Abstract
The sulfate reducing bacterium Desulfovibrio desulfuricans inhabits both the human gut and external environments. It can reduce nitrate and nitrite as alternative electron acceptors to sulfate to support growth. Like other sulphate reducing bacteria, it can also protect itself against nitrosative stress caused by NO generated when nitrite accumulates. By combining in vitro experiments with bioinformatic and RNA-seq data, metabolic responses to nitrate or NO and how nitrate and nitrite reduction are coordinated with the response to nitrosative stress were revealed. Although nitrate and nitrite reduction are tightly regulated in response to substrate availability, the global responses to nitrate or NO were largely regulated independently. Multiple NADH dehydrogenases, transcription factors of unknown function and genes for iron uptake were differentially expressed in response to electron acceptor availability or nitrosative stress. Amongst many fascinating problems for future research, the data revealed a YtfE orthologue, Ddes_1165, that is implicated in the repair of nitrosative damage. The combined data suggest that three transcription factors coordinate this regulation in which NrfS-NrfR coordinates nitrate and nitrite reduction to minimize toxicity due to nitrite accumulation, HcpR1 serves a global role in regulating the response to nitrate, and HcpR2 regulates the response to nitrosative stress.
Similar content being viewed by others
Introduction
Many strains of sulfate reducing bacteria such as Desulfovibrio vulgaris that have been isolated from natural environments outside warm blooded animals lack genes for nitrate reduction, but with few exceptions, they are able to reduce nitrite to ammonia1,2,3,4,5,6,7,8. Nitrite reduction is catalysed by the periplasmic NrfHA nitrite reductase4,9,10,11. Amongst the exceptions is D. alaskensis strain G20, which lacks the nrfHA genes and is extremely sensitive to nitrite toxicity2,7. Although originally there were conflicting reports concerning whether nitrite reduction is constitutive or inducible, it is now well established that nrfHA is expressed in the absence of nitrate or nitrite at a level that is sufficient to provide protection against the toxicity of nitrite produced, for example, by other bacteria that share their environment2,3,4,12,13. This background level increases significantly in the presence of nitrite11,14. Nitrite induction in D. vulgaris was recently shown to be dependent upon nitrite activation of a σ54-dependent two component regulatory system, NrfS-NrfR11,15. Boinformatic analysis revealed that the same is likely to be true for D. desulfuricans 11.
Unlike D. vulgaris, D. desulfuricans is able to use nitrate as an alternative to sulfate as the terminal electron acceptor to support growth2,12,13,16. Nitrate reduction is catalysed by a periplasmic nitrate reductase encoded in the napCMADGH genes14. Expression of the nitrate reductase operon in the most studied type strain, D. desulfuricans 27774, is induced during growth in the presence of nitrate but repressed by sulfate, even in the presence of nitrate12,14. This implies that nitrate reduction is regulated by at least two mechanisms, one for nitrate induction, the other for sulfate repression. We and others have noted the presence of potential binding sites for the NrfS-NrfR two-component regulatory system in the nap regulatory region11,16. No other potential binding sites for NrfR were found in the D. desulfuricans genome17.
Nitric oxide is an obligate intermediate during denitrification, but bacteria that reduce nitrate to ammonia also generate small quantities of NO, which in turn activates a protective nitrosative stress response. The main source of NO for free-living bacteria is nitrite generated either as a product of their own metabolism, or by other bacteria that share their environment. Bacteria that live in the bodies of warm-blooded animals are also exposed to NO generated from arginine as part of the host defence mechanisms. Homologues of genes that protect bacteria from nitrosative stress can be identified in the genomes of sulfate reducing bacteria15,16,18,19,20,21. They include transcription factors such as HcpR and genes that they regulate such as hcp encoding the hybrid cluster protein, Hcp. We recently showed that Hcp in enteric bacteria is a high affinity nitric oxide reductase that protects cytoplasmic proteins from nitrosative damage by NO generated as a side product of nitrite reduction to ammonia22. There is evidence that the same is true for Hcp in sulfate reducing bacteria23.
Sulfate reducing bacteria vary in that while some include single genes encoding Hcp and its transcription regulator, HcpR, others encode two or even more copies of these genes. For example, in D. vulgaris there are two copies of the hcp gene and also two copies of the gene for the flavodiiron protein ROO24. Both have been implicated in the protection against nitrosative stress6,24,25,26. Synthesis of Hcp2 is strongly induced by exposure to NO, and deletion of hcp2 results in increased sensitivity to NO27.
Unlike D. vulgaris, strains of D. desulfuricans and its close relatives are commonly found in the bodies of warm blooded animals and can readily be isolated from human feces28,29. Two of the 5 transcription factors of the Crp-FNR family encoded in the D. desulfuricans 27774 genome, HcpR1 and HcpR2, are both predicted to regulate the response to nitrosative stress. Correlations were noted between the ability of sulfate reducing bacteria to reduce nitrate, their ability to survive in the human body, and the presence of genes for both HcpR1 and HcpR230. A fascinating result from the previous study was that although NO induced expression of the hcp and hcpR1 genes, HcpR2 was shown to be the transcription factor that regulates Hcp synthesis, but hcpR2 transcription is not induced by NO.
HcpR1 binds to a perfect inverted repeat sequence, 5′-TGTGA-N6-TCACA, which is identical to the consensus binding site for the c-AMP receptor protein, CRP, in Escherichia coli 14,19,30. Binding of HcpR1 to this site regulates hcpR1 transcription19,30. There are also inverted repeat sequences predicted to be the binding sites for HcpR1 immediately upstream of the napC promoter, suggesting that HcpR1 might regulate nitrate reduction as well as its own synthesis. This suggests that HcpR1 regulates the synthesis of enzymes that enable D. desulfuricans both to reduce nitrate and protect itself from toxic products generated during nitrate and nitrite reduction. The combined data from these earlier studies indicate that multiple transcription factors are likely to be involved in the regulation of nitrate, nitrite and NO reduction. They suggest that while HcpR1 serves a more global role, NrfS-NrfR is a dedicated system that senses the presence of nitrite or nitrate and coordinates nitrate and nitrite reduction to minimize toxicity due to nitrite accumulation. The first aims of the current work were to determine how expression of nitrate and nitrite reductase genes are regulated in response to substrate availability, and demonstrate that HcpR1 binds to the regulatory region of the nap gene cluster.
Although attempts by us and other laboratories to isolated specific mutants of D. desulfuricans have been unsuccessful, global transcriptome analysis has been used successfully to analyse how other sulphate reducing bacteria respond to oxidative stress, heat shock and electron acceptor availability31,32,33,34. We have therefore used RNA-seq based transcriptome analysis to determine the global response of D. desulfuricans 27774 to nitrate or NO.
Results
Conserved nap gene clusters and their regulatory motifs in Desulfovibrio species
Many genomes of sulfate reducing bacteria have been sequenced since the original report of the structure and sequence of the napCMADGH gene cluster of D. desulfuricans 2777414. This NapM sequence was used to identify similar nap gene clusters in other bacteria. These blast searches revealed additional Desulfovibrio species with similar clusters, despite their widely different genome sizes and GC contents. Similar clusters were found in sulfite reducing Bilophila species35. Some of the bacteria with similar NapM sequences lack the napC gene, but retain the napMADGH sequences (Supplementary Table S1). The napM gene is present in all nap gene clusters found in sulfate reducing bacteria.
We previously showed that HcpR1 binds to a DNA fragment containing the sequence immediately upstream of the D. desulfuricans 27774 hcpR1 gene, suggesting that HcpR1 regulates its own synthesis30. We also reported the presence of three similar sequences in the napC regulatory region of this strain14. Potential HcpR1 binding sites were found in the regulatory regions of eight out of ten scanned napM sequences, although the location of these sites varies in different bacteria (Supplementary Table S1). The position of the sites in Desulfovibrio species is consistent with HcpR1 acting as a class 3 transcription activator, a mechanism in which the binding of transcription factors to two sites is required for optimal activation of gene expression36. In contrast, the predicted HcpR1 site in Deferribacter overlaps the translation start codon, consistent with a repression function. Binding sites similar to those bound by the two-component regulatory system NrfS-NrfR were also located upstream of nap gene clusters in these various sulphate reducing bacteria (Supplementary Table S1).
Binding of purified recombinant HcpR1 to inverted repeat sequences in the regulatory region of the nap gene cluster
To confirm that HcpR1 binds to the inverted repeat sequences, different concentrations of purified HcpR1 were incubated with a 32P end-labelled DNA fragment covering the napC promoter and regulatory region. Protein-DNA complexes were separated from unbound DNA by non-denaturing PAGE and visualized by autoradiography (Fig. 1(a)). A single high affinity complex was detected even at the lowest concentration of HcpR1 protein. At the highest concentration, a second band due to a low affinity complex was detected. For comparison, the napC upstream fragment was also incubated with increasing concentrations of E. coli CRP plus 200 μM c-AMP. In contrast to HcpR1 binding, two band shifts were readily detected even at relatively low protein concentrations. Thus although CRP binds with relatively high affinity to two sites in the napC regulatory region, HcpR1 binds only one site. The concentrations of HcpR1 used in these experiments were in the same range as both the estimated concentrations of other transcription factors in vivo and the concentrations of transcription factors used in previous in vitro experiments. We therefore assume that the observed binding of HcpR1 to promoter DNA is physiologically relevant37,38.
Comparison between the DNA binding affinities of HcpR1 and CRP for the nap promoter fragment and demonstration of binding to the pnap DNA fragment by DNaseI footprinting assays. (a) DNA binding affinities were assessed by EMSA. 32P-labelled nap promoter DNA fragment was incubated with increasing concentrations of HcpR1 alone, or CRP protein in the presence of 200 µM cAMP and then resolved by non-denaturing PAGE. Herring sperm DNA was also included in the incubation mixtures to act as non-specific competitor DNA. Free DNA, DNA-HcpR1 and DNA-CRP complexes are marked with arrows. (b) 32P-end-labelled nap promoter DNA fragment was incubated with increasing concentrations of HcpR1 protein and digested with DNaseI. Digest mixtures were then resolved by denaturing electrophoresis on urea acrylamide gels. Sequences were identified by including Maxam-Gilbert sequencing reactions (GA) on gels. Protected regions are marked by boxes and the positions of the three IR sequences are marked with pink arrows. Samples in tracks from left to right were incubated with 0, 5, 10, 20, 40, 80, 160, 360 and 720 nM HcpR1.
DNA footprinting was used to confirm that HcpR1 binds specifically to the site identified from the bioinformatics analysis (Fig. 1(b)). The high affinity DNA-binding site marked with a red box around all tracks containing HcpR1 corresponds to IR1, the inverted repeat sequence closest to the transcription start site. At higher HcpR1 concentrations, the second binding site IR2 marked with the upper red box was protected.
Effect of electron acceptor during growth on expression at the napC and nrfA promoters
RNA was isolated from cultures growing exponentially in the presence of sulfate, sulfite, nitrate or nitrite. Levels of nap and nrf transcripts were assayed by qRT-PCR. The napC mRNA was 80-fold more abundant during growth with nitrate than in the sulfate control, and 20-fold higher during growth in the presence of nitrite, but less abundant in the sulfite cultures than in the sulphate cultures (Fig. 2a).
qRT-PCR of genes involved in nitrate and nitrite reduction in D. desulfuricans. RNA was purified from cells grown on medium containing nitrate, nitrite, sulfate or sulfite as the sole terminal electron acceptor. RNA was reverse-transcribed with random hexamers. Transcript levels were normalised against polA levels. Expression levels are derived from three biological replicates and are normalised to those given by sulfate grown cells. Stars indicate data derived from ΔCt values statistically significantly different to that for sulfate-grown cultures.
Transcription at the nrfA promoter was also induced 6-fold during growth in the presence of nitrate, in agreement with the RNA-seq data, but far more highly induced, 18-fold, during growth with nitrite than with nitrate (Fig. 2b). Levels of nrfA transcripts in cultures growing with sulfate or sulfite were similar. These results demonstrated that, although expression of both nap and nrf are regulated in response to the availability of nitrate or nitrite, they are not regulated coordinately.
Global response of the D. desulfuricans transcriptome to nitrate and NO
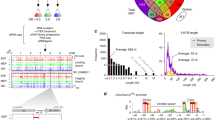
RNA-seq was used to determine the full extent of changes in the transcriptome in response to replacing sulfate by nitrate as the terminal electron acceptor to support growth. Some of the many anticipated changes would be direct effects on genes required for nitrate reduction or for energy conservation. Secondary consequences were anticipated due to the production of nitrite, which is toxic. Finally, some nitric oxide is always generated as a side product during nitrite reduction to ammonia. This would result in a nitrosative stress response. As a first step to distinguish between responses to nitrate or to nitrosative stress, RNA was isolated from bacteria during growth under four conditions: sulfate as the only terminal electron acceptor (control); nitrate as the only electron acceptor; sulfate-grown cultures supplemented with 7.5 μM NO; and nitrate-grown cultures supplemented with 7.5 μM NO. Consistent with previous reports, the yield in the nitrate cultures was almost double that of the sulfate control, but there was a lag in growth before growth on nitrate commenced2,9,14,39. As the addition of NO also caused a lag in growth, bacteria for RNA analysis were harvested after growth had resumed, which was 4 h after the addition of NO when most of the NO had been reduced. Quantitative RT-PCR data had previously shown that similar transcription responses were detected for more than 20 h after NO addition30, so the 4 h exposure allowed the response to NO to be determined. Note, however, that nitrite was generated in cultures supplemented with nitrate, some of which was converted to NO, though the majority was reduced to ammonia. Supplementary Table S2 indicates the overall number of reads per sample at the different steps of the analysis (sequencing, genome mapping, gene assignment). The analysis of inter-sample correlation (Supplementary Fig. S1) shows a consistent grouping according to the electron acceptor (Nitrate or Sulfate). However, within the Sulfate cluster, samples SN1 and S1 are separated from the others. They were therefore considered as outliers and discarded from the analysis of differential expression. Additional statistical analysis that justifies the omission of these data are provided as a technical report at the end of the Supplementary Information. The combined data allowed 4 sets of differential analyses to be completed (Table 1).
Response of the D. desulfuricans transcriptome to growth in the presence of nitrate instead of sulfate
Expression of 310 genes was significantly higher and expression of a further 362 genes was lower during growth in the presence of nitrate compared with growth in the presence of sulphate (Supplementary Table S3). For simplicity, these differences in gene expression are subsequently referred to as induction and repression without implying specific mechanisms, which are unknown. It suggests that switching from sulfate to nitrate respiration involves a rather large transcriptomic change (variation of expression of about 27% of the genes). Note that these groups should include genes specifically induced or repressed not only by nitrate, but also by nitrite and NO generated from nitrate, and therefore should be the largest groups of genes found to be differentially regulated in the study. As this list will include many transcripts that respond to secondary or even tertiary consequences of nitrate reduction, transcripts most strongly induced or repressed by nitrate or its reduction products are listed in Tables 2 and 3, respectively. Comparison of the COG distribution of the genes differentially expressed with the D. desulfuricans whole genome COG distribution revealed that the C (energy production and conversion) and N (cell motility) categories were significantly over-represented in the differentially expressed gene lists compared to the whole genome (Fig. 3 and Supplementary Table S4), showing that these categories were likely important for switching from sulfate respiration to nitrate respiration.
