Abstract
Legionella pneumophila infects human alveolar macrophages and is responsible for Legionnaire’s disease, a severe form of pneumonia. L. pneumophila encodes more than 300 putative effectors, which are translocated into the host cell via the Dot/Icm type IV secretion system. These effectors highjack the host’s cellular processes to allow bacterial intracellular growth and replication. Here we adopted a multidisciplinary approach to investigate WipB, a Dot/Icm effector of unknown function. The crystal structure of the N-terminal domain at 1.7 Å resolution comprising residues 25 to 344 revealed that WipB harbours a Ser/Thr phosphatase domain related to the eukaryotic phospho-protein phosphatase (PPP) family. The C-terminal domain (residues 365–524) is sufficient to pilot the effector to acidified LAMP1-positive lysosomal compartments, where WipB interacts with the v-ATPase and the associated LAMTOR1 phosphoprotein, key components of the lysosomal nutrient sensing (LYNUS) apparatus that controls the mammalian target of rapamycin (mTORC1) kinase complex at the lysosomal surface. We propose that WipB is a lysosome-targeted phosphatase that modulates cellular nutrient sensing and the control of energy metabolism during Legionella infection.
Similar content being viewed by others
Introduction
Legionella pneumophila is a facultative intracellular pathogen responsible for a severe form of pneumonia termed Legionnaire’s Disease1. Infection occurs via the inhalation of aerosols, followed by phagocytosis of the bacterium by alveolar macrophages. Within the macrophage, the bacterium avoids the endosomal pathway by modifying the early entry vesicle into a specialised replicative compartment termed the “Legionella-Containing Vacuole” (LCV), which progressively acquires endoplasmic reticulum-derived membranes, studded with ribosomes2.
During infection, L. pneumophila utilizes a type IV secretion (T4S) system termed the Dot/Icm secretion system to translocate effector proteins into the host cell3,4,5,6,7. Over 300 potential effectors are translocated by the Dot/Icm system. These effectors are encoded on the L. pneumophila chromosome and their genes comprise approximately 10% of the bacterium’s genome8, 9. About 20% of the effectors have been linked to a cellular pathway, and of these only half have an ascribed eukaryotic target or biochemical function2. Although the deletion of the T4S secretion system itself impairs intracellular growth, bacterial mutants lacking individual effectors only exhibit mild defects in intracellular replication. Indeed, several studies have demonstrated that multiple, redundant effectors target the same cellular process2. These factors complicate the dissection of Legionella effector activities.
IcmW-Interacting Protein B (WipB) is a 60 kDa Legionella effector of unknown function originally identified amongst a cohort of proteins that interact with the Dot/Icm T4S chaperone complex IcmS/IcmW10. Legionella deletion mutants lacking wipB do not exhibit a replicative defect in macrophages, unless this is combined with a second mutation in lidA, a gene encoding another T4S effector capable of binding Rab GTPases. L. pneumophila wipB − lidA − double mutants exhibit attenuated intracellular replication in macrophages and increased host cell death, although the significance of this remains unclear2, 11. Given this characteristic lack of attenuation in the single gene deletion mutant, a hallmark of functional redundancy, we adopted an alternative structural and biochemical approach to explore the properties and target/s of WipB. Our data reveal that WipB is a modular effector, with an N-terminal Ser/Thr phosphatase domain related to the eukaryotic phospho-protein phosphatase (PPP) family, and a C-terminal domain sufficient to pilot WipB to the acidified LAMP1-positive lysosomal compartments in eukaryotic cells, where WipB interacts with the v-ATPase and the associated LAMTOR1 phosphoprotein, key components of the lysosomal nutrient sensing (LYNUS) apparatus.
Results and Discussion
The structure of the WipB N-terminal domain reveals a phosphatase domain
WipB was purified following over-expression in E. coli as described in Materials and Methods. However, we observed rapid breakdown of the protein into shorter fragments (Fig. 1a), the shortest of which was identified by mass spectrometry to contain the first 364 residues of the protein. A construct encoding this apparently stable fragment was generated and the protein expressed and purified (Fig. 1a). This fragment, WipB1-364, was stable but did not crystallise. Thus, the construct was further modified to exclude the regions of the N- or C-terminal sequence predicted to have no secondary structures (regions 1–24 and 345–364). The resulting fragment, WipB25-344, also yielded a stable protein, which in contrast to WipB1-364 crystallised readily. The crystals diffracted to a resolution of 1.7 Å (Table 1). The structure was solved using the Molecular Replacement method, with the recently solved structure of WipA (a protein with 34% identity at amino acid sequence level (Fig. S1)) serving as a search model12. Attempts to similarly produce the C-terminal domain of WipB failed, as this region of the protein appears to be unstable when expressed in bacteria. Thus, WipB appears to contain two domains, a structured N-terminal domain and a possibly less stable C-terminal domain.
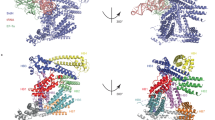
Domain structure of WipB and WipB25-344 structure (a) SDS-PAGE analysis of WipB and WipB fragments. Lane 1: full-length WipB. Lane 2: WipB1-364. Lane 3: WipB25-344. (b) Topology diagram of WipB25-344. β-strands and α-helices are shown as arrows and cylinders, respectively. The central β-sandwich is shown in blue and yellow. The flanking α-helices on the side of the blue or yellow β-sheets are shown in cyan and red, respectively. The three helices at the top are shown in green. (c) Three-dimensional diagram of WipB25-344. The structure is shown in cartoon representation with secondary structures color-coded as in panel B.
WipB25-344 crystallizes as a dimer in the asymmetric unit. While the two chains superpose very well (root-mean-square deviation (RMSD) in Cα atoms of 0.5 Å), chain A exhibits overall lower B-factors (Fig. S2a). In chain A, density for residues 332–344 is missing while the same region is resolved in chain B (Fig. S2b), This region in chain B interacts with chain A, probably an artefact of crystal packing as WipB25-344 is monomeric in solution.
WipB25-344 forms a compact structure consisting of a central β-sandwich and 11 α-helices, 4 α-helices on each side of the β-sandwich (in cyan (α1-4) and red (α8-11), respectively, in Fig. 1b,c) with three more α-helices (α5-7) capping the structure (in green in Fig. 1b,c). The structure can be further subdivided in an N-terminal lobe consisting of α1-4 and β1-3 (cyan and blue in Fig. 1b,c) on the one hand and a C-terminal lobe consisting of α8-11 and β4-6 (in red and yellow in Fig. 1b,c) on the other hand. The structure of WipB25-344 is very similar to that of WipA24-435 determined previously (the structures align with a root-mean-square deviation in Cα atoms of 1.9 Å over 279 WipB residues; Fig. S2c). However, WipA differs from WipB in including a very large α-helical hairpin (Fig. S2c).
A search for similar structures using the DALI server13 highlighted several hydrolases among the highly scored homologous structures, including the catalytic domain of a number of serine/threonine (Ser/Thr) protein phosphatases including PP1, PP2A, PP2B, and PP5, important members of the larger phospho-protein phosphatase (PPP) family of proteins14 Structural superposition of WipB25-344 with these eukaryotic phosphatases (see Fig. S1 for sequence alignment with a number of PPPs, and Figs S2d and 2a for structural superposition with PP2B) reveals a common structural core aligning with an RMSD in Cα atoms of 2 Å. This core consists of the central β-sandwich together with the flanking helices α1, α4, and α6 on one side and α8 and α11 on the other. Importantly, these regions harbour signature motifs conserved in PPPs, notably Ser/Thr phosphatases (Fig. 2a). In the PPP family, the DxH, GDxxDR, and NHE signature motifs are strictly conserved and comprise most of the residues that form the catalytic site14 WipB exhibits similar motifs, namely GDLHA, GDELVDR, and NHG (Fig. 2a and S1), and these motifs cluster in one particular region of WipB25-244, on the loops connecting β1 and α1 (GDLHA), β2 and α4 (GDELVDR), and β3 and α5 (SNG). In this region, the side chains of Asp 32 and His 34 (in GDLHA), Asp118 and Asp122 (in GDELVDR) and of His 151 (in NHG) form a site which in PP2B and other Ser/Thr phosphatase has been shown to be the catalytic site of these enzymes (Fig. 2b). Two additional residues are located in this cluster, His 231 and Asp 331, both conserved and known to be important for activity in PP2B and other PPPs (Fig. 2b).
WipB is related to the PPP phosphatases (a) Structure-based alignment of the core part of the structure conserved between the human phosphatase PP2B (grey, pdb code 5C1V) and WipB (purple). Both structures are in cartoon representation. The three phosphatase motifs, GDLHA, GDELVDR, and NHG, are mapped onto the structure and color-coded in orange, green and blue, respectively. Orientation is the same than in Fig. 1c. (b) Active site of WipB and PP2B. The two proteins are shown as in panel A in the same orientation. Residues of the active site of PP2B conserved in WipB are shown in ball-and-stick representation, with oxygen and nitrogen atoms color-coded in blue and red, respectively. For residues in the three phosphatase motifs GDLHA, GDELVDR, and NHG, carbon atoms are color-coded in orange, green or blue, respectively. (c) Activity assay of WipB1-364. Left panel: release of free phosphate over time from the RRA(pT)VA peptide incubated at 37 °C with 30 nM of WipB1-364. Right panel: phosphate release measurements using increasing concentrations of WipB1-364 wild-type or mutant incubated for 2 minutes at 37 °C with either the RRA(pT)VA (solid line) or the END(pY)INASL (dashed line) peptide. Results with the wild-type protein establish that WipB is a Ser/Thr protein phosphatase while results obtained with the mutants ascertain the catalytic role of residues conserved in the structural alignment with PPPs.
