Abstract
Ascoviruses are double-stranded DNA viruses that mainly infect noctuid larvae, and are transmitted by the parasitoid wasp Microplitis similis Lyle. Ascovirus-parasitoids wasp-noctuid insects constitute the dissemination system. Selection of suitable reference genes for the dissemination system could play an important role in elucidating the pathogenic molecular mechanisms of ascovirus. Unfortunately, such studies on potential reference genes in the dissemination system of ascoviruses are lacking. In the present study, we evaluated 11 candidate reference genes: β-actin1 (ACT1), β-actin2 (ACT2), elongation factor 1 (EF1), elongation factor 2 (EF2), ribosomal protein L10 (L10), ribosomal protein L17A (L17A), superoxide dismutase (SOD), 28S ribosome (28S), Tubulin (TUB) and 18S ribosome (18S). The samples were originally from various virus concentrations and points-in-time of experimental treatments using RefFinder and four algorithms. The results showed that EF1 was the most stable internal gene in S. exigua and M. similis and that EF2 was the most stable in the IOZCAS-Spex-II-A cell line, and the stability of reference genes were confirmed via the expression levels of two inhibitor of apoptosis-like (iap-like) genes from Heliothis virescens ascovirus 3 h (HvAV-3h). This study provides a crucial basis for future research that explores the molecular mechanisms of the pathogenesis of ascoviruses.
Similar content being viewed by others
Introduction
Ascoviruses (Ascoviridae) are circular double-stranded DNA viruses of insects that attack the most common lepidopteran species of the Noctuidae, Crambidae and Plutellidae families1,2,3. Heliothis virescens ascovirus 3h genome length is 190, 519 bp and is a strain of the species Heliothis virescens ascovirus 3a4, 5. HvAV-3h has strong pathogenicity to S. exigua, Spodoptera litura and Helicoverpa armigera 6,7,8. However, ascoviruses must be transmitted by parasitoid wasps in the field. A potential application value of ascoviruses for bio-control was predicted because of the dissemination system and acute pathogenic features of ascoviruses, which was different from other insect viruses, e.g., nucleopolyhedrovirus (NPV). Research on the pathogenic molecular mechanisms is relatively deficient for ascoviruses, and research on implementing a stabile reference gene for ascoviruses has not been performed. RT-qPCR is usually used in research on pathogenic molecular mechanisms. To achieve RNA quantitation and data normalization with different samples, we selected an optimal reference gene to correct the reliability and accuracy of quantitative results. Due to its simple operation, high sensitivity, low pollution and good repeatability, RT-qPCR is widely used in basic science research9. However, diverse samples have different RNA efficiencies of extraction, RNA quality, and reverse transcription efficiency of the product10. Therefore, it is important to ensure a high quality of RNA in RT-qPCR11. Generally, housekeeping genes are considered to be relatively stable expressed in all cell types and physiological states12. However, some studies have indicated that the expression quantity of housekeeping genes in different cell types, tissues, experimental conditions and under different physiological states is inconsistent13, 14. For example, ACT was unsuitable as a reference gene when human cell lines were infected by cytomegalovirus, human herpesvirus-6, camelpox virus, SARS coronavirus or yellow fever virus, and TBP and PPI were the most stable reference genes15. When Spodoptera frugiperda cells were infected with Autographa californica multiple nucleopolyhedrovirus (AcMNPV), the results indicated that ECD was a reliable reference gene for RT-qPCR and was better than 28S as a reference gene for these experiments16. Incorporation of the 28S reverse primer in oligo-dT-primed cDNA synthesis showed lower and less variable cycle thresholds in cells infected by viruses17. PPIA was set as the single, most-optimal internal reference gene for Israeli Acute Paralysis Virus (IAPV) infection experiments in Bombus terrestris 18. In various experimental settings and different tissues, rRNA genes were unsuitable as references gene because their transcription was significantly regulated19. 18S RNA and ACT have been commonly employed as reference genes in Hymenoptera studies16, 20, Meanwhile, a suitable and stable reference gene was significant for the calibration of the qRT-PCR data.
Moreover, iap-like1 and iap-like2 in HvAV-3h were chosen as the target genes which in order to better verify the stability of the optimal internal gene predicted by the different algorithms and softwares. IAPs are a kind of widely distributed endogenous apoptosis suppressor protein, which plays an important role in inhibitor apoptosis in many species21. Therefore, iap-like1 and iap-like2 are probably associated with the molecular mechanism of rapid pathogenesis and chronic death in larvae. The iap genes are detectable in the most of the baculovirus genomes, such as AcMNPV, CpGV (Cydia pomonella granulovirus), OpMNPV (Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus) and BmNPV (Bombyx mori nuclear polyhydrosis virus)22, 23. In this study, the stability of reference genes was assessed. The results could be used as internal controls in mRNA expression studies in ascovirus-infected S. exigua larvae, fat body cells (IOZCAS-Spex-II-A), and the parasitic wasp M. similis.
Materials and Methods
Insects, insect cell lines and viruses
The population of S. exigua larvae was originally collected from the vegetable fields of Huazhong Agriculture University in 2014. The insects were reared on artificial diets and maintained in a thermostatic chamber at 28–30 °C and 60–70% RH (14 L: 10D)24. Adults were fed with a 10% honey solution.
Microplitis similis samples were collected in an experimental cotton field of Hunan Agricultural University, Changsha, Hunan, China, and then reared under laboratory conditions25. The genders of newly emerged parasitoid adults were determined by recognizing the presence of the ovipositor under the microscope. Males and females were fed with a 30% honey solution. Each pair was provided with third-instar S. exigua larvae for propagation26.
The S. exigua fat body cell line (IOZCAS-Spex-II-A) was maintained at 28 °C in Grace’s Insect Medium (Sigma) supplemented with 10% fetal bovine serum. HvAV-3h, a strain of the species Heliothis virescens ascovirus 3a, was used in this study, and the hemolymph containing virion of HvAV-3h was collected from S. exigua larvae infected with HvAV-3h, as described previously4. The titer of hemolymph containing virion of HvAV-3h was 5.6 × 108 pfu/ml, which was determined with the TCID50 method27.
Sample collection
The third instar larvae molted after 24 h, were then injected with different concentrations of hemolymph containing the virion of HvAV-3h (100-, 102-, 104-, 106 and 108-fold), and were harvested at 2, 4, 6, 8, and 10 days post-infection (p.i.)7. The collected samples were preserved in microcentrifuge tubes (1.5 ml) and instantaneously frozen using liquid nitrogen followed by storage at −80 °C.
The IOZCAS-Spex-II-A cell line was infected with different concentrations of sterile hemolymph containing viruses (102-, 104-, 106 and 108-fold) and harvested at 1, 3, 5, and 7 days p.i.6. The collected samples were preserved in microcentrifuge tubes (1.5 ml) and stored at −80 °C after being washed with PBS twice.
According to Li26, the female parasitoid wasp acquires the virus on the ovipositor and this can persistent for 4.1 ± 1.4 days. In the present study, different concentrations (100- to 103-fold) of the virus were used to infect the third-instar S. exigua larvae, which had molted after 24 h. Then they were exposed to female parasitoids for 24 h. All experiments were conducted in a controlled temperature and humidity environment (27 ± 2 °C, humidity 70 ± 10%, L14: D10). Determination of viruliferous vectors was conducted following Tillman28.
RNA extraction and cDNA synthesis
Total RNA was isolated according to the Trizol RNA isolation kit manufacturer’s protocol. A total of 50 μl DEPC water was used to dissolve the RNA sediment. The A260/A280 ratio and A260/A230 ratio of the RNA were determined using a UV-1800 Spectrophotometer (SHIMADZU) and DU 730 nucleic acid/protein analyzer (BECKMAN COULTER). RNA samples with an A260/A280 ratio ranging from 1.8 to 2.0 and an A260/A230 ratio >2.0 were used for cDNA synthesis29. The first-strand complementary DNA was synthesized from 1 μg of total RNA with a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China) in a total volume of 20 μl. According to the manufacturer’s protocol, in the first step, 10 μl mixture was incubated for 2 min at 42 °C, and then 10 μl of master mix was added and incubated for 15 min at 37 °C and 5 s at 85 °C. The cDNA was preserved at −80 °C until further use.
Reference gene selection and primer design
According to a previous study30, the PCR primer sequences of S. exigua and the IOZCAS- Spex-II-A cell line were used for quantification of the expression of the genes encoding ACT1, ACT2, EF1, EF2, L10, L17A, SOD, TUB, 18S and 28S, as shown in Table S1. The primers for M. similis (Table S2) were designed via Beacon Designer 8.0 software with the following settings: primer melting temperature, 60 ± 1 °C; primer GC content, 40–60%; primer length, 18–24 bp; and amplicon length, 100–200 bp. Other parameters were set by default. The lengths of the PCR-amplified specific products and not dimers of PCR products were assessed using gel electrophoresis. A 10-fold dilution series of cDNA was employed as a standard curve, and the reverse-transcription qPCR efficiency was determined for each gene and each treatment using the linear regression model31. According to the equation: E = (10[−1/slope] −1) × 100, the corresponding qRT-PCR efficiencies (E) were calculated32. After detecting the efficiencies of the chosen primers, the primers that displayed a coefficient of correlation greater than 0.99 and efficiencies between 90% and 115% were selected for the next qRT-PCR (Table 1).
Quantitative real-time PCR
Quantitative real-time PCR was performed using SsoFastTM EvaGreen® Supermix (Bio-Rad, Singapore) via a MyiQTM 2 Two Color Real-Time PCR Detection System (Bio-Rad). Each reaction was performed in a 20 μl total volume with 10 μl SsoFastTM EvaGreen® Supermix, 1 μl of cDNA template, 1 μl of 10 μM of each primer and 7 μl of nuclease-free water in an iQTM 96-well PCR plate (Bio-Rad). The program was set as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 10 s. At the end of the reactions, a melting curve analysis from 65 °C to 95 °C was used to ensure amplified product consistency and specificity. All reactions were performed in triplicate.
Statistical analysis
A stable level of each reference gene was statistically analyzed with four software packages: geNorm33, NormFinder34, BestKeeper35, delta cycle threshold (Ct) method36, and Online software RefFinder (freely available at: http://fulxie.0fees.us/?type = reference). Application of the geNorm, NormFinder, and BestKeeper tools was based on the Microsoft Excel program. When geNorm and NormFinder tools performed a stable analysis of the data, the cycle threshold (Ct) was converted into a linear scale (the highest relative quantity for each gene was set to 1). The geNorm algorithm calculated an expression stable value (M) for each gene and then compared the pair-wise variation Vn/Vn + 1. The gene with the lowest M value represented the most stable expression. A ratio of Vn/Vn + 1 below 0.15 indicated that the use of an additional reference gene would not significantly improve normalization33. NormFinder combined the interclass variance and intraclass variance to calculate a stable value. The assessment of the reference gene stability was dependent on the size of the stable value34. The raw data of cycle threshold (Ct) values (CP values) and PCR efficiency (E) of the reference genes were determined as the best fitted standards by BestKeeper. The cardinal principle for identification of stably expressed reference genes by Bestkeeper was that the expression levels of suitable reference genes should be highly correlated. Therefore, the correlation between each candidate gene and the index was calculated, describing the relation between the index and the contributing candidate reference gene by the highest R value, lowest SD and CV values (<1) and the P value35. We also used the online software RefFinder, which integrates the above-mentioned four algorithms (geNorm, Normfinder, BestKeeper, and the delta Ct method) to compare and rank the examined candidate reference genes. According to the results of RefFinder, candidate genes with the top rankings were considered to be the most stably expressed under the tested experimental conditions and thus could be selected as optimal reference genes. Every gene was sorted by the five different statistical approaches separately.
Evaluation the stability of selected reference genes
The third instar larvae molted after 24 h and IOZCAS-Spex-II-A cells were infected with 102-fold concentrations of hemolymph containing the virion of HvAV-3h, and then were harvested at 2 and 3 days p.i. The relative expression levels of inhibit apoptotic-like (iap-like) genes were measured using the most two stable reference genes and least two reference genes via Livak method37, respectively.
Results
Selection of candidate reference genes
To investigate the eight commonly used reference genes ACT1, ACT2, EF1, EF2, L10, L17A, SOD, TUB, and 28S from S. exigua and the IOZCAS- Spex-II-A cell line and six reference genes, including 28S, 18S, EF1, TUB, SOD, and ACT, from M. similis (Table 1), we determined the correlation coefficient (R2) values of all candidates that varied from 0.990 to 0.998 across the cDNA diluted points and, concurrently, the PCR efficiency values of all pair-primers that varied between 90.3% and 114.5% (Table 1).
Expression profiles of candidate reference genes
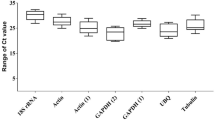
It is well known that the threshold cycle (Ct) can reflect the expression level of candidate reference genes to a certain extent. In ascovirus-infected M. similis (Fig. 1A), 18S RNA with a Ct value of 7.92 had the highest expression level and was more fluctuant than the other candidate reference genes. According to the original Ct value of S. exigua (Fig. 1B), the highest expression reference gene was 28S with a Ct value of 12.44, and the maximal fluctuating amplitude was 6.75. ACT1 was the least variable compared to the other candidate reference genes. In ascoviruses-infected IOZCAS-Spex-II-A cell line samples, the Ct values of the candidate reference genes under the same threshold value for fluorescence ranged from 13.67 for 28S to 27.07 for EF1, which represented the highest and lowest expression levels, respectively. The fluctuation showed no significant difference with each gene (Fig. 1C).
Range of Ct values in the transmission system of HvAV-3h. The above plots show expression levels of 6 candidate reference genes in (A) all M. similis samples, 9 candidate reference genes in (B) all S. exigua samples and (C) all IOZCAS-Spex-II-A cell line samples. Values are given as Ct values from the mean of duplicate samples. Bars indicate the standard error of the mean.
Analysis of gene expression stability in ascoviruses-infected M. similis
The comprehensive gene ranking of the most stable to least stable genes was EF1, SOD, TUB, 28S, ACT and 18S. All four programs identified EF1 as the most stable gene in ascoviruses-infected M. similis samples (Table 2). Based on geNorm analysis, the four genes should not be used as reference genes for normalizing gene expression data for all samples (Fig. 2F). From the point of view of different ascovirus concentrations, except for the 102-fold treatment, EF1 was the most stable gene according to the geomean of ranking value (Table S4).
Validation of 6 candidate reference genes with these samples under ascovirus-infected M. similis using geNorm. Virus initial concentration (A), virus concentration diluted 10 multiples (B), virus concentration diluted 100 multiples (C), virus concentration diluted 1,000 multiples (D), and all samples set (E). (A,B,C,D,E) represent average expression stability values (M) of 6 candidate genes, and (F) shows the determination of the optimal number of candidate genes for normalization by geNorm analysis.
Analysis of gene expression stability in ascovirus-infected S. exigua larvae
The stability rankings generated by NormFinder were consistent with those generated by the Delta Ct method and geNorm. However, the gene stability rankings by BestKeeper analysis were different from the other three methods. In all programs except for BestKeeper, EF1, L17A, and EF2, showed the most stable genes (Table 3). According to the Geomean of Ranking value by Reffinder, the stability rankings from the most stable to the least stable gene in all ascovirus-infected S. exigua were EF1, L17A, EF2, ACT1, L10, SOD, TUB, ACT2, and 28S (Table 3). Based on the geNorm algorithm (Fig. 3F), the gene pair EF2/L17A was the most stably expressed in all samples. Moreover, the inclusion of additional reference genes did not lower the V value below the proposed 0.15 cut-off until the ninth gene was added at 100- and 102-fold dilutions in all samples (Fig. 3G). From the point of different ascovirus concentrations, L10 was the most stable gene in 100- and 108-fold dilutions, and ACT2 and EF2 were the first positions in 102- and 104-fold dilutions, while L17A was the most stable in the 106-fold dilution (Table S5).
Validation of 9 candidate reference genes with these samples under different concentrations of ascoviruses in S. exigua using geNorm. Virus initial concentration (A), virus concentration diluted 100 multiples (B), virus concentration diluted 10,000 multiples (C), virus concentration diluted 1,000,000 multiples (D), virus concentration diluted 100,000,000 multiples (E), and all samples set (F). (A,B,C,D,E,F) represent average expression stability values (M) of 8 candidate genes, and (G) shows the determination of the optimal number of candidate genes for normalization by geNorm analysis.
Analysis of gene expression stability in the ascovirus-infected IOZCAS-Spex-II-A cell line
The stability rankings generated by the Delta Ct method, NormFinder, and BestKeeper showed that EF2 and L17A were the most stable genes, and gene stability ranked by the Delta Ct method, BestKeeper, and NormFinder were different regarding the results generated by the geNorm method (Table 4). As shown for M value and the optimal number for geNorm, all of the values were far below 1.5 (Fig. 4F). Individually, the gene pairs ACT1/L10, ACT1/EF2, EF1/TUB and ACT1/EF1 were the most suitable genes in 102-, 104-, 106- and 108-fold dilutions, respectively. EF1/L10 was the best pair across all samples. According to the RefFinder results, the stability rankings from the most stable to the least stable gene in the ascovirus-infected IOZCAS-Spex-II-A cell line samples were as follows: EF2, L17A, ACT2, SOD, EF1, L10, TUB, ACT1 and 28S (Table 4). As for different ascovirus concentrations, ACT1 was the most stable gene in the 102-fold dilution and SOD was the most stable in the 106-fold dilution. EF2 was in the first position in the 104- and 108-fold dilutions (Table S6).
Validation of 9 candidate reference genes with these samples under different concentrations of ascoviruses in the IOZCAS-Spex-II-A cell line using geNorm. Virus concentration diluted 100 multiples (A), virus concentration diluted 10,000 multiples (B), virus concentration diluted 1,000,000 multiples (C), virus concentration diluted 100,000,000 multiples (D), and all samples set (E). (A,B,C,D,E) represent average expression stability values (M) of 9 candidate genes, and (F) shows determination of the optimal number of candidate genes for normalization by geNorm analysis.
Evaluation of selected reference genes
The results of the relative expression analysis of iap-like1 and iap-like2 (Table S3) using the two most stable reference genes EF1 and LA17A in the S. exigua were shown in Fig. 5A,B. Additionally, 28S and ACT2 predicted as the two least stable genes, were applied for normalization to further verify whether the use of unstable reference gene can lead to an inaccurate relative expression (Fig. 5C,D). At the same time, the results of the relative expression analysis of iap-like1 and iap-like2 using the two most stable reference genes EF2, L17A and the two least stable reference genes 28S and ACT1 in the IOZCAS-Spex-II-A cell line were shown in Fig. 5E–H. In this two samples, the fold changes of the two iap-like genes normalized with stable reference gene showed consistent results.
The evaluation of the selected reference genes. The relative expression of inhibitor of apoptosis-like genes normalized with the two most stable reference genes EF1, L17A (A,B) and the two least stable reference genes 28S, ACT2 (C,D) in S. exigua larvae, the two most stable reference genes EF2, L17A (E,F) and the two least stable reference genes 28S, ACT1 (G,H) in the IOZCAS-Spex-II-A cell line.
Discussion
Ascoviruses are insect-specific double-stranded circle DNA viruses that attack lepidopterans, most commonly species in the family Noctuidae6. HvAV-3h has been recently isolated from S. exigua 4. Li found that the early instars of S. exigua were significantly easier to infect with HvAV-3h compared to the later instars, using 10-fold serial dilutions (0 to 7) of HvAV-3h-containing hemolymph to infect S. litura larvae. There were no significant differences in larval mortalities from 100- to 103-fold dilutions; however, significant declines were observed at the 104-fold dilution and above7. Compared to the healthy larval population, the typical symptoms and survival times of the diseased larval population were considerably extended, food intake was significantly reduced, and the body weight remained fairly constant in the 3rd and 4th instar larvae, which happened after inoculation with the ascovirus. However, the corrected mortality rates for the 1st through 5th instar inoculated per os were very low8. Therefore, the ascovirus mode of dissemination, which relies on the parasitoid wasp M. similis, served as a vector when the female parasitoid wasp acquired the virus. The ovipositor with the virion viability was 4.1 ± 1.4 days, and infected host larvae were still acceptable for egg laying by parasitoids. The parasitoids thereafter transmitted the virus to healthy hosts26. It follows that ascoviruses, with this dissemination system, have probably been regarded as significant for long-term pest control. Moreover, there are few studies that have elucidated the selection of reference genes for ascovirus infection dissemination systems. Therefore, stable expression of reference gene (s) is important for understanding the molecular mechanism of rapid pathogenesis and chronic death.
RT-qPCR is now the most sensitive method to study low-abundance mRNA from various tissue samples and experimental conditions. Thus, it is necessary to precisely determine normalization strategies38. Previous studies demonstrated that when reference genes were selected, the geometric mean of multiples should be used to ensure more accurate results33.
Spodoptera exigua would lead to several disordered phenomena due to the viral infection of S. exigua. There is no knowledge on the molecular mechanism. Thus, to gain a clear understanding of the pathogenic mechanisms, the screening of reference genes for S. exigua and different concentrations of ascovirus has provided a foundation for future research. At present, according to differences between physiological stages and different tissues in S. exigua, their suitability as reference genes was diverse with each treatment. In general, SOD, ACT2, ACT1, EF1 and GAPDH were stably expressed in all developmental stage sample sets. L10, EF2, L17A and EF1 were ranked highest in all tissue sample sets30, 39. The expression of these internal genes is considerably different in different experimental conditions. We therefore reassessed the stability before RT-qPCR testing. Our results indicated that EF1 ranked hi4ghest in all sample sets by RefFinder, Delta Ct and Normfinder, whereas the BestKeeper method ranked ACT1 as the best reference gene and EF1 ranked at the fifth position. The subtle differences in ranking among the top order reference genes could be imputed to differences in algorithms of the employed software programs and sensitivities towards the co-regulated reference genes.
A previous study showed that after the parasitoid M. similis possessed the virus, the virus just stayed in the ovipositor, and the virus could only be spread in a mechanical pathway26. Therefore, we inferred that the parasitoid possession of the virus should barely affect the transcription factors. In most studies of Hymenoptera, 18S or ACT has been commonly employed as the reference gene40,41,42. At the same time, none of the studies contributed to a comprehensive selection of internal control genes for Hymenoptera (Braconidae). In our study, although 18S RNA had the highest expression of all the candidate reference genes, it was not the most stable. When the parasitic wasp carried the virus, the most stable reference gene was EF1 in all sample sets by RefFinder, Delta Ct, geNorm and Normfinder. In this regard, the result was different from other studies that used 18S RNA as an internal gene.
HvAV-3e was replicated in three noctuid cell lines from Sf9 and Helicoverpa zea (BCIRL-Hz-AM1 and FB33). However, HvAV-3e did not replicate in the Pieris rapae (Pieridae) cell line, which was non-noctuid43. This means that ascoviruses were likely to impact the IOZCAS-Spex-II-A cell line, which was derived from the fat body of S. exigua. Consequently, stable reference genes can be helpful in further research on the cytopathic effect of ascoviruses. This study showed that the IOZCAS-Spex-II-A cell line was susceptible to infection by ascoviruses. AcMNPV could also easily infect the IOZCAS-Spex-II-A cell line16, 44. It has been reported that with the selection of reference genes in the Sf21 cell line infected by ascoviruses, DNA-free RNA was used as a template with a combination of the Sf21 cell line 28S gene-specific reverse primer and the oligo-dT primer for first strand cDNA synthesis. The results indicated that the Ct values were significantly higher and more variable during the course of viral infection when only the oligo-dT primer was used in the cDNA synthesis step than when the 28S-R primer in conjunction with the oligo-dT primer was used16, although the stability of the reference genes was not analyzed by the geNorm, NormFinder, BestKeeper, and delta cycle threshold (Ct) method and Online software RefFinder. In our study, we applied the conventional synthesis method of cDNA using RefFinder, Delta Ct, geNorm, Normfinder and BestKeeper algorithms to analyze the Ct values. As a consequence, the results indicated that 28S was the least suitable gene in the IOZCAS-Spex-II-A cell line across all samples, while the EF2 gene was expressed most stably in comparison with 8 other candidate internal genes. Generally, the most stable reference gene of the IOZCAS-Spex-II-A cell line and S. exigua should provide the same results. We acquired EF2 and L17A, which were relatively stable in screening of the reference genes for S. exigua and the IOZCAS-Spex -II-A cell line. However, in S. exigua, EF1 was the top-ranked by RefFinder, Delta Ct and NormFinder but was in the fifth positon by BestKeeper and the third position by geNorm. This phenomenon was probably due to the virus being able to directly impact the cell line; however, virus attack of insects may be influenced by other factors such as pH values, environmental temperature and host larvae with different instars.
Pairwise variation analysis with the geNorm applet suggested the use of two or more reference genes for attaining better accuracy in normalization for most of the experimental conditions30. The gene pairs EF2/L17A and EF1/L10 were considered the most suitable pairs of genes to normalize samples in S. exigua and the IOZCAS-Spex-II-A cell line, respectively, across all samples. However, conditions such as changes in M. similis would require normalization by three or more reference genes because the values of pairwise variations were above the cut-off range of 0.15. Thus, across all of the samples in S. exigua and the IOZCAS-Spex-II-A cell line, we advise the use of two reference genes under different experimental conditions.
To verify stability of candidate reference genes predicted by the RefFinder and four other algorithms, the most stable and least stable genes were applied for normalization the two IAP genes. Based on the sequence similarity to known IAPs, the iap-like genes have been chosen as the target genes. In the genomes of HvAV-3h, IAP genes have evolved mechanisms to reduce formation of apoptosis to guarantee the propagation of HvAV in host cells45. When the most stable reference genes (EF1, L17A and EF2) were employed to calibrated the data of gene expression in the S. exigua and IOZCAS-Spex-II-A cell line, the expression levels of the two iap-like genes revealed no significant changes. However, using the least stable reference genes 28S, ACT1and ACT2 to analyze the expression levels of the two iap-like genes, the results showed significant variation between two calculations which used the least stable reference genes. Therefore, it is necessary to use an appropriate stable reference gene for calibration of gene expression.
In summary, keeping in view the ecological control importance of the HvAV-3h-parasitic wasp (M. similis)-insect (S. exigua) dissemination system and pathogenic molecular mechanisms of ascoviruses, gene expression studies should continue to constitute a meaningful part of basic research with ascoviruses. Hence, establishing a best reference gene for RT-qPCR in dissemination systems will benefit researchers in this research arena. To the best of our knowledge, this is the first comprehensive report on the identification and validation of optimal candidate reference genes for accurate transcript normalization of gene expression in studies using RT-qPCR in the dissemination system of ascoviruses under various experimental conditions. We recommend the use of EF1 in S. exigua and M. similis and EF2 in the IOZCAS- Spex-II-A cell line. This study offers a way forward for the study of the pathogenic molecular mechanism of ascoviruses.
References
Stasiak, K., Renault, S., