comprises an open reading frame (ORF) that encodes 135-amino acids. The nucleotide and amino acid sequences of the precursor protein are shown in Fig. 1a. Analysis of the N-terminal sequence of the deduced protein with the SignalP program (www.cbs.dtu.dk/services/SignalP/) revealed the presence of a 24-amino acid signal peptide (Fig. 1a). The predicted 83-amino acid residues of the small protein are flanked at the C-terminus by a dibasic Arg-Arg motif, which consists of a potential proteolytic processing site. The precursor protein possesses a Gly residue at its C-terminal end that contributes to amidation (Fig. 1a). Therefore, the predicted C-terminal amino acid sequences of the small protein are Gly-Met-NH2. As a result, this small protein was termed neurosecretory protein GM (NPGM). The mature NPGM contains two Cys residues, suggesting an intramolecular disulfide bond formation (Fig. 1a). Mature NPGM in chicken shares a relatively high amino acid sequence identity (54%) with chicken NPGL, but it does not share any amino acid sequence identity with other known neuropeptides (Fig. 1b).
Nucleotide sequence of NPGM precursor and amino acid sequence alignment of NPGM and NPGL precursor proteins. (a) The predicted signal peptide is denoted by a wavy line. The sequence of the predicted 83-amino acid residues of the mature small protein is presented in bold face. The Gly (G) C-terminal amidation signal and the Arg (R)-Arg (R) dibasic processing site are underlined. Two Cys (C) residues are boxed. The asterisk indicates the stop codon (TAG). (b) Amino acid sequence alignment of NPGM and NPGL precursor proteins deduced from chicken cDNA sequence. Black boxes highlight conserved amino acids. Gaps, indicated by hyphens, were inserted to optimize sequence alignment. Predicted mature small proteins are underlined. The conserved Cys (C) residues are indicated by asterisks.
Distribution of the NPGM precursor mRNA and mature protein
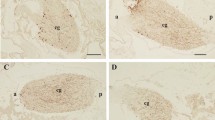
The expression levels of the NPGM precursor mRNA were examined in 1-day-old chicks by real-time PCR using samples from different brain regions, such as the telencephalon, diencephalon, mesencephalon, cerebellum, and hypothalamic infundibulum. NPGM precursor mRNA was expressed only in the hypothalamic infundibulum; its expression in other regions of the brain was not detectable (Fig. 2a). In situ hybridization data indicated that NPGM mRNA-containing cells were localized in the MM and the IN within the hypothalamic infundibulum in 1-day-old chicks (Fig. 2b and c). Additionally, NPGM mRNA was expressed predominantly in the posterior region of the MM (Fig. 2b). No signal was detected with the sense probe (Fig. 2d). Immunohistochemical analysis indicated presence of NPGM-immunoreactive cells and fibers in the MM and the IN; this result was replicated in terms of mRNA expression, assessed via in situ hybridization (Fig. 2b,e and g). The immunoreactive signals were blocked by the neutralization of the primary antibody pre-incubated with synthetic mature NPGM (Fig. 2f and h).
Localization of NPGM mRNA and the mature protein in the hypothalamic infundibulum. (a) Real-time PCR analysis of the NPGM precursor mRNA concentrations in different brain regions. Each value represents the mean ± SEM (n = 5). The asterisk indicates a statistically significant difference (One-way ANOVA with Tukey’s test as a post-hoc test: **P < 0.01). (b–d) Schematic representation and photomicrographs illustrating the distribution of cells expressing NPGM precursor mRNA in the hypothalamic infundibulum. (b) The location of the hypothalamic infundibulum is shown in the coronal brain illustration. Cells expressing the NPGM precursor mRNA and mature NPGM in the medial mammillary nucleus (MM) and the infundibular nucleus (IN) within the hypothalamic infundibulum are represented by dots in the illustration. (c) Cells expressing the NPGM precursor mRNA in the MM and the IN were obtained from 1-day-old chicks. (d) No signals were detected by the sense probe. (e–h) Photomicrographs of NPGM-immunoreactive cells and fibers. NPGM-immunoreactive perikarya in the MM (e) and the IN (g). No signals were detected by the antibody adsorption test (f,h). Scale bar = 100 µm.
Expression of NPGM and NPGL mRNA during post-hatch development
To investigate the common and/or different features between NPGM and NPGL, we analyzed the NPGM and NPGL precursor mRNA levels in the hypothalamic infundibulum including the MM and the IN, during post-hatch development by real-time PCR using 1-, 8-, and 15-day-old chicks. NPGM mRNA expression levels gradually decreased during development (Fig. 3a). In contrast, NPGL mRNA expression levels were elevated in 8-day-old chicks compared to 1-day-old (Fig. 3b). In situ hybridization also showed that NPGM mRNA was highly expressed in the MM and the IN after hatching, and that this expression decreased from day 1 to day 15 (Fig. 4a,b,e and f). In contrast, NPGL mRNA expression in the MM and the IN increased from day 1 to day 15 (Fig. 4c,d,g and h). Although NPGM and NPGL mRNA showed opposite expression patterns during post-hatch development, both mRNAs were expressed in similar locations in the MM at the same developmental stage.
Expression level of NPGM and NPGL mRNA during post-hatch development using real-time RT-PCR. Real-time RT-PCR analysis of NPGM (a) and NPGL (b) precursor mRNA in the hypothalamic infundibulum in 1-, 8-, or 15-day-old chicks. Data are expressed as mean ± SEM (n = 5–6). The asterisk indicates a statistically significant difference versus 1-day-old (One-way ANOVA with Tukey’s test as a post-hoc test: *P < 0.05, ***P < 0.005 vs. D1).
Localization of NPGM and NPGL mRNA-expressing cells in 1- and 15-day-old chicks using in situ hybridization. (a–h) Photomicrographs of cells expressing the NPGM (a,b,e,f) and NPGL (c,d,g,h) precursor mRNA in the medial mammillary nucleus (MM) (a–d) and the infundibular nucleus (IN) (e–h) were obtained after performing in situ hybridization with coronal brain sections from 1-day-old (a,c,e,g) and 15-day-old (b,d,f,h) chicks. Scale bar = 100 µm.
Co-localization of NPGM and NPGL
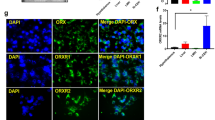
By immunohistochemical analysis with specific antibodies (Supplementary Fig. S1), we further investigated whether NPGM and NPGL were produced in the same neurons in the hypothalamic infundibulum. Approximately 30–40% NPGM-immunoreactive cells were co-localized with NPGL-immunoreactive cells in the MM in 1-day-old chicks (Fig. 5a and c and Supplementary Fig. S2a). Almost all NPGM-immunoreactive cells were co-localized with NPGL-immunoreactive cells in the MM in 15-day-old chicks (Fig. 5b and d and Supplementary Fig. S2b). Although some NPGM-immunoreactive cell bodies were detected in the IN in 1-day-old chicks, there were no NPGL-immunoreactive cell bodies in the IN (Fig. 5e and g). By 15 days of age, a few NPGM-immunoreactive cells had become co-localized with NPGL-immunoreactive cells in the IN (Fig. 5f and h and Supplementary Fig. S2c). In addition, many dense NPGL-immunoreactive fibers were distributed around immunoreactive cells in the MM and the IN in 15-days-old chicks (Fig. 5d and h and Supplementary Fig. S2b and c). Taken together, NPGM-producing cells also produced NPGL in the MM in 1- or 15-day-old chicks, and the ratio of co-localization between NPGM and NPGL increased during post-hatch development. In contrast, in the IN, almost no cells produce both NPGM and NPGL in either 1- or 15-day-old chicks.
Localization of NPGM and NPGL neurons in 1- and 15-day-old chicks using immunohistochemistry. (a–h) Photomicrographs of NPGM- (a,b,e,f) and NPGL- (c,d,g,h) immunoreactive cell bodies and fibers in the medial mammillary nucleus (MM) (a–d) and the infundibular nucleus (IN) (e–h) were obtained after performing immunohistochemistry with coronal brain sections from 1-day-old (a,c,e,g) and 15-day-old (b,d,f,h) chicks. Arrow-heads indicate NPGM- and NPGL-coexpressing neurons. Triangles indicate only NPGM-producing neurons. Scale bar = 50 µm.