Distribution of the differentially expressed genes in COG categories (in percentage) during growth with nitrate compared to sulfate (in dark grey) and in the whole D. desulfuricans genome (in light grey). The two filled back bars and asterisks beside the first two entries indicate the COG classes showing significant over-representation (f < 0.001, where “f” stands for Family-Wise Error Rate). Full details of the statistical analysis of the data are provided in the Supplementary Information as a technical report and Supplementary Table S6.
The genes most highly induced during growth with nitrate included the expected periplasmic nitrate reductase, Ddes_0614–0619, and nitrite reductase, Ddes_0081–0082 (Table 2 and Supplementary Table S3). It also includes two clusters, Ddes_1668–1673 and Ddes_1238–1244, potentially encoding respiratory NADH-dehydrogenases that transfer electrons from NADH to the quinone pool. Particularly intriguing is the strong induction by nitrate of a two-component regulatory system in which Ddes_0843 encodes an Rrf2 family sensor protein and Ddes_0844 encodes a receiver domain protein. Why genes for aromatic amino acid biosynthesis and flagella proteins are also induced during growth in the presence of nitrate remains to be determined. Note also the strong induction by nitrate of the fumarate reductase genes. This was surprising because in enteric bacteria expression of the fumarate reductase operon is repressed by nitrate40,41,42.
The genes most significantly repressed in the presence of nitrate included a hypothetical gene transfer island, Ddes_0702–0725, that were repressed up to 100-fold (Supplementary Table S3). Other strongly repressed genes encode proteins for various iron uptake mechanisms, and a cluster of genes for nitrogen fixation that are probably in an operon (Table 3 and Supplementary Table S3). Genes for six putative transcription factors were repressed by nitrate: the AraC family transcription factor encoded by Ddes_0430; the σ54-dependent Fis family transcription factor encoded by Ddes_0219; the ArsR family transcription factor encoded by Ddes_1925; the PAS/PAC sensor histidine kinase encoded by Ddes_0768; the response regulator receiver protein encoded by Ddes_0018; and an XRE family transcription factor encoded by Ddes_2233. This is in agreement with the fact that switching from sulfate to nitrate as terminal electron acceptor requires a fine tuning of the metabolic pathways controlled by several transcription factors. As previously reported the gene for the hybrid cluster protein, hcp (Ddes_1829) was induced in the presence of nitrate (Supplementary Table S3). It was intriguing that genes encoding two enzymes involved in hydrogen peroxide scavenging, catalase (Ddes_1494) and the rubrerythrin (Ddes_0897), were repressed by nitrate.
Global effects of nitric oxide on gene expression
Comparison of data from sulfate grown cultures supplemented with NO with the sulfate control culture revealed that 31 genes were induced specifically by externally supplied NO, and 26 were repressed (Table 4). The two most strongly induced clusters were Ddes-0525 to 0528 and Ddes1828–1829 that are probably in two operons. The first group includes genes for an iron-sulfur protein, an FMN-binding protein, a flavodoxin, and Ddes_0528 encoding the NO-responsive transcription factor, HcpR1. The second group includes Ddes_1829 encoding the hybrid cluster protein (Hcp). Equally significant is the absence of induction of gene Ddes_1827 encoding the transcription factor, HcpR2. This confirmed our recent report that, although HcpR2 regulates Hcp synthesis in response to the availability of NO, hcpR2 expression was not induced by NO30. Two further clusters, Ddes_0288–0289 and Ddes_0334–0338 predicted to encode proteins for heme synthesis and in aromatic amino acid biosynthesis, respectively, were also up-regulated in the presence of NO.
At least two di-iron proteins protect enteric bacteria against nitrosative stress. One is the flavorubredoxin, NorV, with its characteristic β-lactamase fold. The second is the hemerythrin-like di-iron containing domain protein, YtfE (also known as RIC)43,44,45,46,47. An orthologue of NorV has been identified in D. gigas: this is the flavodiiron protein, ROO23,26, which is also the orthologue of Ddes_2012. Surprisingly, until the current work, no orthologue of YtfE has been found in sulphate reducing bacteria. It is therefore significant that expression of D_des 1165 was strongly induced under conditions of nitrosative stress (Table 4). HHpred analysis indicates that the N-terminus of Ddes_1165 is predicted to adopt a fold like that of YtfE from E. coli (probability 99.3%) whilst the C-terminus likely adopts a PAS domain-type fold48. Crucially, Ddes_1165 residues H234, H262 and E239 are equivalent to YtfE residues H129, H160 and E133 which are involved in co-ordination of the di-iron centre49. Analysis of the Ddes_1165 sequence with the tool PHMMER also identifies the presence of hemerythrin-like and PAS domains in this protein. PAS domains are involved in diverse processes but are mediators of intermolecular interactions including protein:protein interactions, a function consistent with the proposed role of YtfE interacting with iron-sulfur cluster proteins43,44,45,46,47. Interestingly, in other studies, the D. vulgaris homolog of Ddes_1165 (DVU2590) was found to be transiently up-regulated by nitrite stress but unaffected by oxidative stress, demonstrating a nitrosative stress-specific response by this gene product50,51. We propose that Ddes_1165 is the equivalent of YtfE in D. desulfuricans and related SRB.
The predicted functions of NO-repressed genes varied greatly, suggesting that many of them might be secondary responses, for example, to nitrosative damage to various enzymes and transcription factors. This rather limited group included Ddes_1643, which is predicted to encode a σ54-dependent transcription factor, and Ddes_2150 encoding the precursor of the “Split Soret” cytochrome c. Note that based upon Northern blot analysis, it was previously reported that expression of Ddes_2150 is more induced during growth with nitrate than with sulfate52. The decreased expression of the large cluster from Ddes_0703 to 0715 encoding a hypothetical gene transfer island might be due either to repression or to gene loss in response to NO exposure (Table 4). With the exception of the Ddes_0663 to Ddes_0666 cluster, genes for ribosomal proteins were absent from the list of NO-repressed genes, suggesting that the differential analysis revealed responses to NO rather than to a decreased growth rate.
The effect of NO on cultures growing with nitrate as the terminal electron acceptor was also determined. Fewer genes were differentially expressed compared to when sulfate was used as terminal electron acceptor: 18 genes were induced, and only 2 were repressed (Supplementary Table S5). This result was consistent with our prediction that some NO will have been generated endogenously from nitrite formed as a product of nitrate reduction. Therefore, some NO-responsive genes would be induced or repressed even during growth in the presence of nitrate alone. This would decrease any additional response to NO when nitrate is also present. Responses to NO were found for the Ddes_0524–0528 gene cluster, confirming that transcription of these genes responds to NO rather than to nitrate, which was present in all of these cultures. Perhaps the most significant result is that the genes of previously unknown function, Ddes_1164 and Ddes_1165, were more strongly induced in the presence of both nitrate and NO than in the absence of nitrate. This strongly supports our proposal above that the diiron protein encoded by Ddes_1165 is the orthologue of YtfE in enteric bacteria.
