Abstract
The fungal communities of different soil compartments in mangrove ecosystem are poorly studied. We sequenced the internal transcribed spacer (ITS) regions to characterize the fungal communities in Avicennia marina root-associated soils (rhizosphere and pneumatophore) and bulk soil compartments. The rhizosphere but not pneumatophore soil compartment had significantly lower fungal species richness than bulk soil. However, bulk soil fungal diversity (Shannon diversity index) was significantly higher than both pneumatophore and rhizosphere soil compartments. The different soil compartments significantly affected the fungal community composition. Pairwise sample analyses showed that bulk soil microbial community composition significantly different from rhizosphere and pneumatophore soil compartments. There was, however no significant difference observed between rhizosphere and pneumatophore soil fungal community composition and they shared relatively more OTUs between them. Further, there was a significant correlation observed between fungal community compositional changes and carbon or nitrogen availability of different soil compartments. These results suggest that few characteristics such as fungal richness and taxa abundance of rhizosphere and pneumatophore soil compartments were significantly different in mangrove ecosystem.
Similar content being viewed by others
Introduction
Mangroves generally grow in latitudes 32°N and 38°S falling in tropical and subtropical climate regions worldwide1. Mangroves are one of the most productive ecosystems that are known to actively participate in various bio-geo chemical processes2, 3. Several factors such as salinity fluctuation, inundation and anoxic nature of mangrove sediments create a stressful environment for the plants1. Avicennia marina is one of the major mangrove plants that generally grow in anoxic mangrove sediments of fringe or coastal mangroves1. Avicennia marina produce lateral roots as well as specialized root modifications called pneumatophores or aerial roots (pencil roots) in order to obtain physical stability in muddy mangrove sediments. Pneumatophores are produced from the lateral roots of the plants and grow vertically away from gravity. Pneumatophores carry out gaseous exchange, respiration and nutrient assimilation necessary for sustenance and growth of A. marina in prevailing atypical conditions of mangrove ecosystem1, 4. In addition, pneumatophores offer protection for A. marina from soil erosion5 and participate in biogeochemical cycles like carbon and nitrogen fixation6,7,8.
Previous studies investigated the diversity and functional characteristics of mangrove fungi on woody parts of trees, decomposing litter and sediments9,10,11,12,13,14,15,16,17. Atypical conditions of magrove roots may create a rich habitat, thus allowing the colonization of different microbial communities5. A very few studies have characterized the pneumatophore-associated (epi- and endosphere) fungi9, 12, 14, 16, 17. However, fungal diversity of lateral root attached soil (hereafter referred to as “rhizosphere”) in mangrove ecosystems is poorly studied18,19,20,21 and pneumatophore attached soil (hereafter referred to as “pneumatophore”) is virtually unknown. The comparative studies on fungal community of root-associated (pneumatophore and rhizosphere) soil compartments would help us in discerning the functional difference between them.
Similar to other ecosystems, fungi are predicted to play an important role in carbon and nitrogen recycling in mangrove ecosystems22,23,24,25. It has been shown that fungal isolates from mangrove ecosystems are able to synthesize the organic matter decomposing enzymes26,27,28,29. It is considered that the fungi initially begin the decomposition of plant materials in mangrove ecosystem30. The yeast and bacteria further complete the decomposition process30. The availability of carbon and nitrogen could influence the fungal community composition across the soil compartments. However, there is no information currently available pertaining to the correlation of microbial community composition to carbon and nitrogen availability in mangrove ecosystem.
Considering the functional specialty of pneumatophore roots in mangrove ecosystems, we hypothesized that root-associated soil compartments, rhizosphere and pneumatophore differ in their fungal richness (number of fungal taxa) and Shannon diversity (a combined measure of richness and the evenness of their abundance). We chose A. marina as focal tree due to its wide spread occurrence in fringe mangroves. In the present study, we tested the following predictions in mangrove ecosystem: (i) fungal richness and diversity are significantly different in rhizosphere, pneumatophore and bulk soil compartments (hereafter referred to as “bulk”), (ii) the selected soil compartments, bulk, pneumatophore and rhizosphere affect fungal community composition significantly and (iii) the availability carbon and nitrogen of different soil compartments affect a part of fungal community composition.