Relationship between NPGM and histamine
We revealed co-localization of NPGM and NPGL, especially in the MM. Furthermore, we investigated the possibility of co-localization with several other bioactive factors expressed in the hypothalamus. From our preliminary and previous experimental data, we considered it likely that the localization of NPGM-containing cells was close to that of the histaminergic neurons in the posterior region of the MM18. It was also clear that NPGM neurons are distinct from neurotensin, gonadotropin-inhibiting hormone (GnIH), and 26RFa (QRFP) neurons, as these are localized in other regions of hypothalamus19,20,21. Therefore, we investigated the relationship between NPGM and histamine by double-labeling using immunohistochemistry and in situ hybridization. We found that NPGM-immunoreactive cells were localized in the MM and the IN (Fig. 6a,c and e), but that the precursor mRNA for histidine decarboxylase (HDC), a histamine-producing enzyme, was only expressed in the MM (Fig. 6b,d and f). All NPGM-immunoreactive cells in the posterior MM were identical to HDC precursor mRNA-containing cells (Fig. 6c and d). Subsequently, we measured the HDC mRNA expression level during post-hatch development. HDC mRNA expression gradually decreased with post-hatch development (Supplementary Fig. S3). Therefore, the expression of HDC mRNA showed a similar developmental pattern to that of NPGM mRNA (Fig. 3a).
Co-localization of NPGM and histamine neurons. (a,c,e) NPGM-immunoreactive cells are localized in the medial mammillary nucleus (MM) (a,c) and the infundibular nucleus (IN) (e). (b,d,f) HDC precursor mRNA expressing cells are localized in the MM (b,d) and the IN (f). Arrow-heads indicate NPGM- and HDC-coexpressing neurons. Scale bar = 100 µm.
Effect of i.c.v. injection of NPGM
We investigated the effect of NPGM on the feeding behavior of 5- or 6-day-old chicks. I.c.v. injection of 1 nmol NPGM immediately decreased food intake for 30 min after injection (Fig. 7a). Additionally, i.c.v. injection of 5 nmol NPGM remarkably decreased food intake up to 90 min after injection (Fig. 7b). These results show that NPGM decreases feeding behavior in a dose-dependent manner (0.2 nmol NPGM had no significant effect).
Effect of intracerebroventricular (i.c.v.) injection of NPGM on food intake in 5- or 6-day-old chicks. (a) Cumulative food intake after i.c.v. injection of vehicle or NPGM (0.2 and 1 nmol) in 6-day-old chicks. (b) Cumulative food intake after i.c.v. injection of vehicle or NPGM (5 nmol) in 5-day-old chicks. Data are expressed as mean ± SEM (n = 8). The asterisk indicates a statistically significant difference versus vehicle (One-way ANOVA with Tukey’s test as a post-hoc test or Student’s t-test: *P < 0.05 vs. Vehicle).
Discussion
After the identification of a cDNA encoding NPGL in the hypothalamus of chicken, we found a paralogous gene, NPGM, in the same animal14. Therefore, in the present study we attempted to characterize NPGM in the chicken hypothalamus. The sequences of NPGM and NPGL were found to be highly conserved, and these small proteins have no homology with previously known peptides. The NPGM precursor mRNA and protein were expressed in the MM and the IN in the hypothalamic infundibulum. Although immunohistochemical analysis indicates the existence of endogenous NPGM protein, we have yet to confirm the presence of mature NPGM using another analysis, such as mass spectrometry. The expression of NPGM mRNA gradually decreased during post-hatching development, in contrast to the expression of NPGL mRNA. Subsequently, we investigated the potential for co-localization of NPGM and NPGL. Co-localization of NPGM and NPGL was observed particularly in the MM, with almost none in the IN. The different developmental patterns and localization of NPGM and NPGL in the IN suggest that they may have different roles. In fact, we previously found that NPGL induces orexigenic activity in chicks17, whereas NPGM was found to exhibit an anorexigenic effect in this study. In mammals, POMC is a precursor protein of α-melanocyte stimulating hormone (α-MSH) and β-endorphin, which have anorexigenic and orexigenic effect, respectively. POMC neurons are generally known as anorexigenic neurons, while it has been reported that POMC neurons promote cannabinoid induced feeding via upregulation of β-endorphin22. In this way, anorexigenic neurons could also produce an orexigenic factor. Although it remains to be elucidated which NPGM neurons in the MM or the IN contribute to control feeding behavior, it is quite possible that anorexigenic and orexigenic activities could be generated from the same NPGM/NPGL neurons. Future studies are necessary to reveal which particular NPGM neurons in the MM or the IN contribute to regulate feeding behavior. Furthermore, because the expression of NPGM alters during development, and because we used chicks of a single age (5- or 6-day-old) for i.c.v. injections in the present study, it would be interesting to investigate the effect of NPGM on food intake in chicks of different ages (1-day-old to adults).