Only two genes, Ddes_1501 encoding a small GTP-binding protein, and Ddes_1581, encoding a hypothetical protein, were significantly repressed by exogenous NO in cultures growing with nitrate. These genes were not found to be differentially expressed in sulfate + NO versus sulfate conditions. Ddes_1501 was up-regulated in nitrate versus sulfate conditions, suggesting that it is important in nitrate respiration but its expression is down-regulated as a stress response to NO (Supplementary Table S3). Note that neither of the genes encoding enzymes involved in hydrogen peroxide scavenging, catalase (Ddes_1494) and the rubrerythrin (Ddes_0897), were differentially expressed when the cultures were challenged with exogenous NO with either nitrate or sulfate as electron acceptor. This suggests that their expression is not dependent on the presence of exogenous NO but rather to reaction products linked to the nature of the electron acceptor.
Only 7 genes responding to the presence of exogenous NO were found in common under sulfate and nitrate conditions. These included the gene cluster Ddes_0524–0528 as well as the genes Ddes_1165 and Ddes_0288. All of these genes were found to be induced in the presence of exogenous NO. Except for Ddes_1165, they were also found significantly up-regulated in nitrate versus sulfate conditions (Supplementary Table S3).
These results indicate that the genes induced by NO are almost certainly required to protect bacteria from nitrosative stress in one of two ways. Some such as Hcp (Ddes_1829) are involved directly as NO scavenging systems. Others such as enzymes involved in amino-acid metabolism might be involved indirectly, for example by limiting toxic effects of NO through mechanisms that need to be determined, or as secondary consequences of nitrosative damage to proteins that regulate their synthesis.
Confirmation that nitrate partially induces a nitrosative stress response
A final comparison of differentially expressed genes during growth with nitrate rather than sulfate when both sets of cultures were also challenged with NO provided independent confirmation of many of the above results (Supplementary Table S6). About 68% of the up-regulated genes are shared between the two conditions, with and without exogenous NO. The smaller number of transcripts induced by nitrate in the presence of NO than in its absence (211 instead of 310) was again consistent with the proposal that some genes were induced by NO produced from nitrite during nitrate reduction. Ddes_1581 was down-regulated during growth with nitrate + NO compared with sulfate + NO, suggesting that this gene responds to exogenous NO only during nitrate respiration (Supplementary Table S6).
Validation of selected RNAseq data by quantitative RT-PCR
To validate the RNA-seq data, expression of hcpR1, hcp, hcpR2, nap and nrf in the presence of nitrate, sulphate and nitric oxide were analysed by qRT-PCR (Supplementary Fig. S2a and b). No significant effects of nitrate or NO on hcpR2 expression were detected by qRT-PCR. The expression profiles of the other targets mirrored those obtained by the RNA-seq analysis, although larger relative changes in expression of napC and hcp were observed in the qRT-PCR analysis. The RNA-seq data were also consistent with our earlier studies14,30 that demonstrated up-regulation of nap, hcpR1, and hcp and no change in expression of hcpR2, in response to nitrate or nitric oxide.
Bioinformatic prediction of the extent of the HcpR1 regulon
The name HcpR was introduced to designate the transcription factor that regulates synthesis of the hybrid cluster protein and an associated electron transfer protein, FrdX18,19. Core regulons for HcpR in D. vulgaris and D. alaskensis were proposed based upon the presence of R, E and R residues in positions 1, 2 and 6 of the DNA recognition helix and hence its similarity to E. coli Crp. These residues are absent from the DNA recognition helix of D. desulfuricans HcpR2, but are present in HcpR1, which is only 24–25% identical to HcpR from the other two species and is not co-located with genes for either FrdX or Hcp.
To investigate the possible global role of HcpR1, a position specific scoring matrix (PSSM) strategy was used to scan the D. desulfuricans ATCC27774 genome for putative HcpR1 binding sites. A seed matrix derived from the HcpR1 consensus sequence TGTGA-N6-TCACA was used to scan the upstream sequences of all genes30 (Supplementary Fig. S3A). Sites having a p-value lower than 10−4 were used to build a “second-generation” matrix (Supplementary Fig. S3B), which in turn was used to scan upstream sequence of all genes to gather putative binding sites with more flexibility than the original seed matrix. This analysis returned 91 genes whose upstream region contains at least one predicted HcpR1 binding site with a p-value < 10− 4 (Supplementary Table S7). Among them are found the genes hcpR1 (Ddes_0528) and napC (Ddes_0614), for which electromobility shift assays (EMSA) showed that HcpR1 effectively binds to the promoter sequences30 with high affinity (Fig. 2). In addition, binding of HcpR1 to promoter sequences of the sat (Ddes_0454) and Ddes_1825 genes with a low affinity was also confirmed by EMSA experiments (Supplementary Fig. S4). However, neither of these two genes was differentially expressed in any of the conditions tested (Supplementary Table S7). Out of the 91 genes, 38 were differentially expressed in the nitrate versus sulfate conditions, 13 being up-regulated (including the napC gene) and 25 down-regulated (Supplementary Table S7). These 38 genes can be considered as an HcpR1 regulon. Only 5 genes were also similarly differentially expressed in sulfate + NO versus sulfate conditions, Ddes_0528 encoding HcpR1 being the only one up-regulated. The other four genes encode two hypothetical proteins (Ddes_1427 and Ddes_1642), a transcription factor (Ddes_1643) and a molybdopterin-containing protein (Ddes_1824). The much lower number of genes in this case could be linked to a dose-dependent response. We previously showed that purified HcpR1 binds heme to give a complex with oxidized and reduced spectra typical of a b-type cytochrome30. Based upon this evidence, it has been proposed that HcpR1 is a heme-containing protein able to react with NO, but how NO modulates the transcriptional regulatory function of HcpR1 is still unknown30.
Discussion
Many aspects of the diversity, environmental importance and potential for biotechnological exploitation of sulfate reducing bacteria for biodegradation or in the oil industry have been studied in depth53,54,55,56. In contrast, knowledge of their physiology, biochemistry, intermediary metabolism and gene regulation is far more limited. There are many gaps in knowledge of how electrons are transferred from primary dehydrogenases to a diverse range of terminal electron acceptors despite the availability of elegant structures of some of the redox protein complexes involved48,57,58,59,60,61,62,63. It is therefore potentially significant that the current study showed that the expression of genes for two NADH dehydrogenases is induced by nitrate, but a third potential NADH dehydrogenase, Ddes_1661, is repressed. This suggests that D. desulfuricans might form large electron transfer complexes in which proteins that reduce specific terminal electron acceptors form complexes with specific dehydrogenases, and that their synthesis might therefore be co-ordinately regulated. As current knowledge of operon structures and transcription start sites is extremely limited, mechanisms of gene regulation remain speculative.
We have previously reported that nitrate reduction by D. desulfuricans 27774 is tightly regulated by nitrate induction, which is over-ridden by sulfate repression14. Nitrate reduction is also strongly inhibited by sulfide generated from sulfate or sulfite reduction. We also proposed that the NO-sensitive transcription factor, HcpR1, is involved in the regulation of nitrate reduction30. In the current paper we demonstrated that, as expected, nitrite reduction is also tightly regulated, presumably by the NrfR-NrfS two-component regulatory system11,15 (Fig. 2). There is an almost perfect NrfR consensus binding site in the napM regulatory region, implying that the NrfR-NrfS system co-ordinately regulates nitrate and nitrite reduction to minimize the accumulation of nitrite. Although the pattern of regulation of nrfA expression is similar to our previously reported regulation of hcpR1 and hcp expression, there are no potential binding sites for HcpR1 or HcpR2 in the nrfHA regulatory region, and while nap and hcpR1 expression are regulated by HcpR1, it is HcpR2 that regulates hcp expression in response to the presence of NO. Clearly there are links between regulation by HcpR1, HcpR2 and NrfS-NrfR that merit further research. Use of alternative promoters appears to be the most likely mechanism for NrfR-dependent induction of the nap and nrfHA genes11. However, superimposed upon regulation of nap gene expression by NrfR is regulation by sulfate, nitrate, HcpR1 and hence by NO. The metabolic signal to which NrfS responds, nitrate or nitrite, remains to be revealed: we suggest it is most probably nitrite because the NrfR- NrfS system is present in many sulphate reducing bacteria that are able to reduce nitrite, but not nitrate. The mechanism by which HcpR2 activates hcp synthesis in response to the presence of NO also remains to be determined. Detailed molecular biological experiments will be required to reveal the mechanism of how HcpR1 regulates expression of the nap and hcpR1 genes, or how NO induces HcpR2-dependent expression of the hcp operon. What is becoming clear is that there are widely different levels of complexity in gene regulation in this bacterium. While this manuscript was in review, a paper describing the proteomic response of D. desulfuricans 27774 to nitrate was published on line. Only proteins soluble in a low ionic strength buffer were analysed, so inevitably there are major differences between the proteomic and our RNAseq data. However, increased accumulation of Nap proteins in response to nitrate was confirmed64.
