Abstract
A recent genome-wide association study (GWAS) identified four genetic variants rs78726293, rs191260602, rs17035816 and rs7688285 in GLRB gene to be associated with panic disorder (PD) risk. In fact, GWAS is an important first step to investigate the genetics of human complex diseases. In order to translate into opportunities for new diagnostics and therapies, we must identify the genes perturbed by these four variants, and understand how these variant functionally contributes to the underlying disease pathogenesis. Here, we investigated the effect of these four genetic variants and the expression of three nearby genes including PDGFC, GLRB and GRIA2 in human brain tissues using the GTEx (version 6) and Braineac eQTLs datasets. In GTEx (version 6) dataset, the results showed that both rs17035816 and rs7688285 variants could significantly regulate PDGFC and GLRB gene expression. In Braineac dataset, the results showed that rs17035816 variant could significantly regulate GLRB and GRIA2 gene expression. We believe that these findings further provide important supplementary information about the regulating mechanisms of rs17035816 and rs7688285 variants in PD risk.
Similar content being viewed by others
Introduction
Panic disorder (PD) is a kind of anxiety disorder, which is prevalent in a 2–3% life-time, and could cause a huge burden of disease1. Deckert et al. recently performed a genome-wide association study of PD/agoraphobia (AG) using large-scale sample size1. They successfully identified four genetic variants rs78726293, rs191260602, rs17035816 and rs7688285 in GLRB gene1. They further conducted an expression quantitative trait loci (eQTL) analysis to detect the functional effect of these four variants on the expression of GLRB and neighbor genes using the Genotype-Tissue Expression (GTEx) eQTL database1. Meanwhile, they evaluated the potential association between rs7688285 and GLRB mRNA expression levels using the post-mortem brain samples of 76 individuals1. Their results showed that none of these four genetic variants could regulate the expression of nearby genes in the GTEx database1. In post-mortem brain samples, Deckert et al. reported significant association between rs7688285 A allele and the increased mean expression of GLRB with beta = 0.498 and P = 0.013 in the midbrain, but not in the forebrain with P = 0.421 or in the amygdalae with P = 0.4871.
Deckert et al. selected the online GTEx eQTL database to evaluate the potential association between these four genetic variants and gene expression of their nearby genes1. In fact, the online GTEx eQTL database only included the all significant variant-gene pairs with a genome wide false discovery rate (FDR) threshold of 0.052. The suggested association between these four genetic variants and gene expression of their nearby genes may have been adjusted by the genome wide FDR threshold, and may not be included in the online GTEx eQTL database. It is important to evaluate these findings using all the original SNP-gene associations in GTEx database.
Meanwhile, evidence shows that the effect of genetic variants on gene expression may occur in disease-relevant tissue types3,4,5,6,7,8,9. It is still necessary to investigate these potential expression associations in other human brain tissues, as Deckert et al. did using the human brain samples. Here, we investigated the effect of these four genetic variants rs78726293, rs191260602, rs17035816 and rs7688285 and the expression of three nearby genes including PDGFC, GLRB and GRIA2 using two eQTLs datasets.
Results
eQTLs analysis in GTEx dataset
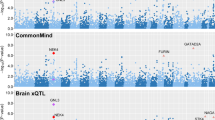
In GTEx (version 6) dataset, we found that rs78726293 and rs191260602 were not available in the. We then focused on the two genetic variants including rs17035816 and rs7688285 and three genes including PDGFC, GLRB and GRIA2 in the following analysis. The results indicated that both rs17035816 and rs7688285 variants could significantly regulate nearby gene expression in human brain tissues (significance threshold 0.05)10. In brief, rs17035816 variant could significantly regulate PDGFC gene expression in cerebellar hemisphere tissue (P = 8.70E-03), putamen basal ganglia tissue (P = 4.14E-03) and cerebellum tissue (P = 3.17E-02), and regulate GLRB gene expression in cerebellar hemisphere tissue (P = 1.50E-03). The rs7688285 variant could significantly regulate PDGFC gene expression in hippocampus tissue (P = 2.03E-02) and putamen basal ganglia tissue (P = 1.37E-02), and regulate GLRB gene expression in hypothalamus tissue (P = 2.63E-02). We further performed a multiple testing correction using a FDR threshold of 0.05 in these 10 brain tissues. Interestingly, rs17035816 variant still significantly regulates nearby gene expression after the multiple hypothesis test correction. More detailed results are described in Table 1.