WipB is a Ser/Thr phosphatase
The structural homology of WipB25-244 with phosphatases of the PPP family suggests that WipB might contain a Ser/Thr phosphatase activity. To test this hypothesis, we investigated the ability of the WipB N-terminal domain (residues 1-364) to catalyse dephosphorylation reactions in vitro using a model phosphothreonine peptide with sequence RRA(pT)VA, a peptide derived from the rat liver pyruvate kinase and routinely used to monitor Ser/Thr phosphatase activities15. As shown in Fig. 2c, WipB1-364 clearly exhibits phosphatase activity. Biochemical quantification of the initial rates at 21 °C revealed a Km of 0.5 ± 0.3 mM and a Vmax of 0.042 ± 0.02 mM.s−1 for the dephosphorylation reaction using 30 nM of WipB1-364.
Since WipB and WipA are structurally similar and WipA exhibits tyrosine protein phosphatase activity, we next tested the activity of WipB against a phosphotyrosine-containing peptide (Fig. 2c). The model peptide END(pY)INASL (a peptide derived from the T cell phosphatase sequence16 and generally used to assay tyrosine phosphatase activities) was used as substrate in the presence of 0, 10, 30 and 90 nM of WipB1-364 and the released free phosphate concentration was measured after incubation at 37 °C for 2 minutes (Fig. 2c). The same experiment was carried out using the RRA(pT)VA peptide for comparison. We observed that WipB1-364 is able to release phosphate from the phosphothreonine, not the phosphotyrosine, peptide. Another phosphotyrosine-containing peptide routinely used to monitor tyrosine dephosphorylation was also used, with sequence DADE(pY)LIPQQG derived from the EGFR protein17, and the lack of activity against phosphotyrosine-containing sequences was confirmed (data not shown).
In order to assess whether the active site identified from comparison with the structure of PP2B is responsible for the measured phosphatase activity, we expressed and purified three derivatives of WipB1-364 each containing an individual point mutation within the putative active site: Asp118 to Ala (WipB1-364;D118A; in GDLHA motif) and Arg123 to Ala (WipB1-364;R123A; in the GDELVDR motif) and His151 to Ala (WipB1-364;H151A; in the NHG motif). The three mutants exhibited a loss in phosphatase activity confirming that the catalytic domain predicted from our structure is indeed functional (Fig. 2c).
To understand the differences between WipB and WipA specificity, we compared the structure of their catalytic sites (Fig. S2e). Both are essentially similar. However, four residues differ in position: Arg185 (123 in WipA), Asp30 (31 in WipA), Asp396 (331 in WipA) and Asp310 (369 in WipA). In WipA, the first three are involved in catalysis while Arg369 of WipA was hypothesized to be involved in pTyr recognition12. In WipB, Arg310 (equivalent of Arg369 in WipA), is conformationally restrained away from the active site and therefore cannot assume a role in substrate recognition. We hypothesize that this is the reason why WipB is inactive against pTyr-containing peptides.
Altogether, these data demonstrate that WipB harbours a functional N-terminal phosphatase domain that can hydrolyse Ser/Thr phosphorylated peptides in vitro and is catalytically independent from the C-terminal domain.
WipB locates to the lysosomal compartment and its C-terminal domain determines its cellular localisation
Having established that WipB is a Ser/Thr protein phosphatase, we next investigated the localisation of WipB when expressed in cultured mammalian cells.
Cultured HeLa cells were transiently transfected with plasmids encoding GFP fusions of WipB, either at the C-terminus (WipB-GFP) or the N-terminus (GFP-WipB). After 24 h, immunoblotting of transfected cell lysates using an anti-GFP antibody revealed the presence of a single species at the expected molecular weight of WipB-GFP and GFP-WipB (Fig. S3a), verifying that in contrast to expression in E. coli, the fusion proteins were not truncated or degraded, irrespective of the location of the GFP.
When transfected HeLa cells were examined using confocal microscopy, GFP-WipB and WipB-GFP predominantly formed puncta in the cytoplasm, with a minor fraction also evident at the plasma membrane (Fig. 3a for GFP-WipB and Fig. S3b for WipB-GFP). Equivalent localisation was observed when cultured retinal pigment epithelial (RPE-1) cells were transiently transfected with GFP-WipB (Fig. S3c).
