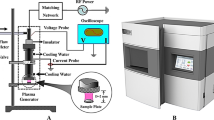
Abstract
Both gamma rays and atmospheric pressure plasma are known to have anticancer properties. While their mechanism actions are still not clear, in some contexts they work in similar manner, while in other contexts they work differently. So to understand these relationships, we have studied Myoglobin protein after the treatment of gamma rays and dielectric barrier discharge (DBD) plasma, and analyzed the changes in thermodynamic properties and changes in the secondary structure of protein after both treatments. The thermodynamic properties were analyzed using chemical and thermal denaturation after both treatments. We have also studied the action of gamma rays and DBD plasma on myoglobin in the presence of osmolytes, such as sorbitol and trehalose. For deep understanding of the action of gamma rays and DBD plasma, we have analyzed the reactive species generated by them in buffer at all treatment conditions. Finally, we have used molecular dynamic simulation to understand the hydrogen peroxide action on myoglobin with or without osmolytes, to gain deeper insight into how the osmolytes can protect the protein structure from the reactive species generated by gamma rays and DBD plasma.
Similar content being viewed by others
Introduction
Cancer is a complex disease that holds the ability to metastasize into different organs of the body, which results in the increase of cancer. Cancer cases worldwide are predicted to almost double in the next 20–40 years1, 2. A huge proportion of cancer patients suffer deterioration for years or spans later3, 4 causing a therapeutic challenge. For the clinical management of cancer, radiation remains by far the most utilized treatment for patients with localized malignant tumors, and it is applied in a course of multiple fractions for several weeks to decrease the toxicity in normal cell5,6,7. Low and high linear energy transfer (LET) radiations are using for radiation therapy to efficiently kill the tumor cells at minimum dose, in order to control the toxicity towards normal cells8. Gamma rays, X-rays, and charged particles are the utmost types of radiation delivered by a machine outside the body, or irradiated through radioactive material placed near to the cancer cells/tissue in the body for cancer treatment. However, ionizing radiations are also used in agriculture, pharmaceutical, and other technological processes, rather than treatment of biological systems. Among all other ionizing radiations, Gamma ray use is more economical and effective, because of its easy availability and penetration power9. Moreover, the change in structural, morphological, and biological systems depends on the strength and duration of exposure of gamma doses. Over the last few years, non-thermal atmospheric pressure (NTP) plasma has been studied as the alternative to ionizing radiations for cancer treatments.
Plasma medicine is a new field that uses NTP plasma for a diversity of medical applications10,11,12, such as sterilization12, wound healing13, blood coagulation14, 15, and cancer treatment16. NTP plasma interacts with the oxygen, nitrogen, water, etc. in air, to produce various radical and non-radical species, for example, hydroxyl radicals (•OH), superoxide (O2 •−), singlet oxygen (1O2), nitrogen dioxide (NO2), hypochlorite (ClO−), atomic oxygen (O), and nitric oxide (NO). During the plasma–liquid interactions, some relatively long lifetime reactive species are generated in liquid, such as hydrogen peroxide (H2O2), nitrites (NO2 −), and nitrates (NO3 −)17, 18. There are mainly two types of NTP plasma devices, i.e. plasma jet and dielectric barrier discharge, which display significant anti-cancer capacity in several cancer cell lines during in vitro studies19,20,21,22,23, and with several xenograft tumors for in vivo studies24,25,26,27.
Some of the cancer cell lines that are studied using NTP plasma, such as skin cancer28, breast cancer29, colorectal cancer30, lung cancer31, cervical cancer32, leukemia33, hepatoma34, as well as head & neck cancer35, reveal the selective anti-cancer treatment modality of NTP plasma treatment, in contrast to conventional anti-cancer approaches (chemotherapy, radiotherapy, and surgical excision). Similar to radiation treatments, NTP plasma treatments also damage the cells via an increase in the local concentration of reactive oxygen and nitrogen species (RONS). The increase in reactive oxygen species (ROS) also leads to induction of antioxidant defense mechanisms in the cell, such as glutathione peroxidase superoxide dismutase, catalase, superoxide dismutase, glutathione reductase, and glutathione peroxidase, as well as other small molecules, such as like NADPH and GSH and NADPH36, 37. Both the enzymatic defense in the cell cytosol compartment of CuZn-superoxide dismutase (CuZnSOD), and its mitochondrial counterpart of MnSOD37, were activated. A recent study shows that both the decreased and enhanced expression of Cu, Zn-SOD, or Mn-SOD influence the plasma-induced HeLa cell death38. However, the role of osmolytes in cancer treatment during plasma and gamma irradiation is still a mystery.
Most of the proteins adopt a tightly folded structure, signifying the lowest possible free energy of the polypeptide chain and the surrounding solvent under physiological conditions39. It has been reported that osmolytes are engaged in cell volume regulation in mammalian cells40. Some cells use organic osmolytes solutes, in addition to inorganic solutes, because high concentrations of inorganic salts (Na+ or K+) disturb the proteins function; however, organic solutes do not perturb the proteins function41. Under physiological conditions, proper protein conformation plays an important role in cell viability41. Different protein conformations depend upon the cellular and functional needs of the body42. In cells, the organic osmolytes accomplished the protein folding by shifting folding equilibrium, and they can manipulate the structure and conformation of proteins that provides the potential to ameliorate challenging diseases that occur due to misfolding of proteins. Protein misfolding challenges cell homeostasis through protein degradation or protein aggregation. In both cases, deleterious genetic disorders can happen. Therefore, osmolytes can stabilize the protein folding, and protein functions, and also shift the folding equilibrium away from the aggregation and/or degradation of proteins43,44,45,46. Therefore, to understand the action of gamma rays and NTP plasma on the proteins in the presence of organic osmolytes, we have used myoglobin as the model protein.
Myoglobin (Mr 16,700) is an oxygen binding protein that is found within muscle cells. Myoglobin protein crystal structure and its unfolding have been well studied47. The importance of myoglobin has been extensively discussed in most biochemistry texts48. Myoglobin contains a heme prosthetic group, and one polypeptide chain of 153 residues. The heme group resides deep inside the hydrophobic pocket within a protein’s structure. Decrease in the sorbet band has been observed due to denaturing effect of polar aqueous solvents on the heme which in turn results in the denatured myoglobin49.