Unexplained is why genes for aromatic amino acid biosynthesis are strongly induced during growth in the presence of nitrate, but genes for catalase and putative iron uptake are repressed. A possible explanation for the induction of fumarate reductase genes during growth with nitrate might be an increased need for succinyl CoA for heme synthesis, but this is mere speculation. These are just a few of the many research challenges revealed from the genome-wide RNA-seq data available from this study. Many genes were differentially repressed by nitrate compared to sulfate when both groups of culture were challenged with NO.
Despite the absence of genetic systems to test many hypotheses based upon results obtained in the current study, the RNA-seq data have revealed many insights into how D. desulfuricans responds to an alternative electron acceptor such as nitrate and nitrosative stress. One of the most striking results is the very limited overlap between genes induced by NO or by nitrate. Two noteworthy exceptions were genes encoding nitrate reduction and HcpR1 synthesis, both of which were predicted be regulated by HcpR114,19,30. This indicates that part of the nitrosative stress response is coordinated with a primary cause of stress, the generation of nitrite from nitrate. Consistent with this interpretation is the lower number of differentially expressed genes in response to NO in cultures with nitrate rather than sulfate as the primary electron acceptor (Table 1) and the lower induction or repression ratios observed in cultures supplemented with both nitrate and NO compared with sulfate plus NO (compare Supplementary Tables S2 and S3).
A possible model for the regulation of genes for nitrate and nitrite reduction is shown in Fig. 4. The model predicts that in the presence of sulfate but absence of NO (Fig. 4a), HcpR2 is competent for binding the downstream DNA target sequence and so expression of ylbA and hcp is repressed. Expression of nap is repressed by a currently unidentified repressor protein14. In the presence of both sulfate and NO, the Fe-S clusters of HcpR2 are damaged by NO leading to the loss of DNA binding by HcpR2 and derepression of ylbA and hcp. It is likely that NO is also sensed by HcpR1, probably via a heme ligand, leading to increased expression by an unknown mechanism of the hcpR1-wrbA-nimA-Ddes_0525-Ddes_0524 genes (Fig. 4b). The model assumes that the presence of nitrite is sensed by the NrfS-NrfR system leading to up-regulation of the divergent nrfHA operon (Fig. 4c). As NO is produced as a by-product of nitrite reduction, both the hcp and hcpR1 operons are also expressed. The presence of both NrfR and HcpR1 in the absence of sulfate leads to a relatively modest increase in expression of nap. Finally, in the presence of nitrate but absence of sulfate, the unidentified repressor of the nap gene cluster is inactivated, so nap expression is fully de-repressed. It is suggested that co-activation of nap is achieved via the activities of HcpR1 and NrfR. Rapid nitrate reduction would result in a burst of NO production leading to high levels of HcpR1 synthesis. HcpR1 could also function as a repressor of nap expression at high concentrations, providing the regulation of the nap cluster with a negative feedback mechanism. However, this is a minimal model as roles for neither sigma-54 nor Rex have been included. The nap and nrf promoters have sigma-54 sequence determinants and Rex binds to sites identical to HcpR1 and so these two regulators are also likely to influence expression of these loci65.
A model of the regulation of key genes involved in the response to nitrate or NO in D. desulfuricans. (a) In the presence of sulfate alone, HcpR2 binds as a repressor to the downstream DNA target sequence downstream of the hcpR2 gene, Ddes_1828, so expression of ylbA and hcp (Ddes_1827 and Ddes_1826, respectively) is repressed. Expression of the nap gene cluster, Ddes_0614–0619, is repressed by an unidentified repressor protein14. (b) In the presence of sulfate and NO, the Fe-S cluster of HcpR2, is damaged by NO leading to the loss of DNA binding by HcpR2 and de-repression of ylbA and hcpI (Ddes_1827 and Ddes_1826). NO is also sensed by HcpR1, probably via a heme ligand, leading to up-regulation by an unknown mechanism of the hcpR1-wrbA-nimA-Ddes_0525-Ddes_0524 (genes Ddes_0524-Ddes_0528). (c) The presence of nitrite is sensed by the NrfS-NrfR system leading to up-regulation of the divergent nrfHA operon (genes Ddes_0081 and Ddes_0082). Both the hcp and hcpR1 operons are expressed since NO is produced as a by-product of nitrite reduction. The presence of both NrfR and HcpR1 in the absence of sulfate leads to a relatively modest increase in expression of nap. (d) The presence of nitrate in the absence of sulfate leads to full de-repression of the nap gene cluster (Ddes_0614 – Ddes_0619) via the action of an unidentified repressor. Co-activation of nap is achieved via the activities of HcpR1 and NrfR. Rapid nitrate reduction results in a burst of NO production leading to high levels of HcpR1 synthesis. HcpR1 could also function as a repressor of nap expression at high concentrations, providing the regulation of the nap cluster with a negative feedback mechanism. Note that this is a minimal model as no roles for sigma-54 or Rex have been included65.
Methods
Media and growth conditions
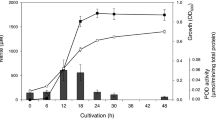
D. desulfuricans strain 27774 was stored at −80 °C as glycerol stocks. Cultures were initially grown in sealed serum bottles at 30 °C in Postgate medium B, which contains both sulfate and ferrous salts30,66. For growth and RNA experiments, exponential phase cultures of D. desulfuricans were sub-cultured in Postgate Zero medium (NaH2PO4.2H2O, 3 g l−1; KH2PO4, 0.5 g l−1; NH4Cl, 0.5 g l−1; CaCl2, 10 mg l−1; MgCl2.6H2O, 25 mg l−1; FeCl2.4H2O, 1.5 mg l−1; sodium lactate, 9 ml l−1; citric acid, 0.2 g l−1; Yeast extract, 1 g l−1; pH 7.0). Nitrate, sulfate and sulfite were added to a final concentration of 7.5 mM. NO was added to a final concentration of 7.5 μM. We have previously shown that 7.5 mM nitrite completely inhibits growth, but this strain is able to grow in the presence of sequential additions of nitrite to a final concentration of 2.5 mM, which was therefore the concentration of nitrite used in the growth experiments14.
Electromobility shift assays
DNA fragments containing promoter proximal DNA sequences were amplified from genomic DNA by PCR and cloned in pGEM T Easy vector (Promega). T Easy-derived plasmids containing promoter fragments were purified from E. coli, digested with appropriate restriction enzymes, typically EcoRI and HindIII, and then treated with CIP. The excised promoter fragments were purified by electroelution, phenol-chloroform extraction and ethanol precipitation.