Results
We investigated fungal communities of bulk, rhizosphere, and pneumatophore soil compartments (Fig. 1) in a mangrove ecosystem. The root-associated soil compartments (rhizosphere and pneumatophore) were collected from A. marina trees. High-throughput sequencing of internal transcribed regions (ITS) was yielded 117, 688 good quality sequences (Supplementary file: Supplementary Table 1). The removal of rare OTUs (OTUs with less than five sequences) reduced the total number of sequences to 115 021 (Supplementary file: Supplementary Table 1). Good’s coverage (https://www.mothur.org/wiki/Coverage) scores were high across the compartments ranging from 0.97 to 0.99 indicating that the sequencing depth was adequate to dependably describe the fungal microbiome (Supplementary file: Supplementary Table 1). A total of 21 different fungal classes were identified across the three different compartments (Fig. 2). Out of which 20, 19 and 18 were found in bulk, rhizosphere and pneumatophore compartments, respectively.
The fungal richness and diversity (Shannon) was affected by compartment
To gain insights into the effect of compartment on fungal richness and diversity (Shannon), we performed one way analysis of variance (ANOVA) with compartment as fixed factor. The analyses showed that both OTU richness and Shannon diversity index were significantly affected by compartments (Supplementary file: Supplementary Table 2).
Furthermore, in order to identify the significant difference between compartments, comparisons of observed OTU richness and Shannon diversity index were carried out using communities retrieved from bulk, rhizosphere and pneumatophore samples (Fig. 3). The pneumatophore accounted for the highest OTU richness followed by bulk. The rhizosphere accounted for the lowest OTU richness and it was significantly (p < 0.05) different from pneumatophore and bulk compartments (Fig. 3A). The bulk samples accounted for the highest Shannon diversity index and significantly (p < 0.05) different from pneumatophore and rhizosphere (Fig. 3B).
Fungal community composition was affected by compartment
To gain insights into the effect of compartment on fungal community composition, we performed unconstrained principal coordinate analysis (PCoA) to identify the clustering pattern of microbial communities. The bulk samples clustered separately from pneumatophore and rhizosphere compartments in global PCoA (Fig. 4). Permutational Multivariate Analysis of Variance (PERMANOVA) confirmed that the observed clustering pattern in PCoA was statistically significant (p < 0.05; Supplementary file: Supplementary Table 3). A total of 40.7% [(Factor sum of squares)/(total sum of squares) * 100] of the variations was explained by the compartment as fixed factor in PERMANOVA (Supplementary file: Supplementary Table 3).
Clustering pattern of fungal community composition in bulk, pneumatophore and rhizosphere soil compartments. The C and N in the figure indicate the carbon and nitrogen content of the different soil compartments, respectively. The significant correlation was observed between fungal community composition and carbon or nitrogen content of the soil compartments. OTUs were clustered at 97% similarity level. The weighted and constrained principal-coordinate analysis (PCoA) based on Bray–Curtis dissimilarity on standardized and square-root transformed data was performed in PRIMER software v6.
We performed pairwise PCoAs of bulk Vs rhizosphere, bulk Vs pneumatophore, and rhizosphere Vs pneumatophore (Supplementary file: Supplementary figure 1). The PERMANOVA analyses showed that bulk Vs rhizosphere and, bulk Vs pneumatophore clustering patterns were statistically significant (p < 0.05; Supplementary file: Supplementary Table 4). But, rhizosphere Vs pneumatophore clustering pattern was not significantly different (p > 0.05; Supplementary file: Supplementary Table 4).
We performed Kruskal–Wallis test with log transformed (log [X + 1]) relative abundance data to identify the OTUs that are responsible for community separation in different compartments. To exclude rare OTUs, we included OTUs with at least 1% abundance in minimum of three samples, which resulted in 25 fungal OTUs. There were 13 and 11 OTUs that displayed significant (p < 0.05) difference between bulk Vs pneumatophore and bulk Vs rhizosphere compartments, respectively (Supplementary file: Supplementary Table 5). There was no significant difference observed in any OTUs between pneumatophore and rhizosphere soil compartments (Supplementary file: Supplementary Table 5).