WipB is targeted to lysosomes by its C-terminal domain where it interacts with components of the lysosomal nutrient sensing system (a) HeLa cells expressing GFP-fusion proteins of WipB, WipBD118A, WipB1-364 and WipB365-524. Scale bar, 10 µm. (b) HeLa cells expressing GFP-WipB (green) stained using an anti-LAMP-1 antibody (Magenta) after permeabilisation and fixation. Scale bar, 10 µm. (c) HeLa cells expressing GFP-WipB (green) were incubated with Lysotracker (red) for 15 min before fixation. Scale bar, 10 µm. (d) SDS-PAGE of fractions following co-immunoprecipitation of GFP, GFP-WipB or the indicated GFP-WipB derivatives from HeLa cell lysates using anti-GFP antibody and immunoblotting with anti-v-ATPase A, -v-ATPase B or LAMTOR1 antibodies. A cropped blot is here displayed and the corresponding full-length blot is included in the supplementary information. (e) SDS-PAGE of fractions following co-immunoprecipitation of v-ATPase A, v-ATPase B or LAMTOR1 from lysates of HeLa cells expressing GFP or GFP-WipB and immunoblotting with anti-GFP antibody. A cropped blot is here displayed and the corresponding full-length blot is included in the supplementary information.
To investigate whether the GFP-WipB puncta were targeted to any specific cellular compartment or whether GFP-WipB expression influenced the distribution or morphology of cellular organelles, GFP-WipB transfected cells were fixed and co-stained with markers characteristic of different subcellular compartments and organelles. GFP-WipB did not co-localise or disrupt the morphology of the Golgi apparatus (giantin), mitochondria (MitoTracker®) or the rough endoplasmic reticulum (calreticulin) (Fig. S4). However, GFP-WipB was strongly enriched around LAMP-1 positive compartments (Fig. 3b). Similarly, when transfected cells were loaded with LysoTracker® that specifically labels acidified compartments, LysoTracker-positive compartments also coincided with WipB puncta (Fig. 3c). In addition, when cells expressing GFP-WipB were loaded with TRITC-Dextran 10,000, which accumulates in endosomal compartments, partial co-localisation with the WipB puncta was also evident (Fig. S3d). Indeed, GFP-WipB could be frequently and reproducibly visualised as a defined ring of fluorescence surrounding LAMP-1 positive, LysoTracker-positive and TRITC-Dextran loaded compartments in transfected cells (Fig. 3c and S3d, zoom panels), indicative of protein recruitment. We next assessed the extent of colocalisation by calculating Manders coefficients to allow an unbiased evaluation of the degree of overlap between GFP-WipB and the various cellular markers. The resulting quantitative analysis strongly reinforced our qualitative observations, demonstrating statistically significant associations between WipB-GFP and LysoTracker, WipB-GFP and TRITC-Dextran, and most strikingly between WipB-GFP and LAMP1, but not between WipB-GFP and giantin (Fig. S3e). Correspondingly, the reciprocal analysis confirmed identical relationships when co-localisation was considered with respect to the individual cellular markers and GFP-WipB (Fig. S3e). These data reveal that the Ser/Thr phosphatase WipB is recruited to acidified LAMP1 compartments, characteristic of lysosomes.
To establish whether the Ser/Thr phosphatase activity influenced WipB localisation, cells were equivalently transfected with the catalytically dead GFP- WipBD118A derivative. As with active GFP-WipB, GFP- WipBD118A was stable after expression (Fig. S3a), and adopted a localisation indistinguishable from GFP-WipB (Figs 3a, S3d and S3f), demonstrating that Ser/Thr phosphatase activity is not a determinant of WipB localisation in eukaryotic cells.
To identify the region of WipB responsible for lysosomal targeting, GFP fusions of the isolated catalytic domain (GFP-WipB1-364) and C-terminal domain (GFP-WipB365-524) were equivalently transfected. Analysis of protein expression by immunoblotting of transfected cell lysates showed that each derivative was stably expressed as a fusion of the expected molecular weight (Fig. S3a). GFP-WipB1-364 and GFP-WipB365-524 were then visualised following transfection of HeLa cells using confocal microscopy (Fig. 3a). In contrast to GFP-WipB, GFP-WipB1-364 distributed uniformly throughout the cytoplasm, mirroring the localisation of control GFP. Strikingly, GFP-WipB365-524 exhibits a localization similar to GFP-WipB, demonstrating that the C-terminal domain is sufficient for lysosomal targeting.
WipB interacts with components of the eukaryotic LYNUS apparatus
The targeting of the WipB Ser/Thr phosphatase to lysosomes by the C-terminal domain, suggested that WipB might target host phosphoproteins within the endo-lysosomal system. To identify potential host targets of WipB, we performed co-immunoprecipitation experiments with anti-GFP antibodies from transfected cell lysates, followed by mass spectrometry analysis, using HeLa cells expressing GFP-WipB, the catalytically dead derivative GFP-WipBD118A, or GFP alone as a control. The proteins identified under each condition were compared and purged of those interacting with GFP alone. The proteins remaining in the GFP-WipB and GFP-WipBD118A datasets were strikingly enriched for lysosomal proteins with 23% of the lysosomal membrane proteome represented amongst the data18. Amongst the common targets, we identified lysosomal proteins present within the top 50 hits that are also predicted to contain Ser/Thr phosphorylation sites according to the UNIPROT database. This identified flotillin-1, LAMTOR1 (p18), and two subunits of the vacuolar H+-ATPase (v-ATPase), subunits B and d1 (Table 2). Three of these four proteins are functionally linked, as LAMTOR1 and the two v-ATPase subunits interact as part of a larger macromolecular assembly termed the lysosomal nutrient sensing (LYNUS) apparatus that controls the mammalian target of rapamycin (mTORC1) kinase complex at the lysosomal surface19. LAMTOR1 is a phosphoprotein that links the Ragulator complex to the lysosomal surface, which is also involved in mTOR signalling and is activated via the v-ATPase20. Therefore, we focussed on these three WipB interactors.
Firstly, we verified our mass spectrometry results by immunoblotting the anti-GFP immunoprecipitation samples using antibodies specific for subunits A and B of the v-ATPase and against LAMTOR-1 (Fig. 3d and Fig. S5a). These data show that GFP-WipB and GFP-WipBD118A, but not GFP alone, can indeed immunoprecipitate the three target proteins.
Secondly, equivalent immunoprecipitations with N-terminal GFP-WipB1-364 and C-terminal GFP-WipB365-524 were performed (Fig. 3d and Fig. S5a). While GFP-WipB365-524 interacts with the two v-ATPase subunits and LAMTOR1, GFP-WipB1-364 only binds the v-ATPase subunits. This reveals that while both domains of WipB can engage with the v-ATPase subunits, only the C-terminal domain can interact with LAMTOR1.
Finally, we performed reverse co-immunoprecipitation using antibodies against the v-ATPase A and B subunits and LAMTOR1 (Fig. 3e and Fig. S5b). Cells were transfected with either GFP alone or GFP-WipB and lysates incubated in the presence of beads coupled to antibodies against the v-ATPase A or B subunits, or LAMTOR1. Immunoprecipitated proteins were then analysed by SDS-PAGE and Western blotting using anti-GFP antibody. As shown in Fig. 3e, all three proteins were able to pull-down GFP-WipB, but not GFP alone.
Altogether, these results show that the WipB Ser/Thr phosphatase targets key components of the host LYNUS complex on the lysosome, when expressed in mammalian cells.
Discussion
In this study, we determined the structure and characterized the catalytic domain of the Legionella pneumophila effector WipB. We showed that this domain belongs to the PPP family of eukaryotic phosphatases and has a Ser/Thr phosphatase activity in vitro 21.
Ser/Thr phosphatases regulate a wide number of processes in eukaryotic cells, including targeting signalling cascade components to membrane receptors14. Their substrate(s) specificity often resides within their regulatory domains/subunits. Similarly, we showed that WipB has a specific subcellular localisation in HeLa cells controlled by its C-terminal domain, which targets it to lysosomes.
We demonstrated that WipB can interact with subunits of the v-ATPase and LAMTOR1 when expressed in cultured cells. The v-ATPase is a transmembrane complex that controls the acidification of cellular organelles, comprised of 17 subunits22, which harbour several phosphorylation sites that regulate pump function23. Legionella may modulate the function of the v-ATPase since part of the LCV membrane is derived from the lysosomal compartment24, 25, yet the lumen remains at neutral pH26. Indeed, the Legionella effector SidK binds the v-ATPase regulatory subunit VatA, resulting in the inhibition of the complex and organelle acidification26. As with wipB mutants, a sidK mutant did not exhibit defects in intracellular replication26. Consequently, it is possible that redundant functions of SidK and WipB converge to repress the activity of the host v-ATPase through direct binding, promoting bacterial growth.
Recently, mTOR-driven metabolic reprogramming was found to be important for intracellular replication by Legionella 27. An unknown T4S effector/s was proposed to activate mTOR to promote LCV expansion, a process that required a functional Dot/Icm secretion system and the IcmS chaperone, in addition to host phosphatidylinositol-3-kinase. Lysosomes play an important role in coordinating nutrient sensing and signalling pathways involved in cell metabolism and growth. LAMTOR1 is a key phosphoprotein of this system20, directing the nucleation of the multi-component LYNUS apparatus including mTOR and v-ATPase on the lysosomal surface19, although its mode of regulation are not yet clearly understood. Although further investigation is required, it is tempting to speculate that WipB is a T4S effector involved in the metabolic reprogramming of Legionella-infected cells.
Using transposon site hybridization (TraSH), several “functional groups” of effectors were proposed to concomitantly act on cellular pathways: while individual effector deletion has no effect, their combined deletion altered L. pneumophila growth in host cells11. While single deletion of wipB and lidA are phenotypically silent, a wipB/lidA double mutant exhibited defects in the stability of the LCV. During infection, LidA promotes the recruitment of ER-derived vesicles to the LCV through its binding to the AMPylated form of the Rab1 GTPase28. In this manner, LidA contributes to replication-niche formation, essential for L. pneumophila to gather the resources it needs for cell division. On the other hand, escaping the host defence mechanisms by repressing the lysosomal pathway, a process WipB could be involved in, appears crucial to bacterial survival. Thus, impairing both LidA and WipB function would alter two main complementary virulence strategies, eventually leading to intracellular growth defects.
A recent genetic study conducted on the genomes of 41 Legionella species remarkably showed that effector repertories share some conserved domains within and between species29. Those domains, catalytic, binding or uncharacterized domains, are shuffled to generate effector pools, which consist of nearly 6000 effectors in the whole sequenced genus. Using the L. pneumophila effectors, the authors define 608 orthologue groups of proteins sharing one or several conserved domains, representing 80% of the 6000 putative proteins. One orthologue group is based on the WipB sequence (group number LOG_00109), which according to their findings, could have orthologues in 32 out of the 41 sequenced Legionella species. In addition, a search of homologues to the WipB catalytic domain using BLAST30 also identifies hypothetical proteins effectors in Coxiella and Fluoribacter species that could then share the same enzymatic activity. Thus our findings on L. pneumophila WipB might have implications for understanding a whole family of effectors from the entire Legionella genus.
L. pneumophila secretes a wide range of protein effectors during the infection that leads to Legionnaire’s disease. Assigning the functions of these effectors is often complex given the high redundancy they exhibit. Here we demonstrate how combining structural biology and cellular biology is a powerful alternative route to identifying the role and targets of Legionella effectors.
Methods
Cloning
The WipB DNA (AAU28775) encoding the wild type protein (Q5ZS02_LEGPH, lpg2718) was cloned in a modified pETM14 vector (EMBL) or in empty pEGFP-C2 and -N2 vectors using a PCR-based in-fusion HD cloning system (Clontech Laboratories). For the pETM14-derived constructs, the expression cassette contained an N-terminal deca-histidine tag followed by a 3 C protease cleavage site. Site directed mutagenesis was performed using standard molecular biology protocols.
Expression and purification of WipB25-344 and WipB1-364
All recombinant proteins were over-expressed in C43(DE3) bacterial strains. The cells were harvested by centrifugation (6000 g, 15 min) and resuspended in a lysis buffer containing 25 mM Tris-HCl pH 7.5, 0.3 M NaCl, 5 mM β-mercaptoethanol (βME), 10 mM imidazole, 5% glycerol, a tablet of protease inhibitors (Complete, EDTA-free by Roche) and 0.25 mg/ml lysozyme. Cells were lysed in an EmulsiFlex-C3 homogeniser (Avestin) and the crude extract was centrifuged at 50,000 g for 45 min. The supernatant was loaded onto a 5 ml HisTrap column (GE Healthcare) equilibrated with the lysis buffer, connected on an AKTA purifier (GE Healthcare). Washing steps were performed with extended volumes of lysis buffer though the column as well high salt buffer (25 mM Tris-HCl pH 7.5, 1 M NaCl, 5 mM βME, 10 mM imidazole, 5% glycerol). The protein was eluted using an imidazole gradient (Elution buffer: 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM βME, 0.6 M imidazole, 5% glycerol). The eluted protein was dialyzed overnight against buffer A (25 mM Tris-HCl pH 7.5, 5% glycerol) in presence of 3C-protease fused to GST to a molar ratio protein/protease of 50:1. The cleaved protein was collected in the flowthrough of two 1 ml HisTrap and GST-Trap columns (GE Healthcare) connected in sequence to remove uncleaved proteins and the protease. It was then loaded on a resource Q column (GE healthcare) and eluted with a gradient of NaCl at a concentration of about 0.1 M NaCl. The eluted protein was further concentrated and loaded to a superdex200 16/60 column (GE healthcare) equilibrated in 25 mM Tris-HCl pH 7.5, 0.15 M NaCl, and 5% glycerol.
Phosphatase activity assay
Assays were conducted on purified protein after His-tag removal using the Tyrosine and Ser/Thr Phosphatase Assay Systems kits (Promega). Briefly, the protein was diluted in its purification buffer containing 1 mM of MnCl2 and incubated at either 21 °C or 37 °C with the provided peptide before stopping the reaction with a buffer containing molybdate and malachite green to measure free phosphate at 600 nm in a 96-well microplate. One phosphothreonine-containing peptide (sequence RRA(pT)VA) and two phosphotyrosine-containing peptides (END(pY)INASL and DADE(pY)LIPQQG) were provided in the kits and were used as per manufacturer’s instructions.
Crystallization, data collection and processing
Initial crystallization screens were performed using the sitting-drop vapor-diffusion technique, by mixing equal volumes (0.2 μl) of protein solution (13 mg/ml) at 16 °C. Crystals in the shape of thin needles appeared after 3-4 days reaching a maximum length of 0.1-0.2 mm against a reservoir solution containing 0.23 M ammonium citrate dibasic and 22% PEG3000. Before data collection, harvested crystals were immersed in a solution containing the precipitant mixture and 10% MDP and cryo-cooled in liquid nitrogen. All data sets were collected at 100 K. Crystals of the WipB25-344 were collected at the PetraIII P13 beam-line (EMBL-Hamburg/DESY P13, Germany). The data set was indexed, processed and scaled using the XDS package31, (Table 1).
Structure determination and refinement
The WipB25-344 crystals belonged to the P 21 space group with a solvent content of 43.12% corresponding to two molecules per asymmetric unit (AU). The structure was determined by molecular replacement using MOLREP32 and the WipA structure (PDB Code 5N72) containing only the α-helices and β-strands of the phosphatase domain as search model. The coordinates were further improved by maximum-likelihood and TLS refinement using the PHENIX suite33 and manual improvements of the model using COOT34. The final model converged to a final Rwork/Rfree of 0.187/0.217 at a resolution of 1.70 Å. The WipB25-344 model covers the WipB amino-acid sequence 24-331 (chain A) and 25–344 (chain B) and contains in addition 484 water molecules.
Antibodies and cellular biology reagents
CHIP grade rabbit anti-GFP (Invitrogen) was used for co-immunoprecipitation and mouse anti-GFP (clontech) for Western-blotting. Rabbit anti-vATPase A (Abcam), anti-vATPase B (Abcam) or anti-LAMTOR1 (Sigma) were used for co-immunoprecipitation and Western blotting. Anti-Sec. 61b (Millipore) was used as a loading control in Western-blotting. HRP-coupled anti-mouse and anti-rabbit (Invitrogen) and TrueBlot anti-rabbit (Rockland) were used for Western-blotting and detected using ECL kit (GE Healthcare). For cell labelling, we used anti-LAMP1 (rabbit, NEB), anti-Giantin (rabbit, Covance), anti-LaminA/C (mouse, Santa Cruz), and MitoTracker, LysoTracker Red, Dextran TRITC 10,000 and Transferrin647 were purchased from Thermofisher.
Cell culture, transfection, labelling and microscopy
Hela cells were routinely cultured, transfected and immunolabeled as previously described35. For in vivo labeling, cells were incubated at 37 °C for 15 minutes with Dextran TITRC 10,000 at 1 mg/ml, MitoTracker at 250 nM, Transferrin-Alexa647 at 0.025 mg/ml or LysoTracker Red at 50 nM in DMEM then washed in HBSS. Transferrin-labeled cells were stripped in stripping buffer (150 mM NaCl, 100 mM Glycine, 5 mM KCl, 1 mM CaCl2, pH4.5).
Cells were imaged using a confocal microscope (TCS Sp5 AOBS; Leica) with an oil-immersion objective (63 × , 1.4 NA; Leica) as previously described35. Images were analyzed using Fiji36.
Mass spectrometry analysis
HeLa cells were sonicated 24 h after transfection in PBS containing protease inhibitors. Lysates were incubated for 30 min at 30 °C with beads coupled to anti-GFP antibodies equilibrated in PBS Tween 0.02%(v/v). After washing, beads were processed in mass spectrometry as described previously35.
Co-immunoprecipitation
Dynabeads protein G (Thermofisher) were incubated with anti-GFP, anti-vATPase A, anti-vATPase B or anti-LAMTOR1 antibodies in a buffer PBS Tween 0.02% (v/v) 10% BSA (w/v) for 10 min. Cell lysates were incubated with the washed beads for 1 h in the presence of 5% BSA to reduce non-specific interactions. After washing, proteins were eluted with 75 mM Tris HCl pH 7.4, 0.5 M EDTA, 0.05% SDS and analyzed by western blotting.