Purified DNA fragments were end-labelled with using T4 polynucleotide kinase (New England Biolabs, UK). The end-labelling reaction consisted of 2 µl polynucleotide kinase buffer, 1 µl γ 32P-ATP, 10–16 µl of DNA fragment, 1 µl T4 polynucleotide kinase and sterile distilled water to a total volume of 20 µl. Labelling reactions were incubated at 37 °C for 1 hour and then excess γ 32P-ATP was removed by passing the reaction mixture through two 200 µl bed volumes of Sephadex G-50 which had be pre-equilibrated with 1 x Tris-EDTA buffer pH 8.0.
EMSA incubation mixtures were prepared to a final volume of 10 µl and included: 0.2–1 µl of radiolabelled DNA fragment (2–4 ng of DNA fragment per EMSA incubation), 1 µl of 10 x binding buffer (20 mM Tris-HCl, pH 8.0; 100 mM KCl; 2 mM MgCl2; 10% glycerol (w/v)), 0.5 µl 4 mM spermidine, 0.5 µl 400 ng/µl herring sperm DNA, 0.5 µl 1 mg/ml bovine serum albumin and 1 µl of HcpR1 or HcpR2 at an appropriate concentration. Incubation mixtures were also supplemented with additional additives where noted. The CRP protein used in some of these experiments was kindly provided by Dr. David Lee, University of Birmingham. EMSA mixtures were incubated at 25 °C for 30 minutes and then loaded onto 6% (w/v) acrylamide gels prepared with 0.25 x TBE and 0.2% (v/v) glycerol. Electrophoresis was in 0.25 x TBE at 160 V for 90–180 minutes. Following electrophoresis, gels were fixed in 10% (v/v) acetic acid and 10% (v/v) methanol for 10 minutes. Fixed gels were transferred to 3 mm Whatman filter paper and dried under vacuum. Dried gels were stored in cassettes with a Fuji Imaging Phosphor screen overnight which were then visualised with the Bio-Rad Molecular FX Imager System and QuantityOne software (BioRad).
DNaseI footprinting assays
Fragments for DNaseI footprinting were liberated from the plasmid pTnapWT by digestion with HindIII and then treated with CIP. The digest mixture was purified by phenol-chloroform and ethanol precipitation and then digested with EcoRI. The resulting fragment was purified by electroelution, phenol-chloroform extraction and ethanol precipitation prior to labeling with γ-32P-ATP as described previously.
Maxam-Gilbert G + A sequencing reactions were prepared as standards for DNaseI footprinting assays. End-labeled DNA fragments were treated with formic acid for 90 seconds at room temperature and the reaction was stopped by ethanol precipitation. The purified DNA was re-suspended in 1 M piperidine and incubated at 90 °C for 30 minutes. DNA was purified by ethanol precipitation and the DNA pellet re-suspended in loading buffer (20 mM EDTA, 0.05% (w/v) bromophenol blue, 0.05% (w/v) xylene cyanol, dissolved in deionized formamide). G + A reactions were stored at −20 °C until required.
End-labeled DNA fragments were used in in vitro DNaseI footprinting assays to map HcpR binding. Incubation mixtures were prepared as described for EMSAs using end-labeled DNA fragments in volumes of 20 µl. After 30 minutes at 25 °C the incubation mixtures were supplemented with a dilution series of DNaseI (Roche Applied Science) ranging from 0–0.003 U and then incubated for an additional 40 seconds prior to inactivation of DNaseI with 200 µl of 500 mM EDTA. Digest mixtures were purified by phenol-chloroform extraction and ethanol precipitation. Purified DNA pellets were resuspended in denaturing gel loading buffer and stored at −20 °C until required.
DNaseI footprinting reactions and G + A sequencing reactions were resolved by electrophoresis on 6% (w/v) acrylamide denaturing urea gels prepared using the SequaGel UreaGel system (National Diagnostics) according to the manufacturer’s instructions. Samples were heated to 90 °C prior to loading onto gels. Electrophoresis was achieved in 1xTBE buffer (89 mM Tris base, 89 mM boric acid, 2.5 mM EDTA) at 60 W for ~2 hours. Gels were fixed, dried and visualised as described for electromobility shift assays.
RNA isolation and reverse transcription into cDNA
RNA was isolated from 5 to 15 ml samples of cultures in the exponential phase of growth that had been sedimented by centrifugation at 10,000 g for 5 min. Pellets were resuspended in 3 ml of RNAlater (Ambion), snap frozen in liquid nitrogen and stored for up to 2 weeks at −80 °C. Total RNA was purified with the QIAGEN RNeasy mini kit according to the manufacturer’s instructions with the inclusion of at least one on-column DNase digestion step to eliminate any contaminating DNA. RNA was eluted from the RNeasy spin columns with 30 μL RNase–free water and stored at −80 °C for up to 1 month. RNA integrity was assessed with a NanoDrop ND1000c Spectrophotometer (Labtech., UK). Samples with an A260/A230 ratio of less than 1.8 were rejected, as were samples less concentrated than 200 ng/μL.
RNA was reverse transcribed to cDNA with random hexamer primers using the Tetro cDNA Synthesis kit (BIOLINE, London, UK) and 2 ng of total RNA as template. Multiple RT-PCR reactions were prepared from each sample to provide a cDNA pool.
Quantitative RT-PCR
A Stratagene Mxp3005 machine set to detect SYBR green fluorescence and 96-well plates capped with optical strip tops was used for qRT-PCR. Reaction mixes were prepared using Brilliant III Ultra-Fast SYBR Green QPCR mastermix kit (Agilent). Gene specific primers to a final concentration of 400 nM and cDNA at a concentration of 5 to 50 ng / μL were added. Cycling parameters were as recommended by the manufacturer except that samples were denatured for 20 s and annealed for 20 s at 56 °C. The dissociation curve for each qPCR reaction was calculated using the pre-programmed Mcp3005 protocol and allowed assessment of PCR efficiency for each reaction. Primer specificity was also checked by agarose gel electrophoresis. Transcript levels were quantified by the ΔΔCt method using the levels of polA mRNA as a reference67. For each growth condition tested, RNA was prepared from at least three biological replicates and each qPCR reaction was run in triplicate. Thus each qRT-PCR measurement was the result of at least 9 replicates.
Preparation of RNA samples for RNA-seq analysis
RNA was prepared as described above for up to 20 independent cultures for each growth condition: these were culture growing with sulfate, sulfate plus NO, nitrate, or nitrate plus NO. Each RNA sample was treated twice to remove any traces of DNA. Samples were checked by qPCR for DNA contamination without reverse transcription. The 5 samples with highest concentration and A260/A230 ratio were transferred on dry ice to Oxford Gene Technologies (UK) for further quality control testing before RNA-seq analysis. Three samples from each growth condition were depleted for ribosomal RNA before reverse transcription.
Analysis of the RNA-seq data
The automated workflow for the RNA-seq analysis was implemented in the snakemake programming language, and is available on the supporting website. The workflow includes the following sequential steps: i > read quality control with fastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/); ii > read mapping with bowtie2 in non-oriented paired-ends mode, with at most 1 mismatch per read, and all other options left to their default values68,69. The genome sequence (fasta-formatted) and features (GTF-formatted) were downloaded from EnsemblGenomes Bacteria release 32 (http://bacteria.ensembl.org/index.html) (strain identifier Desulfovibrio_desulfuricans_subsp_desulfuricans_str_atcc_27774.ASM2212v1); iii > assignment of reads per genes in each sample by using feature Counts (from the subread suite)70. Finally the detection of “differentially expressed genes” (DEG) relied on the R library edgeR71. We applied a threshold of 0.05 on the adjusted p-value, and of 1.5 on the fold change to select for DEG. Table 1 gives the number of differentially expressed genes for each comparison between growth conditions (Sulfate + NO vs Sulfate; Nitrate + NO vs Nitrate; Nitrate vs Sulfate; Nitrate + NO vs Sulfate + NO).
A detailed technical report of all the statistical treatments (data exploration, differential analysis, cross-species functional enrichment) is available on the supporting website.
Analysis of regulatory sequences
The software suite Regulatory Sequence Analysis Tools (RSAT, http://rsat.eu/) was used to analyse regions upstream of genes in the reference genome Desulfovibrio_desulfuricans_ATCC_27774_uid59213, which was downloaded from NCBI.
The tool retrieve-seq was used to retrieve promoter sequences up to 400 bp upstream from position −1 relative to the translation start codon up to the nearest neighbouring gene.
The HcpR1 (Ddes_0528) binding sequence TGTGA-N6-TCACA, where N6 denotes a succession of 6 undefined nucleotides, was used to construct a seed matrix by setting an arbitrary weight of 8 on each residue of the binding site, and 0 on other nucleotides (Supplementary Fig. S2). The upstream sequences of all genes were scanned with matrix-scan to collect sequences matching this seed matrix with a p-value < 10−4. The 125 resulting sites were aligned to build a second-generation matrix with the tool convert-matrix. This second-generation matrix was used to rescan all promoters with matrix-scan, and sites with a p-value < 10−4 were identified.
References
Barton, L. L., Legall, J., Odom, J. M. & Peck, H. D. Energy coupling to nitrite respiration in the sulfate-reducing bacterium Desulfovibrio gigas. J. Bacteriol. 153, 867–871 (1983).
Mitchell, G. J., Jones, J. G. & Cole, J. A. Distribution and regulation of nitrate and nitrite reduction by Desulfovibrio and Desulfotomaculum species. Arch. Microbiol. 144, 35–40 (1985).
Pereira, I. A. C., LeGall, J., Xavier, A. V. & Teixeira, M. Characterization of a heme c nitrite reductase from a non-ammonifying microorganism, Desulfovibrio vulgaris Hildenborough. Biochim. Biophys. Acta 1481, 119–130 (2000).
Greene, E. A., Hubert, C., Nemati, M., Jenneman, G. E. & Voordouw, G. Nitrite reductase activity of sulfate-reducing bacteria prevents their inhibition by nitrate-reducing, sulphide-oxidizing bacteria. Env. Microbiol 5, 607–617 (2003).
Haveman, S. A., Greene, E. A. & Voordouw, G. Gene expression analysis of the mechanism of inhibition of Desulfovibrio vulgaris Hildenborough by nitrate-reducing, sulfide-oxidizing bacteria. Env. Microbiol. 7, 1461–1465 (2005).
He, Q. et al. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Env. Microbiol. 72, 4370–4381 (2006).
Korte, H. L. et al. Independence of nitrate and nitrite inhibition of Desulfovibrio vulgaris Hildenborough and use of nitrite as a substrate for growth. Environ. Sci. Technol. 49, 924–931 (2015).
Moura, I., Bursakov, S., Costa, C. & Moura, J. J. G. Nitrate and nitrite utilization in sulfate-reducing bacteria. Anaerobe 3, 279–290 (1997).
Liu, M. C. & Peck, H. D. The isolation of a hexaheme cytochrome from Desulfovibrio desulfuricans and its identification as a new type of nitrite reductase. J. Biol. Chem. 256, 13159–13164 (1983).
Pereira, P. M., Teixeira, M., Xavier, A. V., Louro, R. O. & Pereira, I. A. The Tmc complex from Desulfovibrio vulgaris Hildenborough is involved in transmembrane electron transfer from periplasmic hydrogen oxidation. Biochemistry. 45, 10359–10367 (2006).
Rajeev, L. et al. Regulation of nitrite stress response in Desulfovibrio vulgaris Hildenborough, a model sulfate-reducing bacterium. J. Bacteriol. 197, 3400–3408 (2015).
Keith, S. M. & Herbert, R. A. Dissimilatory nitrate reduction by Desulfovibrio. FEMS Microbiol. Lett. 18, 55–59 (1983).
Seitz, H. J. & Cypionka, H. Chemolithotrophic growth of Desulfovirbio desulfuricans with hydrogen coupled to ammonification of nitrate or nitrite. Arch. Microbiol. 146, 63–67 (1986).
Marietou, A., Griffiths, L. & Cole, J. Preferential reduction of the thermodynamically less favorable electron acceptor, sulfate, by a nitrate reducing strain of the sulfate reducing bacterium, Desulfovibrio desulfuricans 27774. J. Bacteriol. 191, 882–889 (2009).
Rajeev, L. et al. Systematic mapping of two component response regulators to gene targets in a model sulfate reducing bacterium. Genome Biol. 12, R99 (2011).
Dalsgaard, T. & Bak, F. Nitrate reduction in a sulfate-reducing bacterium, Desulfovibrio desulfuricans, isolated from rice paddy soil: sulphide inhibition, kinetics, and regulation. Appl. Env. Microbiol. 60, 291–297 (1994).
Cadby, I. T., Busby, S. J. W. & Cole, J. A. An HcpR homologue from Desulfovibrio desulfuricans and its possible role in nitrate reduction and nitrosative stress. Biochem. Soc. Trans. 39, 224–229 (2011).
Rodionov, D. A., Dubchak, I. L., Arkin, A. P., Alm, E. J. & Gelfand, M. S. Reconstruction of regulatory and metabolic pathways in metal-reducing delta proteobacteria. Genome Biol. 5, R90 (2004).
Rodionov, D. A., Dubchak, I. L., Arkin, A. P., Alm, E. J. & Gelfand, M. S. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comp. Biol. 1, e55 (2005).
da Silva, S. M. et al. An HcpR paralogue of Desulfovibrio gigas provides protection against nitrosative stress. FEBS Open Bio. 5, 594–604 (2015).
Zhou, A. et al. Functional characterization of Crp/Fnr-type global transcriptional regulators in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 78, 1168–1177 (2012).
Wang, J. et al. The roles of the hybrid cluster protein, Hcp, and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol. Microbiol. 100, 877–892 (2016).
Rodrigues, R. et al. Desulfovibrio gigas flavodiiron protein affords protection against nitrosative stress in vivo. J. Bacteriol. 188, 2745–2751 (2006).
Johnson, S. et al. A genomic island of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough promotes survival under stress conditions while decreasing the efficiency of anaerobic growth. Environ. Microbiol. 11, 981–991 (2009).
Wildschut, J. D., Lang, R. M., Voordouw, J. K. & Voordouw, G. Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris Hildenborough under microaerophilic conditions. J. Bacteriol. 188, 6253–6260 (2006).
Yurkiw, M. A., Voordouw, J. & Voordouw, G. Contribution of rubredoxin: oxygen oxidoreductases and hybrid cluster proteins of Desulfovibrio vulgaris Hildenborough to survival under oxygen and nitrite stress. Environ. Microbiol. 14, 2711–2725 (2012).
Figueiredo, M. C. O. et al. Hybrid cluster proteins and flavodiiron proteins afford protection to Desulfovibrio vulgaris upon macrophage infection. J. Bacteriol. 195, 2684–2690 (2013).
Jia, W., Faulkner, M., Cadby, I. & Cole, J. Why was Desulfovibrio fairfieldensis not found in faecal DNA from patients with gastric disease? Path. Dis. 67, 3 (2013).
Jia, W. et al. Diversity and distribution of sulfate-reducing bacteria in human faeces from healthy subjects and patients with inflammatory bowel disease. FEMS Immunol. Med. Microbiol. 65, 55–68 (2012).
Cadby, I. T. et al. Regulation, sensory domains and roles of two Desulfovibrio desulfuricans ATCC27774 Crp family transcription factors, HcpR1 and HcpR2, in response to nitrosative stress. Mol. Microbiol. 102, 1120–1137 (2016).
Chhabra, S. R. et al. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188, 1817–1828 (2006).
Lobo, S. A., M, A. M. P., Carita, J. N., Teixeira, M. & Saraiva, L. M. The anaerobe Desulfovibrio desulfuricans ATCC 27774 grows at nearly atmospheric oxygen levels FEBS Microbiol. Lett. 581, 433–436 (2007).
Zhang, W. et al. Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie Leeuwenhoek 89, 221–237 (2006).
Zhang, W., Culley, D. E., Hogan, M., Vitiritti, L. & Brockman, F. J. Oxidative stress and heat-shock responses of Desulfovibrio vulgaris by genome wide transcriptomic analysis. Antonie Leeuwenhoek 90, 41–55 (2006).
Warren, Y. A., Citron, D. M., Merriam, C. V. & Goldstein, E. J. C. Biochemical differentiation and comparison of Desulfovibrio species and other phenotypically similar genera. J. Clin. Microbiol. 43, 4041–4045 (2005).
Beatty, C. M., Browning, D. F., Busby, S. J. W. & Wolfe, A. J. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 185, 5148–5157 (2003).
Ishihama, A. et al. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in. Escherichia coli. J Bacteriol. 196, 2718–2727 (2014).
Browning, D. F., Cole, J. A. & Busby, S. J. W. Suppression of FNR-dependent transcription activation at the Eschericha coli nir promoter by Fis, IHF and H-NS: modulation of transcription activation by a complex nucleo-protein assembly. Mol. Microbiol. 37, 1258–1269 (2000).
McCready, R. G. L., Gould, W. D. & Cook, F. D. Respiratory nitrate reduction by Desulfovibrio sp. Arch. Microbiol. 135, 182–185 (1983).
Iuchi, S. & Lin, E. C. C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in. Escherichia coli. J. Bacteriol. 84, 3901–3905 (1987).
Jones, H. M. & Gunsalus, R. P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169, 3340–3349 (1987).
Constantinidou, C. C. et al. A reassessment of the fumarate and nitrate reduction regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL and NarQP as Escherichia coli adapts from aerobic to anaerobic growth. J. Biol. Chem. 281, 4802–4808 (2006).
Justino, M. C., Almeida, C. C., Goncalves, V. L., Teixeira, M. & Saraiva, L. M. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulphur clusters. J. Biol. Chem. 282, 10352–10359 (2007).
Overton, T. W. et al. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 190, 2004–2013 (2008).
Todorovic, S. et al. Iron-sulfur repair YtfE protein from Escherichia coli: structural characterization of the di-iron center. J. Biol. Inorg. Chem. 13, 765–770 (2008).
Justino, M. C., Baptista, J. M. & Saraiva, L. M. Di-iron proteins of the Ric family are involved in iron-sulfur cluster repair. Biometals 22, 99–108 (2009).
Nobre, L. S. et al. Escherichia coli RIC is able to donate iron to iron-sulfur clusters. PLoS ONE 9, e95222 (2014).
Henry, J. T. & Crosson, S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Ann. Rev. Microbiol. 65, 261–286 (2011).
Lo, F. C. et al. Crystal Structure Analysis of the Repair of Iron Centers Protein YtfE and Its Interaction with NO. Chemistry 22, 9768–9776 (2016).
He, Q. et al. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl Environ Microbiol 72, 4370–4381 (2006).
Pereira, P. M. et al. Energy metabolism in Desulfovibrio vulgaris Hildenborough: insights from transcriptome analysis. Antonie Leeuwenhoek 93, 347–362 (2008).
Abreu, I. A. et al. A novel iron centre in the split-soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774. J. Biol. Inorg. Chem. 8, 360-370.
Widdel, F. & Pfennig, N. Studies of dissimilatory sulfate-reducing bacteria that decompose fatty acids. II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch. Microbiol. 131, 360–365 (1982).
Xue, Y. & Voordouw, G. Control of microbial sulfide production with biocides and nitrate in oil reservoir simulating bioreactors. Front. Microbiol. 6, 1387 (2015).
Barton, L. L & Hamilton, W. A. Eds Sulfate-Reducing Bacteria: Environmental and Engineered Systems. Cambridge University Press. New York (2007).
Rabus, R. et al. A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulfate-reducing prokaryotes. Adv. Microb. Physiol. 66, 55–321 (2015).
Aragão, D. et al. Reduced hybrid cluster proteins (HCP) from Desulfovibrio desulfuricans ATCC 27774 and Desulfolvibrio vulgaris (Hildenborough). X-ray structures at high resolution using synchrotron radiation. J. Biol. Inorg. Chem 8, 540–548 (2003).
Dolla, A., Pohorelic, B. K., Voordouw, J. K. & Voordouw, G. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174, 143–151 (2000).
Dolla, A., Fournier, M. & Zorah, D. Oxygen defense in sulfate-reducing bacteria. J. Biotechnol. 126, 87–100 (2006).
Keller, K. L. et al. New model for electron flow for sulfate reduction in Desulfovibrio alaskensis G20. Appl. Environ. Microbiol. 80, 855–868 (2014).
Oliveira, T. F. et al. The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J. Biol. Chem. 283, 34141–34149 (2008).
Ramel, F. et al. Membrane-bound oxygen reductases of the anaerobic sulfate-reducing Desulfovibrio vulgaris Hildenborough: roles in oxygen defence and electron link with periplasmic hydrogen oxidation. Microbiology 159, 2663–2673 (2013).
Santos, A. et al. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350, 1541–1545 (2015).
Sousa, J. R. et al. Understanding the response of Desulfovibrio desulfuricans ATCC 27774 to the electron acceptors nitrate and sulfate - biosynthetic costs modulate substrate selection. Biochim. Biophys. Acta 1865, 1455–1469 (2017).
Christensen, G. A. et al. Rex (encoded by DVU_0916) in Desulfovibrio vulgaris Hildenborough is a repressor of sulfate adenylyl transferase and is regulated by NADH. J. Bacteriol. 197, 29–39 (2015).
Postgate, J. R. “The Sulphate-Reducing Bacteria”. Cambridge University press. 17–18 (1984).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 24, 402–408 (2001).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Langmead, B. & Saltzberg, S. L. Fast gapped-read alignment with Bowtie2. Nature Methods 9, 357–359 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomics features. Bioinformatics 30, 923–930 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 159–160 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing.“Journal of the royal statistical society”. Series B (Methodological) 57, 289–300 (1995).
Author information
Authors and Affiliations
Contributions
The project was designed and coordinated by J.A.C., who drafted the manuscript. RNA-seq experiments were completed by M.F. and J.A.C.; RNA isolation was optimised by M.T.; I.T.C. completed the qRT-PCR, E.M.S.A. and D.N.A.ase footprinting experiments; RNA-seq data were analysed by A.D., J.v.H., J.C. and J.L.; figures and text were prepared by all authors; the draft of the manuscript was revised by J.v.L., A.D., I.T.C., M.T. and J.A.C.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cadby, I.T., Faulkner, M., Cheneby, J. et al. Coordinated response of the Desulfovibrio desulfuricans 27774 transcriptome to nitrate, nitrite and nitric oxide. Sci Rep 7, 16228 (2017). https://doi.org/10.1038/s41598-017-16403-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-16403-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.