eQTLs analysis in Braineac dataset
In Braineac dataset, we found that rs78726293, rs191260602 and rs7688285 were not available. We then focused on the rs17035816 variant and three genes including PDGFC, GLRB and GRIA2 in the following analysis. The results showed that rs17035816 variant could significantly regulate nearby gene expression in human brain tissues (significance threshold 0.05). In brief, rs17035816 variant could significantly regulate GLRB and GRIA2 gene expression in cerebellar cortex tissue with P = 1.49E-02 and P = 3.49E-02, respectively. More detailed results are described in Table 2.
Meta-analysis
We further performed a meta-analysis in the same brain tissues including Cerebellum, Hippocampus, Frontal cortex, and Putamen. The results showed that rs17035816 variant could significantly regulate GLRB gene expression in cerebellum tissue. More detailed results are described in Table 3.
Discussion
Deckert et al. highlighted four genetic variants rs78726293, rs191260602, rs17035816 and rs7688285 in GLRB gene to be associated with PD risk1. In fact, GWAS is an important first step to investigate the genetics of human complex diseases as widely described in previous studies11,12,13,14,15,16,17,18,19,20,21,22,23,24,25. In order to translate into opportunities for new diagnostics and therapies, we must identify the genes perturbed by these four variants, and understand how these variant functionally contributes to the underlying disease pathogenesis3,4,5,6,7,8,12,26,27,28,29,30. If a genetic variant is associated with increased or decreased expression of a particular gene, this suggests that the gene on which the variant acts could be in the causal pathway31. However, Deckert et al. revealed no significant cis-eQTL using the online GTEx database1. Here, we successfully identified significant cis-eQTL using all the original SNP-gene summary association results in the GTEx (version 6), even after the multiple hypothesis test correction using FDR threshold of 0.05.
In the GTEx dataset, we confirmed previous findings. Deckert et al. analyzed the post-mortem brain samples of 76 individuals, and identified that the rs7688285 A allele could significantly regulate increased mean expression of GLRB with beta = 0.498 and P = 0.0131. Here, our findings showed that rs7688285 variant A allele could significantly regulate increased PDGFC gene expression in hippocampus tissue (beta = 0.296 and P = 2.03E-02), reduced PDGFC gene expression in putamen basal ganglia tissue (beta = −0.297 and P = 1.37E-02), and increased GLRB gene expression in hypothalamus tissue (beta = 0.192 and P = 2.63E-02). Meanwhile, the results also showed some novel findings. Take rs17035816 variant for example, it could significantly regulate nearby gene expression even after the multiple hypothesis test correction as described in Table 1.
We further evaluated the potential association between these four genetic variants and the expression of three nearby genes including PDGFC, GLRB and GRIA2 in the Braineac dataset including 10 brain regions from 134 neuropathologically normal individuals of European descent32. Interestingly, the rs17035816 could significantly regulate increased GLRB and GRIA2 gene expression in cerebellar cortex. We believe that these findings further provide important supplementary information about the regulating mechanisms of rs17035816 and rs7688285 variants in PD risk in the human brain tissues. Genetic variants may need tissue, cell, region, disease specific factors to exert their influences on gene expression33,34. Here, we identified different results in GTEx (version 6) and Braineac eQTLs datasets. We think that disease status may influence the association between these genetic variants and GLRB and GRIA2 gene expression.
Here, the two variants rs78726293 and rs191260602 are not available in the GTEx dataset. Three variants rs78726293, rs191260602, and rs7688285 are not available in the Braineac dataset. We have used HaploReg (version 4) to identify the proxy SNPs based on the linkage disequilibrium (LD) information in 1000 Genomes Project (EUR) with r2 ≥ 0.82. However all these tagged SNPs are still not available in the GTEx dataset and the Braineac dataset. We think that following studies should further evaluate these potential expression associations using other eQTLs datasets in human brain regions.