The higher order fungal taxa were affected by compartment
We applied linear statistics to identify the fungal classes that differed across the compartments. Out of 21 total classes identified, 11 of them were highly enriched in one or two compartments (Fig. 5). For instance, unclassified Ascomycota was highly enriched (p < 0.05) in rhizosphere and pneumatophore compartments compared to bulk. In contrast, Saccharomycetes was highly enriched (p < 0.05) in bulk and pneumatophore compartments compared to rhizosphere.
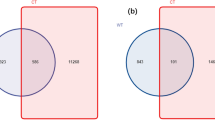
Root-associated compartments shared relatively more OTUs
A total of 446, 582, and 528 OTUs were present in bulk, pneumatophore and rhizosphere compartments, respectively (Fig. 6). A total of 137 OTUs were shared between bulk, pneumatophore and rhizosphere compartments (Fig. 6). A total of 23% of OTUs were shared between bulk and pneumatophore compartments. The rhizosphere and pneumatophore compartments shared relatively higher percentage OTUs (29%) followed by bulk and pneumatophore compartments (23%). The bulk and rhizosphere compartments shared 20% of OTUs between them.
Carbon and nitrogen content of different compartments determined a part of fungal community diversity (Shannon) and composition
The carbon and nitrogen content of pneumatophore were significantly (p < 0.05) higher than bulk (Supplementary file: Supplementary Table 2). In contrast, the carbon and nitrogen content of rhizosphere was comparable to the pneumatophore and bulk.
Pearson correlation analysis showed that no significant (p > 0.05) correlation observed between species richness and carbon or nitrogen content of different compartments (Supplementary file: Supplementary Table 6). However, there was a significant (p < 0.01) correlation observed between Shannon diversity and carbon or nitrogen content of different compartments (Supplementary file: Supplementary Table 6 and Supplementary Figure 3). Similarly, distance based linear model analysis (DistLM) showed that significant correlation between fungal community composition with carbon or nitrogen content of different compartments (p < 0.05; Table 1). The nitrogen and carbon content of the soil compartments explained 15% and 16% of the variation in fungal community composition, respectively (Table 1).
Discussion
The fungal diversity and community composition in different soil compartments of mangrove ecosystem is poorly studied21. To our knowledge, this is the first study to characterize the pneumatophore soil fungal communities in mangrove ecosystem. Furthermore, we compared the fungal communities from bulk, pneumatophore and rhizosphere compartments. Our results showed that the different soil compartments of A. marina affect the fungal richness, diversity (Shannon) and community composition in the present mangrove ecosystem.
We hypothesized that fungal richness and diversity were significantly different in rhizosphere, pneumatophore and bulk compartments. Our results were not consistent with our hypothesis as bulk fungal OTU richness was higher than the rhizosphere compartment. In contrast, the pneumatophore fungal richness was comparable to bulk. The difference in sampling depth and more anaerobic condition in rhizosphere samples could be the potential reason for difference in lower richness of rhizosphere samples compared to pneumatophore. However, Shannon diversity of the bulk was higher than the both pneumatophore and rhizosphere compartments. The bulk fungi were more diverse than rhizosphere soil in other ecosystems31, 32. In contrast, it has been shown that fungal diversity was higher in rhizosphere than bulk in similar mangrove environment21. The difference in sampling depth of rhizosphere samples between the present (30 cm) and earlier (10 cm) studies21 could be the possible reason for contrasting results.
The fungal community composition of bulk was significantly different from root-associated soil compartments (pneumatophore and rhizosphere soils). This is consistent with fungal richness and diversity observations of this study. In addition, the root-associated compartments shared relatively more OTUs between them than individually with bulk. It is well known that the plant compounds released by roots play a major role in selecting the root-associated soil fungal communities32,33,34. In this study, the clustering pattern of different soil compartments showed that within sample variation was more pronounced in root-associated soil compartments than bulk. In contrast to our study, it has been shown that within sample variation was higher in bulk than rhizosphere in another mangrove ecosystem21. The difference in study site and methods (whole metagenomic sequencing Vs targeted amplicon sequencing) could be the potential reason for the dissimilarity between present and earlier21 studies.
The rhizo-deposited photosynthate and other exudates are important source of carbon and nitrogen for the microorganisms in the neighborhood of roots34. In this study, carbon and nitrogen content of root-associated samples were higher than bulk. Interestingly, only a fraction of variation in community composition was explained by carbon and nitrogen content of soil compartments. Also, negative correlation was observed between Shannon diversity and carbon or nitrogen content of the soil compartments. The carbon and nitrogen availability is considered to be one of the determinants of microbial communities in ecosystems35, 36.
The Dikarya (Ascomycota and Basidiomycota) were the most abundant phyla in this study, which is consistent with previous works on marine surface37,38,39 and mangrove ecosystems15, 21, 40. In this study, Dothideomycetes was the most abundant class (31% of total sequences) across the samples. In contrast, Dothideomycetes has been reported as a low abundant class of A. marina rhizosphere and sediments21. In this study, Saccharomycetes abundance was high in bulk compared to rhizosphere. Saccharomycetes has been reported as one of the major abundant fungal classes in anoxic mangrove sediment15. In contrast, Saccharomycetes is shown to be highly abundant in rhizosphere compared to bulk21. Our results and earlier study21 showed that the site-specific characteristics are the main reason for selecting the major root-associated soil fungal communities in mangrove ecosystems.
In this study, Amorosia littoralis (OTU1009) was one of the most abundant species in pneumatophore and rhizosphere samples compared to bulk. Amorosia littoralis has been reported as endophytic fungi of different mangrove trees including A. marina 41. Another major OTU (OTU0486) of this study, classified as Debaryomyces nepalensis was abundant in bulk compared to pneumatophore and rhizosphere soils. Debaryomyces nepalensis is a well-known salinity tolerant yeast species reported in mangrove environments42, 43. The higher presence of D. napelensis in the present mangrove ecosystem is justified due to higher salinity irrespective of seasonal difference44.
In conclusion, few characteristics such as species richness and taxa abundance were significantly different between pneumatophore and rhizosphere soils in this study. Most of the characteristics of the root-associated soil fungal communities (pneumatophore and rhizosphere) were significantly different from bulk soil. This indicates the possible effect of pneumatophore on surrounding soil and could be termed as “pneumatosphere” similar to rhizosphere for main roots. Further targeted studies are needed to understand the functional difference of pneumatosphere and rhizosphere soil fungal communities.
Materials and Methods
Site description and Sample collection
The sampling site is located at the edge of Vellar River and Bay of Bengal (11.49° N 79.76° W). The sampling was conducted during low tide season on 6th December 2014. The samples were collected from five trees separated by 5 to 12 meters distance apart. The soil samples were collected using poly vinyl carbonate (PVC) corer having 5 cm width. All the soil samples are composite of three corers per tree. The samples were collected from the following depths, bulk (0–10 cm), pneumatophore (0–10 cm) and rhizosphere (30 cm below the ground level). The bulk samples were collected 2–3 meters away from base of the tree, where influence of root is not present. Pneumatophore attached soil samples were collected from uprooted pneumatophores with the help of corer. Rhizosphere attached soil samples were collected vertically from base of the tree so as to avoid the collection of pneumatophore.
Carbon and nitrogen content analyses
Carbon and nitrogen content analyses were carried out using C-H-N analyzer (http://www.sticindia.com/saif_instruments.html#chanl) at Sophisticated Test & Instrumentation Centre (Cochin University of Science and Technology), Cochin, India.
Soil DNA extraction and high-throughput sequencing
The DNA was extracted from about 200 mg of soil using Power Soil DNA isolation kit (Mobio, USA) following manufacturer’s protocol with minor modification in the initial lysis step by vortexing at high–speed for 25–30 minutes. The soil DNA was extracted from all the samples within 24 hours of sampling and stored in −80° C. The PCR reaction and ion torrent sequencing was carried out as described in ref. 45. Briefly, we used fITS7 (5′-GTGARTCATCGAATCTTTG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) in order to overcome the long amplicon bias46. The PCR amplifications were performed as follows: one cycle of 5 minutes at 94 °C, followed by 30 cycles of 30 seconds at 94 °C, 45 seconds at 53 °C, and 90 seconds at 72 °C, followed by one cycle of 7 minutes at 72 °C. We used M13 system based library preparation47. The sequencing was carried out using Ion PGM Sequencing 400 Hi-Q Kit (Ion 314 chips; Life Technologies, Thermo Fisher Scientific, Waltham, Massachusetts, USA) following manufacturer’s instructions.
Bioinformatics
The bioinformatics analyses were carried as described in ref. 45 using Mothur software (v. 1. 35. 0)48. In brief, the parameters used were minlength = 200, maxambigs = 0, maxhomop = 8, qwindowaverage = 25, qwindowsize = 50 and bdiffs = 1. We used ITSx 1.0.7 software to extract the ITS2 region for better clustering of the sequences. Chimeric reads were removed using UCHIME 4.2 software (http://drive5.com/uchime). Fungal ITS sequences were clustered at 97% sequence similarity thresholds using CD-HIT 4.6.1 (cd-hit.org). Taxonomic identities were assigned to the sequences using the UNITE + INSD datasets (http://www.mothur.org/wiki/UNITE_ITS_database). Further, we removed the rare OTUs (less than five sequences) and normalized to their lowest number (3 464) of sequences for the further downstream analyses. OTU richness, Shannon diversity indices and venn diagram were calculated in mothur (v. 1. 35. 0)48.
Statistical analyses
The clustering pattern of fungal community composition of different soil compartments was visualized using unconstrained principal coordinate analysis (PCoAs) and the statistical significance of clustering pattern was estimated by Permutational Multivariate Analysis of Variance (PERMANOVA)49. To test the correlation between fungal community composition and carbon and nitrogen content of different soil compartments, we performed distance based linear model (DistLM) statistical analyses. All the above statistical analyses were performed in PRIMER software V650. We used generalized linear statistical model analysis to test the difference in carbon and nitrogen content of different soil compartments after testing the homogeneity of variances using the SPSS software (IBM SPSS v24).
Data availability statement
Raw sequence data of all the 15 samples were submitted to sequence read archive (SRA accession number: SRP102307).
References
Kathiresan, K. & Bingham, B. L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 40, 81–251 (2001).
Jennerjahn, T. C. & Ittekkot, V. Relevance of mangroves for the production and deposition of organic matter along tropical continental margins. Naturwissenschaften 89, 23–30 (2002).
Kristensen, E., Bouillon, S., Dittmar, T. & Marchand, C. Organic carbon dynamics in mangrove ecosystems: a review. Aquat. Bot. 89, 201–219 (2008).
Chapman, V. The functions of the pneumatophores of Avicennia nitida jacq. (Proceedings of the Linnean Society of London Ser. 152, Wiley Online Library, 1940).
Giri, C. et al. Distribution and dynamics of mangrove forests of South Asia. J. Environ. Manage. 148, 1–11 (2015).
Dromgoole, F. I. Carbon dioxide fixation in aerial roots of the New Zealand mangrove Avicennia marina var. resinifera (Note). New Zeal. J. Mar. Freshw. Res. 22, 617–619 (1988).
Pelegri, S. P., Rivera-Monroy, V. H. & Twilley, R. R. A comparison of nitrogen fixation (acetylene reduction) among three species of mangrove litter, sediments, and pneumatophores in south Florida. USA Hydrobiologia. 356, 73–79 (1997).
Lugomela, C. & Bergman, B. Biological N2-fixation on mangrove pneumatophores: preliminary observations and perspectives. Ambio. 31, 612–613 (2002).
Garg, K. Effect of salinity on cellulolytic activity of some pneumatophore-inhabiting fungi of Sunderban mangrove swamps. (1982).
Velho, S. & DeSouza, J. Studies on pectinolytic fungi from the mangrove sediments. Mahasagar 15, 167–173 (1982).
Ananda, K. & Sridhar, K. Diversity of filamentous fungi on decomposing leaf and woody litter of mangrove forests in the southwest coast of India. Curr. Sci. 87, 1431 (2004).
Tariq, M., Dawar, S. & Mehdi, F. S. Isolation of fungi from Avicennia marina. Pak. J. Bot. 38, 805 (2006).
Kathiresan, K., Manivannan, S., Nabeel, M. & Dhivya, B. Studies on silver nanoparticles synthesized by a marine fungus, Penicillium fellutanum isolated from coastal mangrove sediment. Colloids and surfaces B: Biointerfaces 71, 133–137 (2009).
Gilna, V. & Khaleel, K. Diversity of fungi in mangrove ecosystem. Journal of Experimental Sciences 2 (2011).
Arfi, Y., Marchand, C., Wartel, M. & Record, E. Fungal diversity in anoxic-sulfidic sediments in a mangrove soil. Fungal Ecology 5, 282–285 (2012).
Janarthine, S. & Eganathan, P. Plant Growth Promoting of Endophytic Sporosarcina aquimarina SjAM16103 Isolated from the Pneumatophores of Avicennia marina L. International journal of microbiology 2012 (2012).
Sarma, V. V. & Vittal, B. Biodiversity of mangrove fungi on different substrata of Rhizophora apiculata and Avicennia spp. from Godavari and Krishna deltas, east coast of India. Fungal Diversity 5, 23–41 (2000).
Lee, B. K. & Baker, G. E. Fungi associated with the roots of red mangrove, Rhizophora mangle. Mycologia, 894-906 (1973).
Thiripurasundari, G., Usharani, G., Parthasarathi, R. & Neelakandan, T. Microbial diversity of the rhizosphere soil of Avicennia marina and Avicennia officinalis collected from mangrove forest. Actinomycetes 10, 104cfu (2010).
Selvakumar, V., Panneerselvam, A., Vijayakumar, N., Savery, M. & Thajuddin, N. Diversity of endophytic and rhizosphere soil fungi of Avicennia marina in Maravakadu Mangrove Forest. IOSR J Pharm Biol Sci 9, 24–28 (2014).
Simões, M. F. et al. Soil and rhizosphere associated fungi in gray mangroves (Avicennia marina) from the Red Sea—a metagenomic approach. Genomics, proteomics &. bioinformatics 13, 310–320 (2015).
Hyde, K. & Lee, S. Ecology of mangrove fungi and their role in nutrient cycling: what gaps occur in our knowledge? Hydrobiologia 295, 107–118 (1995).
Agate, A., Subramanian, C. V. & Vannucci, M. Mangrove microbiology: role of microorganisms in nutrient cycling of mangrove soils and waters. (1988).
Holguin, G., Vazquez, P. & Bashan, Y. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol. Fertility Soils 33, 265–278 (2001).
Reef, R., Feller, I. C. & Lovelock, C. E. Nutrition of mangroves. Tree Physiol. 30 (2010).
Fell, J. W. & Newell, S. Y. Role of fungi in carbon flow and nitrogen immobilization in coastal marine plant litter systems. Mycology series (1981).
Findlay, R., Fell, J., Coleman, N. & Vestal, J. Biochemical indicators of the role of fungi and thraustochytrids in mangrove detrital systems. The biology of marine fungi.Cambridge University Press, Cambridge, 91–104 (1986).
Singh, N. & Steinke, T. Colonization of decomposing leaves of Bruguiera gymnorrhiza (Rhizophoraceae) by fungi, and in vitro cellulolytic activity of the isolates. S. Afr. J. Bot. 58, 525–529 (1992).
Bremer, G. Lower marine fungi (Labyrinthulomycetes) and the decay of mangrove leaf litter (Asia-Pacific Symposium on Mangrove Ecosystems, Springer, 1995).
Matondkar, S., Mahtani, S. & Mavinkurve, S. Studies on mangrove swamps of Goa: 1. Heterotrophic bacterial flora from mangrove swamps. Mahasagar 14, 325–327 (1981).
Smit, E., Leeflang, P., Glandorf, B., van Elsas, J. D. & Wernars, K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65, 2614–2621 (1999).
Gomes, N. C. et al. Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl. Environ. Microbiol. 69, 3758–3766 (2003).
Kowalchuk, G., Hol, W. & Van Veen, J. Rhizosphere fungal communities are influenced by Senecio jacobaea pyrrolizidine alkaloid content and composition. Soil Biol. Biochem. 38, 2852–2859 (2006).
Butler, J. L., Williams, M. A., Bottomley, P. J. & Myrold, D. D. Microbial community dynamics associated with rhizosphere carbon flow. Appl. Environ. Microbiol. 69, 6793–6800 (2003).
Evans, K. L., Greenwood, J. J. & Gaston, K. J. Dissecting the species-energy relationship. Proc. Biol. Sci. 272, 2155–2163 (2005).
Waldrop, M. P., Zak, D. R., Blackwood, C. B., Curtis, C. D. & Tilman, D. Resource availability controls fungal diversity across a plant diversity gradient. Ecol. Lett. 9, 1127–1135 (2006).
Cury, J. C. et al. Microbial diversity of a Brazilian coastal region influenced by an upwelling system and anthropogenic activity. PLoS One 6, e16553 (2011).
Gao, Z., Johnson, Z. I. & Wang, G. Molecular characterization of the spatial diversity and novel lineages of mycoplankton in Hawaiian coastal waters. The ISME journal 4, 111–120 (2010).
Wegley, L., Edwards, R., Rodriguez‐Brito, B., Liu, H. & Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719 (2007).
Li, W., Wang, M., Bian, X., Guo, J. & Cai, L. A High-Level Fungal Diversity in the Intertidal Sediment of Chinese Seas Presents the Spatial Variation of Community Composition. Frontiers in Microbiology 7 (2016).
Li, J., Sun, X., Chen, L. & Guo, L. Community structure of endophytic fungi of four mangrove species in Southern China. Mycology 7, 180–190 (2016).
Kumar, S. & Gummadi, S. N. Purification and biochemical characterization of a moderately halotolerant NADPH dependent xylose reductase from Debaryomyces nepalensis NCYC 3413. Bioresour. Technol. 102, 9710–9717 (2011).
Boonmak, C. et al. Candida siamensis sp. nov., an anamorphic yeast species in the Saturnispora clade isolated in Thailand. FEMS Yeast Res. 9, 668–672 (2009).
Loganathachetti, D. S., Sadaiappan, B., Poosakkannu, A. & Muthuraman, S. Pyrosequencing-based seasonal observation of prokaryotic diversity in pneumatophore-associated soil of Avicennia marina. Curr. Microbiol. 72, 68–74 (2016).
Poosakkannu, A., Nissinen, R., Männistö, M. & Kytöviita, M. Microbial community composition but not diversity changes along succession in arctic sand dunes. Environ. Microbiol. 19, 698–709 (2017).
Ihrmark, K. et al. New primers to amplify the fungal ITS2 region–evaluation by 454‐sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677 (2012).
Mäki, A., Rissanen, A. J. & Tiirola, M. A practical method for barcoding and size-trimming PCR templates for amplicon sequencing. BioTechniques. 60, 88–90 (2016).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Anderson, M. J. A new method for non‐parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001).
Clarke K. R. & Warwick R. M. Change in marine communities: an approach to statistical analysis and interpretation. 2nd Edition.(PRIMER-E Ltd: Plymouth, United Kingdom.) (2001).
Acknowledgements
Author DSL thanks Lady Tata memorial trust, Mumbai for providing junior (2012–2015) and senior research scholarship (2015–2017). A special thanks to The Ella and Georg Ehrnrooth Foundation (awarded to AP) for awarding the sequencing cost.
Author information
Authors and Affiliations
Contributions
The experiments were designed by D.S.L., A.P. and S.M. Data analyses were carried out by D.S.L. and A.P. The manuscript was written by D.S.L. and A.P. The manuscript was revised by D.S.L., A.P. and S.M.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sanka Loganathachetti, D., Poosakkannu, A. & Muthuraman, S. Fungal community assemblage of different soil compartments in mangrove ecosystem. Sci Rep 7, 8560 (2017). https://doi.org/10.1038/s41598-017-09281-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-09281-3
This article is cited by
-
Influence of decomposition agent application and schedule in wheat straw return practice on soil quality and crop yield
Chemical and Biological Technologies in Agriculture (2023)
-
Plant-Microbial Symbioses in Coastal Systems: Their Ecological Importance and Role in Coastal Restoration
Estuaries and Coasts (2022)
-
The importance of conditionally rare taxa for the assembly and interaction of fungal communities in mangrove sediments
Applied Microbiology and Biotechnology (2022)
-
Microbial diversity and ecological interactions of microorganisms in the mangrove ecosystem: Threats, vulnerability, and adaptations
Environmental Science and Pollution Research (2022)
-
Analysis of Fungal Composition in Mine-Contaminated Soils in Hechi City
Current Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.