Therefore, in this work we have treated the myoglobin protein with gamma rays and DBD NTP plasma in ambient air with or without osmolytes (sorbitol (500 mM) and trehalose (500 mM)), and checked the structural and thermodynamic properties, using CD spectroscopy and UV-vis spectroscopy. In addition, we have studied the reactive oxygen and nitrogen species (RONS) generated in phosphate buffer after the irradiation of gamma rays and DBD plasma. Further, we have studied the molecular dynamic simulation of myoglobin and hydrogen peroxide (H2O2) interactions with and without the osmolytes. This study provides information on the role of osmolytes in protein folding during gamma ray and DBD plasma treatments.
Results and Discussion
Protein denaturation studies provide remarkable insight into the kinetics and thermodynamics of protein folding50,51,52,53,54,55,56,57,58,59. Chemical or temperature induced unfolding provides insight into the thermodynamic stability of a protein. Therefore, in the present study we study the chemical and thermal denaturation of Myoglobin proteins after Gamma ray and DBD plasma treatments.
Change in absorbance of the protein after treatment with Gamma rays and DBD plasma in the presence of urea
The interaction of heme with histidine residue in the restricted pocket of fully-folded Myoglobin protein leads to an intense Soret band peak49. To illustrate myoglobin unfolding with increasing urea concentration, we studied the absorbance intensity at 409 nm. The high absorbance intensity is proportional to the concentration of folded protein molecules, and changes of absorbance at 409 nm correlate to the myoglobin unfolding. The absorbance at 409 nm in the spectra for native myoglobin in phosphate buffer is assumed to correspond to 100% folded myoglobin protein (Af) at 0 M urea concentration, and the absorbance at 409 nm with 9.0 M urea is assumed to the unfolded protein (Au).
Here, the A409 is the absorbance at all concentrations of urea (except 0 and 9 M), and this was used to calculate the fraction folded. Figure S1 shows that without treatment with the Gamma rays, 50% fraction of proteins was denatured at 6.5 M urea. But after treatment with gamma rays at the absorbed dose of 228, 424, and 1,136 Gy, the 50% fraction of proteins was denatured at 6, 5.6, and 5 M of urea concentration, respectively. These results reveal that as the gamma rays absorbed dose increases, the myoglobin is denatured at lower urea concentration. The least denaturation occurred at the absorbed dose of 228 Gy, and the maximum denaturation occurred at the absorbed dose of 1,136 Gy as the function of urea concentration. When we added the 500 mM of sorbitol or trehalose to the Myoglobin protein solution, the 50% fraction of protein denaturation occurred at 6.5 M of urea concentration, for both osmolytes. Hence, the fraction of protein denatured in the presence of urea is the same for Myoglobin protein with or without osmolytes. So later, we have treated the protein with gamma rays at different intensity, in the presence of sorbitol and trehalose, respectively. Figure S1 shows that the 50% fraction of protein was denatured at 5.2, 5.8, and 6.1 M of urea in the presence of sorbitol, and 5.6, 6, and 6.3 M of urea in the presence of trehalose, for the absorbed dose of 1,136, 424, and 228 Gy, respectively.
Further, we have treated the Myoglobin protein with DBD plasma in the ambient air, containing 40% humidity. We observed that after treatment of 5, 10, and 20 min of DBD plasma, the 50% fraction of protein was denatured at 6, 5.6, and 4.5 M of urea, as shown in Figure S2. Whereas for the protein solution in the presence of sorbitol (500 mM), when treated with DBD plasma for 5, 10, and 20 min, the 50% fraction of protein was denatured at 6, 5.8, and 5.5 M of urea. On the other hand, in the presence of 500 mM trehalose, the 50% fraction of protein was denatured at 6.2, 6.0, and 5.6 M of urea, after treatment of DBD plasma for 5, 10, and 20 min (Figure S2). This shows that in both treatments, of gamma rays and DBD plasma, the structure of Myoglobin denatured at lower concentration of urea, as the absorbed dose of gamma rays increases, or the treatment time of DBD plasma increases. But in the presence of osmolytes, the structure of protein was denatured at higher concentration of urea as compared to without osmolytes after the treatment with gamma rays and DBD plasma, respectively.
Thermal denaturation of Myoglobin protein after the treatment with gamma rays and DBD plasma
Well-defined three-dimensional structures of proteins commonly occur only within the bounds of specific environmental conditions. Outside the environmental boundaries, proteins display an unfolded state. Many research groups used CD spectroscopy to understand the melting temperature or transition temperature (Tm) of proteins/enzymes56, 57, 60, 61. Therefore, in our study we have used CD spectroscopy to understand the change in Tm of protein in the presence of osmolytes; and also, the impact of gamma rays and DBD plasma on the change of Tm of protein, with or without osmolytes.
Firstly, we have treated the myoglobin with gamma rays, and we observed at absorbed dose of 1,136, 424, and 228 Gy, the Tm of protein was 80, 81, and 82 °C, while the Tm of control protein (without gamma treatment) was 83 °C (Fig. 1a). On the other hand, when we have added the sorbitol (500 mM) to the myoglobin protein, the Tm was increased to 85 °C. Figure 1b shows that after the gamma irradiation on the solution containing the sorbitol and myoglobin, the Tm values were 81, 82.5, and 83 °C, at 1,136, 424, and 228 Gy, respectively. Nevertheless the addition of trehalose to the myoglobin sample solution increased the Tm value to 85.5 °C. Figure 1c shows that the changes in the Tm values of trehalose containing solution after the gamma rays treatment were 83, 83.5, and 84 °C at 1,136, 424, and 228 Gy absorbed dose, respectively. These results show that both osmolytes, i.e. sorbitol and trehalose, can protect the protein structure, even in the presence of gamma rays. But between the osmolytes, the trehalose has more counteraction tendency to control the myoglobin structure, even at high gamma absorbance dose.