NPGM-immunoreactive fibers were mainly observed in the hypothalamus, including the MM and the IN. Within the MM and the IN, some neuropeptides including NPY and α-MSH, derived from POMC, participate in a variety of physiological functions3,4,5,6. To investigate the correlation between NPGM and known factors, we analyzed the localization of several bioactive substances, such as neurotensin, GnIH, 26RFa (QRFP), and histamine through our previous and preliminary experiments18,19,20,21. We found that that NPGM-expressing cells are localized close to histaminergic neurons. Recently, we characterized HDC, which is a rate-limiting enzyme for histamine synthesis in chicken18. As a result, we found that neuronal histamine is produced in the MM and i.c.v. injection of histamine suppresses feeding behavior18. In the present study, morphological analysis showed that NPGM-containing neurons might also produce histamine in the MM. These results suggest that NPGM, histamine, and NPGL are produced in the same cells in the MM in chicks. In mammals, it has been demonstrated that neuronal histamine contributes to various functions, namely, feeding, energy homeostasis, sleep/wakefulness, motor behavior, and cognition23,24,25. For instance, it has been suggested that histaminergic neurons are activated by fasting26. Furthermore, histamine neurons are also activated by NPY and may terminate feeding elicited by NPY in mice27. This result suggests that histaminergic neurons in the MM maintain basal levels of food intake in birds. Consequently, NPGM produced in histaminergic neurons may act to maintain feeding behavior.
The IN in the avian hypothalamic infundibulum is known to be related to regulation of feeding and growth via NPY/AgRP, POMC, GHRH neurons, and a few other candidates4,5,6,28. Therefore, NPGM in the IN may be related to these functions. In this study, we showed that NPGM suppressed feeding, whereas NPGL induced feeding and body growth in chicks14,17. There was almost no co-localization of NPGM and NPGL in the IN. In addition, the expression of NPGM and NPGL mRNAs during post-hatch development showed opposite patterns. Thus, the function, localization and developmental expression pattern of NPGM in the IN is different from that of NPGL. It is well known that food intake in chicks increases in association with body growth. It is possible that a decrease in anorexigenic NPGM expression, and an increase in orexigenic NPGL expression combine to influence post-hatch development. Future studies on the developmental functions of NPGM and NPGL in individual nuclei, the IN, or the MM, will elucidate complex feeding mechanisms in chicks.
In summary, we have characterized a cDNA encoding a novel secretory protein, NPGM, in the chicken hypothalamic infundibulum. We have revealed the distribution and co-localization of NPGM with NPGL and histamine in the MM. NPGM and histamine display a similar expression pattern during post-hatch development and both have anorexigenic action. Although various factors regulating energy homeostasis have been discovered, including NPY, AgRP, α-MSH, and CRH in birds, the mechanisms of energy homeostasis cannot be explained by these factors alone. We expect that a further investigation of NPGM and NPGL will lead to a greater understanding of the mechanisms regulating energy homeostasis.
Methods
Animals
Experimental protocols were approved by the Institutional Animal Care and Use Committee of Hiroshima University (permit number: C13-13) and were in accordance with the Guide for the Care and Use of Laboratory Animals prepared by Hiroshima University.
Male layer chicks (Gallus domesticus, 1-day-old) were purchased from a commercial company (Nihon Layer, Gifu, Japan) and housed in a windowless room, maintained at 28 °C on a 20-hour light (4:00–24:00): 4-hour dark (0:00–4:00) cycle. Chicks were provided food and tap water ad libitum.
RNA and cDNA preparation
Chicks were sacrificed by decapitation. Telencephalon, diencephalon, mesencephalon, cerebellum, and hypothalamic infundibulum were dissected out and snap-frozen in liquid nitrogen for further RNA processing. Total RNA from chicken brain tissues was extracted using TRIzol reagent or an RNAqueous-Micro kit (Life Technologies, Carlsbad, CA) in accordance with the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA using a ReverTra Ace kit (TOYOBO, Osaka, Japan). These experiments were performed according to the manufacturer’s instructions.
Cloning the full-length cDNA of chicken NPGM from the chick hypothalamus
The method of cloning full-length cDNA was similar to that described in a previous report21. We first found the sequence of NPGM as the paralogous gene of NPGL by a BLAST search. We performed molecular cloning of the full-length cDNA. Real-time PCR showed that NPGM mRNA was highly expressed in the hypothalamic infundibulum, as described in Fig. 2a. Therefore, cDNA synthesized from the mRNA extracted from the hypothalamic infundibulum was used as a source for cloning. Primers for cloning full-length cDNA were designed from the predicted nucleotide sequences in the NPGM as follows; sense primer: 5′-ATGGAATTCATGTGGAAGAG-3′ (nucleotide no. 1 to 20 from the ATG initiation codon) and anti-sense primer: 5′-AGCATCTACAGTAAATGCTG-3′ (nucleotide no. 474 to 493). Nucleotide sequences were determined using an ABI PRISM BigDye Terminator Ready Reaction Cycle Sequencing kit (Applied Biosystems, Foster City, CA), an ABI PRISM 310 Genetic Analyzer (Applied Biosystems), and DNASIS Pro software (Hitachi Software Engineering, Kanagawa, Japan).
Quantitative RT-PCR
PCR amplifications were conducted with the THUNDERBIRD SYBR qPCR Mix (TOYOBO) using the following procedure: 95 °C for 20 s, followed by 40 cycles at 95 °C for 3 s and at 60 °C for 30 s using Step One Real-time Thermal Cycler (Applied Biosystems). The primer sequences are listed in Supplementary Table 1-1. Relative quantification for each expression was determined by the 2−ΔΔCt method using β-actin (ACTB) as an internal control29.
In situ hybridization for the detection of NPGM and NPGL
The chicks (1-day or 15-day-old) were killed by decapitation. Their brains were fixed in 4% paraformaldehyde (PFA) solution overnight at 4 °C and then placed in a refrigerated solution (30% sucrose in 10 mM phosphate-buffer, pH 7.4) until they sank. The tissues were embedded in Tissue-Tek OCT Compound (Sakura Finetek, Tokyo, Japan) and sectioned coronally at a thickness of 14 µm with a cryostat at −20 °C. The sections were then mounted on slides. Digoxigenin (DIG)-labeled antisense and sense RNA probes were produced from a section of the small protein precursor cDNA sequence using a DIG RNA Labeling kit with SP6 and T7 polymerases (Roche Diagnostics, Basal, Switzerland). RNA probe labeling was performed according to the manufacturer’s instructions. The primer sequences are listed in Supplementary Table 1-2. Single staining with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) was performed as previously described21. Hybridizations were performed overnight at 60 °C or 55 °C for NPGM or NPGL, respectively. The sections were incubated with blocking solution [1% bovine serum albumin, BSA; 1% normal goat serum; 0.3% Triton-X100 in phosphate buffered saline (PBS)], followed by incubation with alkaline phosphatase-labeled anti-DIG Fab fragments (1: 1000 dilution, 1 093 274; Roche) in blocking solution for 1 h. Signals were detected after immersion of the sections overnight in the NBT/BCIP solution. The DIG labeled sense probe was used as a control to verify specificity.
Production of chicken NPGM and antibodies
The procedure for the production of recombinant NPGM was similar to that described previously30. We prepared NPGM combined with the elongated C-terminal Gly (NPGM-Gly) as a recombinant protein using the Escherichia coli (E. coli) expression system. We transformed E. coli using the pCold-TF expression vector (TaKaRa Bio, Shiga, Japan) containing the NPGM sequence. The recombinant small protein was purified on a Ni2+ column (Qiagen, Venlo, Netherlands), and His6-TF-tag was removed with Factor Xa (New England Biolabs, Ipswich, MA). The disulfide bond bridge was formed by potassium ferricyanide. The small protein was purified by reverse phase high-performance liquid chromatography (RP-HPLC) using C8 column (10 × 150 mm; YMC, Kyoto, Japan) at a flow rate of 0.5 mL/min for 40 min with a linear gradient of 40–60% acetonitrile containing 0.1% trifluoroacetic acid. The solvent was evaporated and lyophilized. The C-terminus of recombinant NPGM-Gly was amidated using an amidating enzyme, and the mature form was finally purified by RP-HPLC.
Rabbit antisera against recombinant NPGM were produced following our published method21. NPGM solution was mixed with Freund’s complete adjuvant and injected into four rabbits. After a booster injection, blood was collected from the rabbits. Optimal serum with the highest titer was selected via a dot-blot analysis and the antibody specificity of NPGM or NPGL was confirmed via enzyme-linked immunosorbent assay (ELISA). The NPGL antibody was prepared in guinea pig as described previously17. Competitive ELISA was performed following our published method21. In brief, different concentrations of synthetic NPGM or NPGL (0.01–1000 pmol) were added with rabbit anti-NPGM antibody (1: 1000 dilution) or guinea pig anti-NPGL antibody (1: 1000 dilution) to each antigen-coated well of a 96-well microplate (96-well microplates PS F-bottom; Greiner Bio-One, Frickenhausen, Germany), and incubated for 1 h at 37 °C. After reaction with alkaline phosphatase-labeled goat anti-rabbit IgG (1: 1000 dilution, AP-1000; Vector Laboratories, Burlingame, CA) or anti-guinea pig IgG (1: 1000 dilution, ab7140; abcam, Cambridge, MA), immunoreactive products were obtained in a substrate solution of p-nitrophenylphosphate, and the absorbance was measured at 405 nm on a microtiter plate reader (Multiskan GO; Thermo Fisher Scientific, Kanagawa, Japan).
Immunohistochemistry (IHC) for the detection of NPGM and NPGL
The chicks (1-day or 15-day-old) were sacrificed by decapitation. The brain fixation method was performed as described above; the brain was sectioned into 14-µm slices. The sections were then mounted on slides and incubated with 0.3% hydrogen peroxide (H2O2) in absolute methanol for 30 min, for the immunoenzyme technique. After blocking with blocking solution (1% normal goat serum and 1% BSA in PBS containing 0.3% Triton X-100) for 1 h, the sections were incubated with rabbit anti-NPGM antibody (1: 1000 dilution) overnight at 4 °C. The primary immunoreaction was executed by incubation with biotinylated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) as a secondary antibody. Immunoreactivity was detected with an ABC kit (VECTASTATIN Elite Kit; Vector Laboratories), followed by diaminobenzidine (DAB) reaction. Preadsorption was performed as a control using the antigen (10 µg/mL). The antigen and primary antibody were preadsorbed overnight prior to the running of immunohistochemistry in the same way as for the non-control slides.
In the case of double immunohistochemical analysis of NPGM and NPGL, rabbit anti-NPGM antibody (1: 1000 dilution) and guinea pig anti-NPGL antibody (1:1000 dilution), as primary antibodies, Cy3-conjugated donkey anti-rabbit IgG (1: 1000 dilution, 711-165-152; Jackson ImmunoResearch, West Grove, PA) for the detection of NPGM, and Alexa Fluor 488-conjugated donkey anti-guinea pig IgG (H + L) (1: 1000 dilution, 706-545-148; Jackson ImmunoResearch) for the detection for NPGL, as secondary antibodies, were used.
Double staining of HDC and NPGM using in situ hybridization and immunohistochemistry
We detected HDC precursor mRNA and mature NPGM using in situ hybridization and immunohistochemistry, respectively. Brain sections were hybridized using DIG-labeled probe overnight at 50 °C for the detection of HDC, as described previously18. The hybridization protocol of in situ hybridization was the same until the hybridization, as described above. The primer sequences are listed in Supplementary Table 1-2. After hybridization, we carried out immunohistochemistry of NPGM with the same method, as described above. After the DAB reaction, DIG was detected by the HNPP Fluorescent Detection Set (Roche).
Intracerebroventricular (i.c.v.) injection
Five or six-day-old chicks were injected i.c.v. with NPGM according to a previously reported method31. The head of the chick was inserted into an acrylic box with a hole in the top plate. The injection coordinates targeting the left lateral ventricle were 3 mm anterior from the parietal bone, 1 mm lateral from the sagittal suture, and 3 mm ventral from the surface of the skull. NPGM (0.2, 1.0, and 5.0 nmol/10 µl) was dissolved in 30% propylene glycol at pH 8.0. The NPGM solution was injected through the hole using a micro-syringe at a volume of 10 µL.
Statistical analysis
Data were analyzed using Student’s t-test for two groups. In addition, data from three or five groups were statistically analyzed using one-way analysis of variance with Tukey’s test as a post-hoc test, as appropriate. The statistical significance level was set at P < 0.05. All results are presented as the mean ± standard error of the mean (SEM).
Change history
13 April 2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
Morton, G. J., Meek, T. H. & Schwartz, M. W. Neurobiology of food intake in health and disease. Nature Rev. Neurosci. 15, 367–378 (2014).
Song, Z., Everaert, N., Wang, Y., Decuypere, E. & Buyse, J. The endocrine control of energy homeostasis in chickens. Gen. Comp. Endocrinol. 190, 112–117 (2013).
Richards, M. P. Genetic regulation of feed intake and energy balance in poultry. Poult. Sci. 82, 907–16 (2003).
Kameda, Y., Miura, M. & Nishimaki, T. Localization of neuropeptide Y mRNA and peptide in the chicken hypothalamus and their alterations after food deprivation, dehydration, and castration. J. Comp. Neurol. 436, 376–388 (2001).
Mirabella, N., Esposito, V., Squillacioti, C., De Luca, A. & Paino, G. Expression of agouti-related protein (AgRP) in the hypothalamus and adrenal gland of the duck (Anas platyrhynchos). Anat. Embryol. (Berl). 209, 137–141 (2004).
Gerets, H. H. J., Peeters, K., Arckens, L., Vandesande, F. & Berghman, L. U. C. R. Sequence and distribution of pro-opiomelanocortin in the pituitary and the brain of the chicken (Gallus gallus). J. Comp. Neurol. 262, 250–262 (2000).
Furuse, M., Matsumoto, M., Saito, N., Sugahara, K. & Hasegawa, S. The central corticotropin-releasing factor and glucagon-like peptide-1 in food intake of the neonatal chick. Eur. J. Pharmacol. 339, 211–214 (1997).
Vijayan, E. & McCann, S. M. Suppression of feeding and drinking activity in rats following intraventricular injection of thyrotropin releasing hormone (TRH). Endocrinology 100, 1727–1730 (1977).
Furuse, M. et al. Intracerebroventricular injection of ghrelin and growth hormone releasing factor inhibits food intake in neonatal chicks. Neurosci. Lett. 301, 123–126 (2001).
Tachibana, T. & Tsutsui, K. Neuropeptide control of feeding behavior in birds and its difference with mammals. Front. Neurosci. 10, 485 (2016).
Qu, D. et al. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature 380, 243–247 (1996).
Nakazato, M. et al. A role for ghrelin in the central regulation of feeding. Nature 409, 194–198 (2001).
Vaccarin, F. J., Bloom, F. E., Rivier, J., Vale, W. & Koob, G. F. Stimulation of food intake in rats by centrally administered hypothalamic growth hormone-releasing factor. Nature 314, 167–168 (1985).
Ukena, K. et al. Identification of a cDNA encoding a novel small secretory protein, neurosecretory protein GL, in the chicken hypothalamic infundibulum. Biochem. Biophys. Res. Commun. 446, 298–303 (2014).
Matsuura, D. et al. Neurosecretory protein GL, a hypothalamic small secretory protein, participates in energy homeostasis in male mice. Endocrinology 158, 1120–1129 (2017).
Iwakoshi-Ukena, E. et al. Neurosecretory protein GL stimulates food intake, de novo lipogenesis, and onset of obesity. eLife 6, e28527 (2017).
Shikano, K. et al. Effects of chronic intracerebroventricular infusion of neurosecretory protein GL on body mass and food and water intake in chicks. Gen. Comp. Endocrinol 256, 37–42 (2017).
Bessho, Y. et al. Characterization of an avian histidine decarboxylase and localization of histaminergic neurons in the chicken brain. Neurosci. Lett. 578, 106–110 (2014).
Masuda, K. et al. Identification of neurotensin and LANT-6 and localization of mRNA encoding their precursor in the chicken brain. Zoolog. Sci. 31, 353–359 (2014).
Ukena, K., Ubuka, T. & Tsutsui, K. Distribution of a novel avian gonadotropin inhibitory hormone in the quail brain. Cell Tissue Res. 312, 73–79 (2003).
Ukena, K. et al. Identification, localization, and function of a novel avian hypothalamic neuropeptide, 26RFa, and its cognate receptor, G protein-coupled receptor-103. Endocrinology 151, 2255–2264 (2010).
Koch, M. et al. Hypothalamic POMC neurons promote cannabinoid-induced feeding. Nature 519, 45–50 (2015).
Masaki, T. et al. Neuronal histamine regulates food intake, adiposity, and uncoupling protein expression in agouti yellow (Ay/a) obese mice. Endocrinology 144, 2741–2748 (2003).
Yanai, K. et al. Behavioural characterization and amounts of brain monoamines and their metabolites in mice lacking histamine H1 receptors. Neuroscience 87, 479–487 (1998).
Takahashi, K., Lin, J. & Sakai, K. Neuronal activity of histaminergic tuberomammillary neurons during wake-sleep states in the mouse. J. Neurosci. 26, 10292–10298 (2006).
Valdés, J. L. et al. The histaminergic tuberomammillary nucleus is critical for motivated arousal. Eur. J. Neurosci. 31, 2073–2085 (2010).
Ishizuka, T. et al. A role of the histaminergic system for the control of feeding by orexigenic peptides. Physiol. Behav. 89, 295–300 (2006).
Thang, L., Cao, J., Wang, Z., Dong, Y. & Chen, Y. Melatonin modulates monochromatic light-induced GHRH expression in the hypothalamus and GH secretion in chicks. Acta Histochem. 118, 286–292 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔC(T) method. Methods 25, 402–408 (2001).
Masuda, K., Furumitsu, M., Taniuchi, S., Iwakoshi-Ukena, E. & Ukena, K. Production and characterization of neurosecretory protein GM using Escherichia coli and Chinese Hamster Ovary cells. FEBS Open Bio 5, 844–851 (2015).
Davis, J. L., Masuoka, D. T., Gerbrandt, L. K. & Cherkin, A. Autoradiographic distribution of L-proline in chicks after intracerebral injection. Physiol. Behav. 22, 693–695 (1979).
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science KAKENHI Grants (JP22687004, JP26291066, and JP15KK0259 to K.U., and JP25440171 and JP16K07440 to E.I.-U.), Grant-in-Aid for JSPS Fellows (15J03781 to K.S.), the Toray Science Foundation (K.U.), the Kieikai Research Foundation (K.U.), and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (K.U.).
Author information
Authors and Affiliations
Contributions
K.U. conceived and designed the experiments. K.S., Y.B., M.K., E.I.-U., S.T., M.F., and T.T. performed the experiments and analyzed the data. G.E.B. and L.J.K. were involved in experimental design and editing of the manuscript. K.U. and K.S. contributed to the preparation and editing of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shikano, K., Bessho, Y., Kato, M. et al. Localization and function of neurosecretory protein GM, a novel small secretory protein, in the chicken hypothalamus. Sci Rep 8, 704 (2018). https://doi.org/10.1038/s41598-017-18822-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18822-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.