References
McDade, J. E. et al. Legionnaires’ Disease. N. Engl. J. Med. 297, 1197–1203 (1977).
Finsel, I. & Hilbi, H. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell. Microbiol. 17, 935–950 (2015).
Segal, G., Purcell, M. & Shuman, H. A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95, 1669–1674 (1998).
Vogel, J. P. Conjugative Transfer by the Virulence System of Legionella pneumophila. Science 279, 873–876 (1998).
Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682 (2002).
Luo, Z.-Q. & Isberg, R. R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101, 841–846 (2004).
Campodonico, E. M., Chesnel, L. & Roy, C. R. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56, 918–933 (2005).
Burstein, D. et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5, e1000508 (2009).
Lifshitz, Z. et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. USA 110, E707–15 (2013).
Ninio, S., Zuckman-Cholon, D. M., Cambronne, E. D. & Roy, C. R. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55, 912–926 (2005).
O’Connor, T. J., Boyd, D., Dorer, M. S. & Isberg, R. R. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science 338, 1440–1444 (2012).
Pinotsis, N. & Waksman, G. Structure of the WipA protein reveals a novel tyrosine protein phosphatase effector from Legionella pneumophila. J. Biol. Chem. doi:10.1074/jbc.M117.781948 (2017).
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–9 (2010).
Shi, Y. Serine/Threonine Phosphatases: Mechanism through Structure. Cell 139, 468–484 (2009).
Donella Deana, A. et al. An investigation of the substrate specificity of protein phosphatase 2C using synthetic peptide substrates; comparison with protein phosphatase 2A. Biochim. Biophys. Acta 1051, 199–202 (1990).
Daum, G. et al. A general peptide substrate for protein tyrosine phosphatases. Anal. Biochem. 211, 50–54 (1993).
Zhang, Z. Y. et al. Substrate specificity of the protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 90, 4446–4450 (1993).
Chapel, A. et al. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell Proteomics 12, 1572–1588 (2013).
Settembre, C., Fraldi, A., Medina, D. L. & Ballabio, A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14, 283–296 (2013).
Nada, S., Mori, S., Takahashi, Y. & Okada, M. in Endosome Signaling Part B (ed. Conn, P. M.) Volume 535, 249–263 (Academic Press, 2014).
Shi, L., Potts, M. & Kennelly, P. J. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiology Reviews 22, 229–253 (1998).
Maxson, M. E. & Grinstein, S. The vacuolar-type H+-ATPase at a glance - more than a proton pump. J. Cell. Sci. 127, 4987–4993 (2014).
Alzamora, R. et al. AMP-activated protein kinase regulates the vacuolar H+ -ATPase via direct phosphorylation of the A subunit (ATP6V1A) in the kidney. Am. J. Physiol. Renal Physiol. 305, F943–56 (2013).
Amer, A. O. & Swanson, M. S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 7, 765–778 (2005).
Urwyler, S. et al. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 10, 76–87 (2009).
Xu, L. et al. Inhibition of Host Vacuolar H+ -ATPase Activity by a Legionella pneumophila Effector. PLoS Pathog 6, e1000822 (2010).
Abshire, C. F., Dragoi, A.-M., Roy, C. R. & Ivanov, S. S. MTOR-Driven Metabolic Reprogramming Regulates Legionella pneumophila Intracellular Niche Homeostasis. PLoS Pathog 12, e1006088 (2016).
Neunuebel, M. R., Mohammadi, S., Jarnik, M. & Machner, M. P. Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J. Bacteriol. 194, 1389–1400 (2012).
Burstein, D. et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175 (2016).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Cryst. D 66, 22–25 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Cryst. D 66, 486–501 (2010).
Dumoux, M., Menny, A., Delacour, D. & Hayward, R. D. A Chlamydia effector recruits CEP170 to reprogram host microtubule organization. J. Cell. Sci. 128, 3420–3434 (2015).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat Meth 9, 676–682 (2012).
Acknowledgements
This work was funded by ERC grant 321630 to G. W. Synchrotron MX data were collected at beamline P13 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We would like to thank Dr. Johanna Kallio for the assistance in using beamline P13.
Author information
Authors and Affiliations
Contributions
M.S.P. performed the biochemistry and cellular experiments and some of the crystallography experiments; M.D. and M.S.P analysed the cellular data; N.P. designed the biochemical experiments, purified, crystallized and solved the structure; G.W. and R.D.H. supervised the research; and all authors wrote the paper.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Prevost, M.S., Pinotsis, N., Dumoux, M. et al. The Legionella effector WipB is a translocated Ser/Thr phosphatase that targets the host lysosomal nutrient sensing machinery. Sci Rep 7, 9450 (2017). https://doi.org/10.1038/s41598-017-10249-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10249-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.