Later, to understand the effect of osmolytes on the Tm values of proteins in the presence of DBD plasma, we have studied the thermal denaturation. We observed that the myoglobin thermodynamics has changed after the treatment with DBD plasma for 5, 10, and 20 min. Figure 2a shows that after treatment with DBD for 5, 10, and 20 min, the Tm value of protein changes to 81, 75, and 74 °C, respectively. Further, we observed that after 5, 10, and 20 min treatment of DBD plasma in the presence of sorbitol, the Tm value of protein changes to 84.5, 83.6, and 83 °C, respectively (Fig. 2b). However, Fig. 2c shows that the addition of trehalose drastically changes the Tm values after treatment of DBD plasma for 5, 10, and 20 min to 84, 83.8, and 83.7 °C, respectively. These results show only a ≈1 °C decrease in the Tm value of protein, even after 20 min DBD plasma treatment in the presence of trehalose.
If we compare the change in Tm of control myoglobin protein (without osmolytes) after gamma rays and DBD plasma irradiations, we observe that in the case of DBD treatment, the changes in Tm values are greater, as compared to the case of gamma ray treatments. However, in the presence of osmolytes (sorbitol and trehalose), the Tm value of protein has increased from 83 to 85 °C. When the gamma rays are irradiated on the samples containing osmolytes, the decrease in Tm values is much less (about ≈4 °C for sorbitol and ≈2 °C for trehalose for the highest studied absorbed dose of 1,136 Gy). Meanwhile, the decrease in Tm values after the DBD plasma treatment for 20 min in the presence of sorbitol is ≈2 °C, and for trehalose is ≈1 °C. These results reveal that osmolytes can control the denaturation action of protein after the DBD treatment better than after the gamma treatments. The possible reason for this process will be discussed in our latter sections.
Structural changes of myoglobin protein after gamma and DBD plasma treatments with or without urea
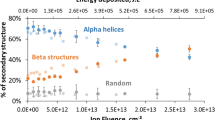
To understand the structural modification in myoglobin protein after treatment with gamma rays and DBD plasma, we further executed CD experiments. Figures 3 and 4 show that the far-UV CD spectrum of myoglobin shows distortion in the secondary structure of protein after different treatments. The displayed CD spectra of myoglobin have two well-pronounced minima at ≈210 and ≈222 nm that resemble those typical for the polypeptide chain that is mostly organized in α-helix conformation. Figure 3, reveals that after the treatment with the gamma rays at absorbed dose of 1,136, 424, and 228 Gy the α-helical structure of myoglobin was 54%, 57%, and 58%, respectively; whereas, the control myoglobin structure has α-helical of 63%. After the addition of sorbitol (500 mM) to the myoglobin solution, the α-helical is increased to 64%. Later, Fig. 3 shows that the percentage of α-helical changes to 55%, 56%, and 58%, after the treatment with gamma rays at the absorbed dose of 1,136, 424, and 228 Gy, respectively. Moreover, the change of α-helical structure in myoglobin with the addition of 500 mM trehalose is 65%. Further, the treatment of myoglobin protein solution in the presence of trehalose with gamma rays at the absorbed dose of 1,136, 424, and 228 Gy shows the change in α-helical structure to 58%, 59%, and 60%, respectively.
Figure 4 shows that, we have treated the myoglobin protein structure with the DBD plasma for 5, 10, and 20 min, and observed the change in α-helical structure of protein. After the treatment of myoglobin protein for 5, 10, and 20 mins, the % of α-helical changes to 60%, 59% and 57%, respectively. Latter, the addition of osmolytes increases the α-helical structure of protein, as seen above. To understand the impact of DBD plasma irradiation on the secondary structure of protein in presence of osmolytes, we observed the change in α-helical structure. The α-helical structure changes to 63%, 60% and 57%, in the presence of sorbitol + Myoglobin + 5, +10 and +20 min DBD plasma treatments, respectively. On the other hand, the addition of trehalose with DBD plasma treatment changes the α-helical structure to 62%, 61% and 59%, after treatment for 5, 10 and 20 min, respectively.
Figures S3 and S4 shows the impact of the various urea concentrations on the α-helical structure of protein at different conditions. We observed the peak at ≈222 nm as the function of urea concentration. So, as the urea concentration increases, the ellipticity value decreases (positive ellipticity). The ellipticity value of myoglobin decrease (positive ellipticity) slightly up to 4 M urea; later, between 5 and 8 M urea, the slope is quite steep; but then becomes constant at 9 M urea (Figure S3). At the absorbed dose of 1,136, 424, and 228 Gy, the ellipticity values are higher than those of the myoglobin protein control sample for 0 to 8 M concentration; at 9 M urea, all of the values meet each other. For Myoglobin + sorbitol and +trehalose, the ellipticity values decrease (positive ellipticity) slightly up to the 5 M urea; but between 5 and 8 M urea, the slope is quite steep; but then becomes constant at 9 M urea. In both cases (Myoglobin + sorbitol and +trehalose), the gamma-treated samples in the presence of sorbitol or trehalose are higher than their corresponding control values. As the absorbed dose (1,136, 424 and 228 Gy) increases, the ellipticity also decreases (positive ellipticity) at all concentrations of urea except for 9 M; this might be because at 9 M urea, the structure of myoglobin denatures completely (as previously measured), so no change in ellipticity observed. The difference of ellipticity among different gamma absorbed dose is least for the trehalose solution, slightly higher for the sorbitol solution, and quite high for the control solution (without osmolytes).
Figure S4 shows that we have checked the impact of the various urea concentrations on the α-helical structure of myoglobin protein in the presence of DBD plasma treatments. We observed a peak at ≈222 nm as a function of urea concentration. So, as the concentration of urea increases, the CD ellipticity values for myoglobin decrease (positive ellipticity), similar to the gamma ray treatments. The ellipticity values decrease (positive ellipticity) slightly up to 5 M urea, but the slopes are quite steep between 6 and 8 M urea; and then become constant at 9 M urea, for DBD plasma treatments at 5, 10, and 20 min (Figure S4). In the presence of osmolytes + protein (Myoglobin + sorbitol and Myoglobin + trehalose) for the DBD plasma-treated samples, the ellipticities at 222 nm are higher than their corresponding control values. As the DBD treatment time increases, the CD ellipticity values for protein decreases (positive ellipticity) at all concentrations of urea, except for 9 M. The difference of ellipticity among different treatment conditions in the presence of different osmolytes is least for the trehalose solution. The impact of DBD plasma treatments as a function of urea concentration doesn’t change the ellipticity values much for the trehalose solutions, as compared to the other solutions. Hence, trehalose solution protects the protein secondary structure from DBD plasma treatments in the best way, as compared to the other conditions of treatment (sorbitol solution or control solutions). Therefore, to understand the reason for the structural change after gamma ray and DBD plasma treatments, we have studied the generation of reactive species in buffer solution.
Analysis of reactive species generated and change in pH in solution after the treatment with gamma rays and DBD plasma
The irradiation of gamma rays and DBD plasma on buffer results in the formation of reactive nitrogen and oxygen species (RNS and ROS). Figures 5 and 6 show the results of the chemical analysis that we have performed to determine the amount of reactive species generated in solution after different treatments (gamma rays and DBD plasma). During this chemical analysis, we have studied the formation of ROS, such as •OH and H2O2, against the RNS, such as NO•, NO2 −, and NO3 −. After the gamma rays treatment on the buffer, we analysed the •OH and NO•. We observed from Fig. 5 that as we increased the gamma radiation absorbed dose from 228, through 424, to 1,136 Gy, the fluorescence intensity also increases for both •OH and NO• radicals. This shows that a greater absorbed dose results in the ability to generate more radicals in buffer solution. Figure 6 show that we have later analyzed the change in fluorescence intensity for both •OH and NO• radicals after DBD plasma treatments for 5, 10, and 20 min. The figure shows similar trends to those of the gamma ray treatments: as the treatment time increases, the fluorescence intensity also increases for both radicals. Hence, the intensity of radicals is directly related to the treatment time; more treatment time results in more generation of •OH and NO• radicals in buffer.
Additionally, we have analyzed the concentration of H2O2, NO2 −, and NO3 − after the gamma irradiation in buffer for the absorbed dose from 228, 424, and 1,136 Gy. We observed that the H2O2 concentration for 228, 424 and 1,136 Gy absorbed dose was ≈18, 36, and 50 μM, respectively. Whereas, Fig. 5 shows that the NO2 − concentration was ≈4.1, 4.6, and 5 μM, and the NO3 − concentration was ≈4.6, 5.1, and 6.2 μM for the absorbed dose of 228, 424, and 1,136 Gy, respectively. Similar to the behavior of •OH and NO• radicals in buffer, the H2O2, NO2 −, and NO3 − concentration also increases as the gamma radiation absorbed dose increases. However, after the same gamma radiation absorbed dose treatment, the concentrations of NO2 − and NO3 − were quite low, as compared to the H2O2 concentration. This might be because of water radiolysis by γ-rays that generate OH radicals62, which react with other OH radicals to produce the H2O2. On the other hand for the NO• radicals, NO2 −, and NO3 − are generated, because of the presence of air molecules during the gamma ray irradiation.
Moreover, we have also analyzed the concentration of H2O2, NO2 −, and NO3 − after the DBD plasma treatment on buffer for 5, 10, and 20 min. We observed that the H2O2 concentration for 5, 10, and 20 min DBD plasma treatment was ≈15, 47, and 72 μM, respectively. Whereas, Fig. 6 shows that the NO2 − concentration was ≈108, 159, and 311 μM, and the NO3 − concentration was ≈130, 210, and 452 μM for 5, 10, and 20 min DBD treatments, respectively. Similar to the gamma ray treatment, the behavior of H2O2, NO2 −, and NO3 − concentration increases as the DBD plasma treatment time increases. However at each DBD plasma treatment time, the concentrations of NO2 − and NO3 − were quite high, as compared to the H2O2 concentration. The generated OH radicals are due to plasma interaction with water molecules in air and on the surface of buffer; when these OH radicals react with each other, they produce H2O2 (similar to the gamma treatments). On the other hand, the NO• radicals, NO2 − and NO3 − ions are generated due to the presence of air. If we compare the H2O2 production in buffer after gamma ray irradiation and plasma irradiation, we observe that in plasma there is slightly more H2O2 produced, than in gamma ray treatment. While more NO2 − and NO3 − ions are generated in plasma than in gamma ray, this might be due to the different environmental conditions for the two treatments. We cannot say anything about the environmental condition in the gamma rays treatment chamber (as we are unable to measure the environmental condition during treatment), while the DBD plasma treatments were performed at atmospheric conditions (air as feeding gas, with humidity of 40%). The difference in environmental condition during treatments might be the reason for the high RNS species generated for DBD plasma, but the amounts of H2O2 produced for gamma rays and DBD plasma are approximately similar to each other. Hence, H2O2 generation during gamma ray and DBD plasma treatment can be the main component for the structural changes of myoglobin protein. Additionally, we have not observed any change in pH and temperature of the solution after treatment from gamma rays or DBD plasma (data not shown). Further, we have studied the molecular dynamic simulation of myoglobin protein with H2O2 in the presence of sorbitol and trehalose, respectively for better understanding the interaction properties.
Molecular dynamics simulation of myoglobin in the presence of H2O2 with or without osmolytes
To understand the influence of osmolytes on the myoglobin protein, and how the presence of osmolytes can affect the denaturation action of hydrogen peroxide (H2O2) on myoglobin protein structure, we analyzed the structural change of these paralogs using MD simulations. We prepared three types of MD simulation systems: with the protein solvated in phosphate buffer (21 mM) and 21% H2O2, with or without osmolytes (sorbitol and trehalose). Through measuring values of the root-mean-square atomic positional deviation (RMSD), we studied the structural stabilities of myoglobin for each environmental condition. Although, H2O2 molecules were uniformly distributed in solutions, but they seemed to condense within 1 Å of the surface of protein in a very short duration. Figure 7 shows that we subsequently measured the RMSD values at all conditions. However, in the presence of 21% H2O2, myoglobin underwent conformational changes, and showed higher RMSD values than for the simulation without H2O2 after the simulation for 50 ns. The RMSD value controlling myoglobin protein was 1.76, but after the treatment with H2O2, it was increased to 2.18, with change in standard deviation value of 0.29, as seen in Table 1. So, we may conclude that the presence of H2O2 destabilizes the myoglobin structure on the basis of these results. Later, we studied the MD simulation to understand the action of the osmolytes sorbitol and trehalose on myoglobin, with or without H2O2. Figure 7 show that Myoglobin in the presence of 21% sorbitol or 21% trehalose with phosphate buffer solution maintained stable conformations for 50 ns. Additionally, we have also measured the RMSD for sorbitol-buffer solution with H2O2. The RMSD value for controlling myoglobin protein with sorbitol was 2.31, but after the treatment with H2O2, it was increased to 2.92, with change in standard deviation value of 0.15 (Table 1). Later, we have checked the RMSD for trehalose-buffer solution with H2O2. We observed that the RMSD value for controlling myoglobin protein with trehalose was 2.47, but after the treatment with H2O2, it was increased to 2.57 with change in standard deviation value of 0.07, as seen in Table 1. Therefore, we conclude that in the presence of H2O2, trehalose can stabilize the protein more than sorbitol can.
Molecular dynamics simulation result of Myoglobin, (a) RMSD plots of the Myoglobin in the phosphate buffer and 21% H2O2; (b) RMSD plots of the Myoglobin in the phosphate buffer, 21% H2O2 + 21% sorbitol; (c) RMSD plots of the Myoglobin in the phosphate buffer, 21% H2O2 + 21% trehalose; (d) Snapshot of Myoglobin with 21% sorbitol + 21% H2O2 (Gray ribbon and wire - myoglobin, Blue - heme group, Green - sorbitol and Red - H2O2).
The protective action of sorbitol and trehalose against the gamma rays and DBD plasma treatments can be explained as the scavenging action of these osmolytes against RONS. In the previously reported articles, it was observed that sorbitol has scavenging activities for •OH inhibition63, 64. And in another study authors reported that sorbitol can prevent the DNA cleavage against the •OH attach, through its •OH-scavenging property65. Additionally, it was observed that trehalose can protect the DNA against the γ and β irradiations66. Another study reveals that trehalose assists as the free radical scavenger, they also observed that trehalose can protect HepG2 cells against the palmitate-induced toxicity67. Although it was also reported that trehalose can protect the cells against the oxygen radicals. And also trehalose can enhance the resistance to cells towards oxidative damage caused by H2O2 68. Hence, these all studies support that both osmolytes (sorbitol and trehalose) have the tendency to protect the biological systems against the RONS action.
Conclusion
In our present study, we have checked the thermodynamics and secondary structure of myoglobin protein with and without osmolytes after the gamma ray and DBD plasma treatment. The thermodynamics of myoglobin protein has been studied as a function of urea concentration, as well as a function of temperature. We observed that the denaturation of protein as a function of urea concentration synchronized with the treatments (gamma rays and plasma). After treatment with gamma rays and plasma, the myoglobin denatures at lower urea concentration. Similarly, the denaturation of myoglobin as a function of temperature also varies after the treatment with gamma rays and DBD plasma. The Tm values of protein decrease more after both treatments without osmolytes. However, the addition of osmolytes, such as sorbitol and trehalose, control the structure of myoglobin, even after the irradiation of gamma and plasma. Among both osmolytes, the trehalose protects the structure of myoglobin against both treatments (gamma and plasma) better than does sorbitol. However, the H2O2 concentrations generated in buffer after gamma and plasma treatments do not have much difference, but the NO2 − and NO3 − concentration values are quite higher for the plasma treatment than for the gamma treatments. But the denaturation still occurs slightly higher for gamma ray treatment, as compared to plasma treatment, even in the presence of osmolytes. Therefore, we conclude that the gamma ray action of treatment is different from that of plasma, in that gamma rays can penetrate the protective action of osmolytes. But still both osmolytes can protect the protein conformation against the gamma and plasma treatments, and trehalose shows higher ability to protect than sorbitol. Moreover, MD simulation results also show that trehalose can better counter the denaturation action of H2O2, than can sorbitol. These findings can play an important role in plasma medicine to understand the treatment mechanism.
Experimental section
Materials
The Myoglobin protein, phosphate buffer (pH = 7.2), sorbitol, and trehalose were supplied by Aldrich Chemical Co. (USA). All chemicals and reagents were used without further purification. The concentrations of OH, NO, H2O2, NO2 −, and NO3 − were measured by methods provided in our previous work69,70,71,72. The NO is detected using 4-amino-5-methylamino-2′,7′-difluorofluorescein (DAF-FM), the excitation/emission maxima is 495/515 nm for the DAF-FM-T (product of the reaction between the NO and DAF-FM). The detection of •OH can be done using terephthalate anions obtained by mixing terephthalic acid (TA) in alkaline aqueous solution. These terephthalate anions react with •OH to create highly fluorescent hydroxyterephthalate ions (HTA) and excitation/emission maxima are 310/425 nm for the formation of HTA. The H2O2 concentration is measured using titanyl ions in the presence of sodium azide to control the H2O2 degradation by nitrites. The NO2 − concentration is measured using Griess reagent supplied by Aldrich Chemical Co. (USA), whereas the NO3 − concentration is obtained using the Acorn Series ION 6 meter (pH/mV/°C Meter), nitrate electrode, from Oakton Instruments, USA.
Dielectric barrier discharge
Experiments were carried out using a scalable DBD device, as described in previous research articles73, 74. The device was set in ambient air at atmospheric pressure. DBD plasmas were generated between the electrodes by supplying a 10 kHz AC high voltage (Logy Electric, LHV-09K). The discharge voltage and current were measured by high-voltage probe (Tektronix, P6015A) and a Rogowski coil (URD, CTL-28-S90–05Z-1R1), respectively. The peak-to-peak discharge voltage and current were 9.2 kV and 0.2 A. The corresponding discharge power density was 1.49 W/cm2, which was deduced from a voltage/charge Lissajous plot.
Circular dichroism spectroscopy
CD spectroscopic studies were performed using J-815 spectrophotometry (Jasco, Japan) equipped with a Peltier system to control the temperature52,53,54,55,56. The samples were pre-equilibrated at the desired temperature for 15 min, and the scan speed was fixed for adaptive sampling (error F 0.01) with a response time of 1 s with 1 nm bandwidth. The secondary myoglobin structures were monitored using a 1.0 mm path length cuvette. The concentration for the secondary myoglobin structure was 0.2 mg/ml, with each spectrum being the average of six spectra. Each sample spectrum was obtained by subtracting the appropriate blank media without myoglobin from the experimental protein spectrum. The percentages of secondary structures were then calculated using Yang’s method69.
Circular dichroism spectroscopy based urea studies
The stability studies were performed by temperature-controlled Jasco J-815 circular dichroism (CD) spectrometry. For each sample, the CD spectra were simultaneously measured from 200 to 250 nm at 25 °C. The ellipticity in the spectrum of native myoglobin in buffer was assumed to correspond to 100% folded protein, and the ellipticity in the spectrum of myoglobin with 9.0 M urea was assumed to correspond to unfolded protein. The displayed CD spectra of myoglobin have two well-pronounced minima at ≈210 and ≈222 nm that resemble those typical for the polypeptide chains that are mostly organized in α-helix conformation, so we have studied the change in ellipticity at 222 nm, to understand the change in α-helix conformation as the function of urea concentration.
Temperature stability studies
Preliminary thermodynamic stability studies were performed by temperature-controlled J-815 spectrophotometry (Jasco, Japan) equipped with a Peltier system. For each sample, CD spectra were simultaneously measured at 222 nm as the function of temperature 25 to 100 °C. The sample was placed in a sealed cuvette to prevent water evaporation. The 222 nm ellipticity in the spectrum of native myoglobin in buffer at 25 °C was assumed to correspond to 100% folded protein, and at 100 °C was assumed to be unfolded protein, and the folded fraction was computed as:
In this method, A222 is the absorbance between 25 and 100 °C, Au is the absorbance of the unfolded protein at 100 °C, and Af is the absorbance of the folded protein at 25 °C. We have studied the change in ellipticity at 222 nm, to understand the change in protein conformation as a function of temperature.
UV-visible spectroscopy
UV-Vis S-3100 Spectrophotometry having a wavelength resolution of 0.95 nm, wavelength accuracy of ±0.5 nm, and wavelength reproducibility of ±0.02 nm was utilized for the analysis. A 0.5 mg/ml concentration of sample was used for UV-vis spectroscopy. The heme moiety in myoglobin exhibits a strong absorbance band at 409 nm that also displays a strong positive signal49. The 409 nm absorbance can be used to quantify the fraction of the folded protein. The fraction of folded protein was computed from the 409 nm absorbance following previous reports59. Briefly, the 409 nm absorbance in the spectrum of native myoglobin in buffer was assumed to correspond to 100% folded protein, and the 409 nm absorbance in the spectrum of myoglobin with 9.0 M urea was assumed to correspond to 0% folded protein (similar to the previous work ref. 75). The folded fraction was computed as:
In this method, A409 is the absorbance, Au is the 409 nm absorbance of the unfolded protein, and Af is the absorbance of the folded protein.
pH and temperature measurement
After the plasma was exposed for 5, 10, and 20 min in buffer, the pH and temperature of solution were measured using Horiba scientific instruments. All measurements were carried out in triplicate.
Molecular dynamics simulation
Crystal structures of the human myoglobin were obtained from the (RCSB) protein data bank website (http://www.rcsb.org; the PDB IDs were 1MBN)69. These structures were treated with the Protein Preparation Wizard of the Schrödinger suite for molecular dynamics (MD) simulations. All water and heat molecules were eliminated, and hydrogens were added and minimized using IMPACT 6.669. MD simulations were performed using Desmond 4.469. Simulation systems were prepared with the Desmond system builder. The box shape of the systems was orthorhombic, and the size was determined by using the 10 Å buffer distance between the solute structures and the simulation box boundary. The systems were solvated with TIP3P model water. Na+ or Cl− ions were added to the systems to neutralize the total charges of the system. Hydrogen peroxide (H2O2) molecules were added to achieve 21% concentration of the systems. The charges of H2O2 were determined from the electrostatic potential (ESP) charge fitting with Jaguar 8.769, version 8.7 (Schrödinger, LLC, New York, NY, 2015), using the basis set and function of B3LYP/6-31 G**. The MD simulations were performed in the (NPT) ensemble with the OPLS2005 all atom force field. A reference temperature of 300 K and pressure of 1 atm were maintained by the Nose-Hoover thermostat and the Martyna-Tobias-Klein barostat. Before performing the main simulations, a series of minimizations and short MD simulations were performed to relax the model system.
Sample preparation
The protein stability was added to 2 ml screw-capped vials in phosphate buffer, at 25 °C and kept for 4 h to attain complete equilibrium after blending the solution. Similarly, the protein was dissolve in the mixture of 500 mM of osmolytes (sorbitol or trehalose) and phosphate buffer at 25 °C, and keep the solution for 4 h to attain complete equilibrium after blending the solution. The samples were treated at 6 mm distances from the plasma for 5, 10, and 20 min, at humidity of 40%, and were then incubated for 4 h at room temperature after plasma treatment. Three samples were treated for each condition to minimize the error.
Statistical analysis
All of the values are represented by the mean ± S.D of the indicated number of replicates.
References
Jemal, A. et al. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers. Prev. 19, 1893–1907 (2010).
Hun, M. J. et al. The global burden of cancer: priorities for prevention. Carcinogenesis 31, 100–110 (2010).
Karrison, T. G. et al. Dormancy of mammary carcinoma after mastectomy. J. Natl. Cancer Inst. 91, 80–85 (1999).
Aguirre-Ghiso, J. A. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer 7, 834–846 (2007).
Burnette, B. & Weichselbaum, R. R. Radiation as an immune modulator. Semin. Radiat. Oncol. 23, 273–280 (2013).
Bentzen, S. M. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat. Rev. Cancer 6, 702–713 (2006).
Moding, E. J. et al. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat. Rev. Drug Discov. 12, 526–542 (2013).
Niemantsverdriet, M. et al. High and low LET radiation differentially induce normal tissue damage signals. Int. J. Radiat. Oncol. Biol. Phys. 83, 1291–1297 (2012).
Moussa, J. P. Role of gamma irradiation in regulation of NO3 level in rocket (Eruca vescaria subsp. sativa) plants. Russ. J. Plant Physiol. 53, 193–197 (2006).
von Woedtke, T. et al. Plasmas for medicine. Phys. Rep. 530, 291–320 (2013).
Yousfi, M. et al. Low-temperature plasmas at atmospheric pressure: toward new pharmaceutical treatments in medicine. Fundam Clin Pharmacol. 28, 123–35 (2014).
Park, J. H. et al. A comparative study for the inactivation of multidrug resistance bacteria using dielectric barrier discharge and nano-second pulsed plasma. Sci. Rep. 5, 13849 (2015).
Masur, K. et al. Human Skin Cell Activity Is Modulated by Cold Atmospheric Pressure Plasma. Wound Repair Regen. 20, A102–A102 (2012).
Ikehara, S. et al. Plasma blood coagulation without involving the activation of platelets and coagulation factors. Plasma Process. Polym. 12, 1348–1353 (2015).
Miyamoto, K. et al. Red blood cell coagulation induced by low-temperature plasma treatment. Arch. Biochem. Biophys. 605, 95 (2016).
Attri, P. et al. Generation mechanism of hydroxyl radical species and its lifetime prediction during the plasma-initiated ultraviolet (UV) photolysis. Sci. Rep. 5, 9332 (2015).
Attri, P. et al. Mechanism and comparison of needle-type non-thermal direct and indirect atmospheric pressure plasma jets on the degradation of dyes. Sci. Rep. 6, 34419 (2016).
Bruggeman, P. & Leys, C. Non-thermal plasmas in and in contact with liquids. J. Phys. D: Appl. Phys. 42, 053001 (2009).
Keidar, M. et al. Cold atmospheric plasma in cancer therapy. Physics of Plasmas 20, 057101 (2013).
Yan, D. et al. The role of aquaporins in the anti-glioblastoma capacity of the cold plasma-stimulated medium. J. Phys. D: Appl. Phys. 50, 055401 (2017).
Kumar, N. et al. Induced apoptosis in melanocytes cancer cell and oxidation in biomolecules through deuterium oxide generated from atmospheric pressure non-thermal plasma jet. Sci. Rep. 4, 7589 (2014).
Kumar, N. et al. The action of microsecond-pulsed plasma-activated media on the inactivation of human lung cancer cells. J. Phys. D-Appl. Phys. 49, 115401 (2016).
Chen, Z. et al. A novel micro cold atmospheric plasma device for glioblastoma both in vitro and in vivo. Cancers 9, 61 (2017).
Vandamme, M. et al. Response of human glioma U87 xenografted on mice to non thermal plasma treatment. Plasma medicine 1, 27–43 (2011).
Brulle, L. et al. Effects of a non thermal plasma treatment alone or in combination with gemcitabine in a MIA PaCa2-luc orthotopic pancreatic carcinoma model. PloS one 7, e52653 (2012).
Keidar, M. et al. Cold plasma selectivity and the possibility of a paradigm shift in cancer therapy. Br. J. Cancer 105, 1295–1301 (2011).
Vandamme, M. et al. ROS implication in a new antitumor strategy based on non-thermal plasma. Int. J. Cancer 130, 2185–2194 (2012).
Lee, H. J. et al. Degradation of adhesion molecules of G361 melanoma cells by a non-thermal atmospheric pressure microplasma. New J. Phys. 11, 115026 (2009).
Wang, M. et al. Cold Atmospheric Plasma for Selectively Ablating Metastatic Breast Cancer Cells. PloS one 8, e73741 (2013).
Ishaq, M. et al. Atmospheric pressure gas plasma-induced colorectal cancer cell death is mediated by Nox2–ASK1 apoptosis pathways and oxidative stress is mitigated by Srx–Nrf2 anti-oxidant system. BBA-Mol Cell Res 1843, 2827–2837 (2014).
Kim, J. Y. et al. Apoptosis of lung carcinoma cells induced by a flexible optical fiber-based cold microplasma. Biosens. Bioelectron. 28, 333–338 (2011).
Tan, X. et al. Single-Cell-Precision Microplasma-Induced Cancer Cell Apoptosis. PloS one 9, e101299 (2014).
Thiyagarajan, M. et al. Induction of apoptosis in human myeloid leukemia cells by remote exposure of resistive barrier cold plasma. Biotechnol. Bioeng. 111, 565–574 (2014).
Zhang, X. et al. Ablation of liver cancer cells in vitro by a plasma needle. Appl. Phys. Lett. 93, 021502 (2008).
Guerrero-Preston, R. et al. Cold atmospheric plasma treatment selectively targets head and neck squamous cell carcinoma cells. Int. J. Mol. Med. 34, 941–946 (2014).
Ames, B. A. et al. Oxidative damage of macromolecules. In: Davis K (Editor), Oxidation Damage and Repair: Chemical, Biological and Medical Aspects. Pergamon Press, Elmsford, NY, USA, 181–187 (1991).
Mates, J. M. et al. Antioxidant enzymes and human diseases. Clin. Biochem. 32, 595–603 (1999).
Nguyen, N. H. et al. Anticancer efficacy of nonthermal plasma dissolved in a liquid, liquid plasma in heterogeneous cancer cells. Sci. Rep. 6, 29020 (2016).
Kresge, N. et al. Precocious Newborn Mice and Epidermal Growth Factor: the Work of Stanley Cohen. J. Biol. Chem. 281, e11–e13 (2006).
Steeves, C. L. et al. The glycine neurotransmitter transporter GLYT1 is an organic osmolyte transporter regulating cell volume in cleavage-stage embryos. Proc. Natl. Acad. Sci. USA 100, 13982–13987 (2003).
Yancey, P. H. et al. Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222 (1982).
Andrew, T. R. et al. Osmolyte effects on kinetics of FKBP12 C22A folding coupled with prolyl isomerization. J. Mol. Biol. 330, 851–866 (2003).
Khan, S. H. et al. Naturally Occurring Organic Osmolytes: From Cell Physiology to Disease Prevention. IUBMB Life 62, 891–895 (2010).
Attri, P. et al. TMAO and Sorbitol Attenuate the Deleterious Action of Atmospheric Pressure Non-Thermal Jet Plasma on α-Chymotrypsin. RSC Adv. 2, 7146–7155 (2012).
Attri, P. & Venkatesu, P. Refolding of urea-induced denaturation of model proteins by trimethylamine N- oxide. Thermochimi. Acta 526, 143–150 (2011).
Attri, P. et al. The influence of osmolytes and denaturants on the structure and enzyme activity, of α-chymotrypsin. J. Phys. Chem. B 114, 1471–1478 (2010).
Puett, D. The Eguilibrium Unfolding Parameters of Horse and Sperm Whale Myoglobin. J. Biol. Chem. 248, 4623 (1973).
Sykes, P. A. et al. Determination of Myoglobin Stability by Visible Spectroscopy. J. Chem. Educ. 76, 1283–1284 (1999).
Jones, C. M. An Introduction to Research in Protein Folding for Undergraduates. J. Chem. Educ. 74, 1306 (1997).
Privalov, P. L. Intermediate States in Protein Folding. J. Mol. Biol. 258, 707–725 (1996).
Attri, P. & Venkatesu, P. Thermodynamic characterization of the biocompatible ionic liquid effects on protein model compounds and their functional groups. Phys. Chem. Chem. Phys. 13, 6566–6575 (2011).
Attri, P. et al. Activity and stability of α-chymotrypsin in biocompatible ionic liquids: enzyme refolding by triethyl Ammonium actetate. Phys. Chem. Chem. Phys. 13, 2788–2796 (2011).
Attri, P. et al. A protic ionic liquid attenuates the deleterious actions of urea on α-chymotrypsin. Phys. Chem. Chem. Phys. 13, 17023–17026 (2011).
Attri, P. & Venkatesu, P. Influence of protic ionic liquids on the structure and stability of succinylated Con A. Int. J. Biol. Macromolec. 51, 119–128 (2012).
Attri, P. & Venkatesu, P. Exploring the thermal stability of α-chymotrypsin in protic ionic liquids. Process Biochem. 48, 462–470 (2013).
Attri, P. & Choi, E. H. Influence of Reactive Oxygen Species on the Enzyme Stability and Activity in the Presence of Ionic Liquids. PLoS ONE 8, e75096 (2013).
Attri, P. et al. Variation in the structural changes of myoglobin in the presence of several protic ionic liquid. Int. J. Biol. Macromolec. 69, 114–123 (2014).
Hill, R. B. et al. De Novo Design of Helical Bundles as Models for Understanding Protein Folding and Function. Acc. Chem. Res. 33, 745–754 (2000).
Fiebig, O. C. et al. Quantitative evaluation of myoglobin unfolding in the presence of guanidinium hydrochloride and ionic liquids in solution. J. Phys. Chem. B 118, 406–412 (2014).
Greenfield, N. J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 1, 2527–2535 (2006).
Dick, M. et al. Trading off stability against activity in extremophilic aldolases. Sci. Rep. 6, 17908 (2016).
LaVerne, J. A. OH radicals and oxidizing products in the gamma radiolysis of water. Radiat Res. 153, 196–200 (2000).
Morelli, R. et al. Fenton-Dependent Damage to Carbohydrates: Free Radical Scavenging Activity of Some Simple Sugars. J. Agric. Food Chem. 51, 7418–7425 (2003).
Hunt, J. V. et al. Hydroxyl radical production and autoxidative glycosylation. Biochem. J. 256, 205–212 (1988).
Shell, T. A. & Lawrence, D. S. A New Trick (Hydroxyl Radical Generation) For An Old Vitamin (B12). J. Am. Chem. Soc. 133, 2148–2150 (2011).
Yoshinaga, K. et al. Protection by Trehalose of DNA from Radiation Damage. Biosci. Biotech. Biochem. 61, 160 (1997).
Leekumjorn, S. et al. Experimental and Computational Studies Investigating Trehalose Protection of HepG2 Cells from Palmitate-Induced Toxicity. Biophys. J. 94, 2869–2883 (2008).
Benaroudj, N. et al. Trehalose Accumulation during Cellular Stress Protects Cells and Cellular Proteins from Damage by Oxygen Radicals. J. Biol. Chem. 276, 24261–24267 (2001).
Park, J. H. et al. Variation in structure of proteins by adjusting reactive oxygen and nitrogen species generated from dielectric barrier discharge jet. Sci. Rep. 6, 35883 (2016).
Attri, P. et al. Interaction studies of carbon nanomaterials and plasma activated carbon nanomaterials solution with telomere binding protein. Sci. Rep. 7, 2636 (2017).
Kumar, N. et al. Influence of Nitric Oxide generated through microwave plasma on L6 skeletal muscle cell differentiation via oxidative signaling pathways. Sci. Rep. 7, 542 (2017).
Choi, S. et al. Structural and function analysis of lysozyme after the treatment with dielectric barrier discharge plasma and atmospheric pressure plasma jet. Sci. Rep. 7, 1027 (2017).
Attri, P. et al. Influence of ionic liquid and ionic salt on protein against the reactive species generated using dielectric barrier discharge plasma. Sci. Rep. 5, 17781 (2015).
Sarinont, T. Effects of plasma irradiation using various feeding gases on growth of Raphanus sativus L. Arch. Biochem. Bioph. 605, 129–140 (2016).
Miller, M. C. et al. Kinetics and mass spectrometric measurements of myoglobin unfolding in aqueous ionic liquid solutions. Int. J. Biol. Macromolec. 85, 200–207 (2016).
Acknowledgements
This work was supported by MEXT KAKENHI Grant Number 24108009 and JSPS KAKENHI Grant Number JP16H03895. P.A. is thankful to FY 2015 Japan Society for the Promotion of Science (JSPS) invitation fellowship. P.A. and E.H.C. acknowledge the SRC program (Grant #2010-0029421) and Leading Foreign Research Institute Recruitment program (Grant # NRF-2016K1A4A3914113) throughout the Basic Science Research Program of the National Research Foundation (NRF) of Korea and in part by Kwangwoon University 2017. A.E.C. and M.K. were supported by NRF Grant No. 2016M3A7B4025405.
Author information
Authors and Affiliations
Contributions
P.A. conceived, wrote the manuscript, designed the experiments and analyzed the data. P.A. and T.S. performed research. M.K. and A.E.C. performed the M.D. simulation work. K.K., M.S. and E.H.C. supervised the study/project. H.S., K.K. and M.S. design the plasma setup.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Attri, P., Kim, M., Sarinont, T. et al. The protective action of osmolytes on the deleterious effects of gamma rays and atmospheric pressure plasma on protein conformational changes. Sci Rep 7, 8698 (2017). https://doi.org/10.1038/s41598-017-08643-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08643-1
This article is cited by
-
Impact of Gamma rays and DBD plasma treatments on wastewater treatment
Scientific Reports (2018)
-
CAP modifies the structure of a model protein from thermophilic bacteria: mechanisms of CAP-mediated inactivation
Scientific Reports (2018)
-
Bacterial inactivation by plasma treated water enhanced by reactive nitrogen species
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.