Materials and Methods
The GTEx dataset
The GTEx (version 6) eQTLs dataset included 53 tissues, 544 donors and 8555 samples35. These 544 donors have several death pathologies including traumatic injury, cerebrovascular disease, heart disease, liver, renal, respiratory, and neurological diseases35. Here, we selected 10 human brain tissues including anterior cingulate cortex, caudate basal ganglia, cerebellar hemisphere, cerebellum, cortex, frontal cortex BA9, hippocampus, hypothalamus, nucleus accumbens basal ganglia, and putamen basal ganglia, which include at least 70 samples10. The GTEx used the RNA-Seq method to measure the gene expression10.
The Braineac dataset
The Braineac eQTLs dataset is from a web server for data from the UK Brain Expression Consortium (UKBEC)32. This dataset includes 10 brain regions from 134 neuropathologically normal individuals of European descent32. The 10 brain regions are cerebellar cortex, frontal cortex, hippocampus, medulla, occipital cortex, putamen, substantia nigra, temporal cortex, thalamus, and intralobular white matter32. The Braineac used the Affymetrix GeneChip Human exon 1.0 ST arrays to measure the gene expression32. The gene expression in transcript level is the Winsorised mean over exon-specific levels32.
eQTLs analysis
In the GTEx dataset, a linear regression analysis was applied to evaluate the SNP-gene expression association using R package Matrix eQTL, assuming an additive model and adjusting for the covariates including genotyping PCs, genotyping array platform, 15, 30 or 35 PEER factors, and the gender10. Here, we downloaded all the SNP-gene association summary results from the GTEx (version 6) database to directly evaluate the potential association between these four genetic variants and gene expression of nearby genes. In the Braineac dataset, a linear regression analysis was also applied to evaluate the potential association between genetic variants and the expression of nearby genes using the R package Matrix EQTL32. Here, we downloaded the gene expression data and the genotype data of genetic variants with 1 Mb upstream of transcription start site and 1 Mb downstream of transcription end site from the Braineac online database32. We then evaluated the potential SNP-gene expression association using the R program. More detailed information is described in a recent study36.
Meta-analysis
In the same brain tissue, we performed a meta-analysis to increase the statistical power. The pooled effect was calculated using the fixed effect model (Mantel-Haenszel) or the random-effect model (DerSimonian-Laird) determined by the heterogeneity3,4,5,6,7,8,12,26,27,28,29,30. A Z test was used to determine the significance of the effect. All tests were computed using the R Package (http://cran.r-project.org/web/packages/meta/index.html).
References
Deckert, J. et al. GLRB allelic variation associated with agoraphobic cognitions, increased startle response and fear network activation: a potential neurogenetic pathway to panic disorder. Mol Psychiatry (2017).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40, D930–934 (2012).
Bao, X. et al. Cell adhesion molecule pathway genes are regulated by cis-regulatory SNPs and show significantly altered expression in Alzheimer’s disease brains. Neurobiol Aging 36(2904), e2901–2907 (2015).
Liu, G., Bao, X. & Wang, R. Expression quantitative trait loci regulate HNF4A and PTBP1 expression in human brains. Proc Natl Acad Sci USA 112, E3975 (2015).
Liu, G. et al. Cis-eQTLs regulate reduced LST1 gene and NCR3 gene expression and contribute to increased autoimmune disease risk. Proc Natl Acad Sci USA 113, E6321–E6322 (2016).
Liu, G., Hu, Y., Jin, S. & Jiang, Q. Genetic variant rs763361 regulates multiple sclerosis CD226 gene expression. Proc Natl Acad Sci U S A 114, E906–E907 (2017).
Liu, G. et al. Convergent Genetic and Expression Datasets Highlight TREM2 in Parkinson’s Disease Susceptibility. Mol Neurobiol 53, 4931–4938 (2016).
Jiang, Q., Hu, Y. & Liu, G. Association of Alzheimer Disease Susceptibility Variants and Gene Expression in the Human Brain. JAMA Neurol 73, 1255 (2016).
Hu, Y., Jin, S., Cheng, L., Liu, G. & Jiang, Q. Autoimmune disease variants regulate GSDMB gene expression in human immune cells and whole blood. Proc Natl Acad Sci USA (2017).
Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Jiang, Q. et al. Alzheimer’s Disease Variants with the Genome-Wide Significance are Significantly Enriched in Immune Pathways and Active in Immune Cells. Mol Neurobiol 54, 594–600 (2017).
Liu, G. et al. Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult Scler 23, 205–212 (2017).
Hu, Y. et al. GAB2 rs2373115 variant contributes to Alzheimer’s disease risk specifically in European population. J Neurol Sci 375, 18–22 (2017).
Liu, G. et al. Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J Neurochem 120, 190–198 (2012).
Liu, G. et al. Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 35, 786–792 (2014).
Liu, G. et al. The CLU gene rs11136000 variant is significantly associated with Alzheimer’s disease in Caucasian and Asian populations. Neuromolecular Med 16, 52–60 (2014).
Liu, G. et al. Analyzing large-scale samples confirms the association between the ABCA7rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol 50, 757–764 (2014).
Shen, N. et al. An Updated Analysis with 85,939 Samples Confirms the Association Between CR1rs6656401 Polymorphism and Alzheimer’s Disease. Mol Neurobiol 51, 1017–1023 (2015).
Zhang, S. et al. CLU rs2279590 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian and Asian populations. J Neural Transm (Vienna) 122, 433–439 (2015).
Chen, H. et al. Analyzing 54,936 Samples Supports the Association Between CD2AP rs9349407 Polymorphism and Alzheimer’s Disease Susceptibility. Mol Neurobiol 52, 1–7 (2015).
Li, X. et al. CD33rs3865444 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Chinese, European, and North American Populations. Mol Neurobiol 52, 414–421 (2015).
Li, Y. et al. CR1rs3818361 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Chinese Population. Mol Neurobiol 53, 4054–4059 (2016).
Liu, G. et al. PICALM rs3851179 Variant Confers Susceptibility to Alzheimer’s Disease in Chinese Population. Mol Neurobiol (2016).
Liu, G. et al. BIN1 gene rs744373 polymorphism contributes to Alzheimer’s disease in East Asian population. Neurosci Lett 544, 47–51 (2013).
Liu, G. et al. PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromolecular Med 15, 384-388 (2013).
Liu, G. et al. Measles contributes to rheumatoid arthritis: evidence from pathway and network analyses of genome-wide association studies. PLoS One 8, e75951 (2013).
Quan, B. et al. Pathway analysis of genome-wide association study and transcriptome data highlights new biological pathways in colorectal cancer. Mol Genet Genomics 290, 603–610 (2015).
Zhao, X. et al. Pathway analysis of body mass index genome-wide association study highlights risk pathways in cardiovascular disease. Sci Rep 5, 13025 (2015).
Wei, J. et al. Multiple analyses of large-scale genome-wide association study highlight new risk pathways in lumbar spine bone mineral density. Oncotarget 7, 31429–31439 (2016).
Shang, H. et al. Pathway analysis of two amyotrophic lateral sclerosis GWAS highlights shared genetic signals with Alzheimer’s disease and Parkinson’s disease. Mol Neurobiol 51, 361–369 (2015).
Surendran, P. et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet 48, 1151–1161 (2016).
Ramasamy, A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17, 1418–1428 (2014).
Allen, M. et al. Gene expression, methylation and neuropathology correlations at progressive supranuclear palsy risk loci. Acta Neuropathol 132, 197–211 (2016).
Zou, F. et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet 8, e1002707 (2012).
The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585 (2013).
Liu, G. et al. Genetic Variants and Multiple Sclerosis Risk Gene SLC9A9 Expression in Distinct Human Brain Regions. Mol Neurobiol (2016).
Author information
Authors and Affiliations
Contributions
Q.J.W., M.F.Y., S.Y.S. and B.L.S. conceived and initiated the project. Q.J.W. and M.F.Y. analyzed the data and wrote the manuscript. P.D.H., C.J.Y., C.J.D., H.X.L. and Y.J.H. reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, Qj., Yang, Mf., Hao, Pd. et al. GLRB variants regulate nearby gene expression in human brain tissues. Sci Rep 7, 13326 (2017). https://doi.org/10.1038/s41598-017-13702-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13702-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.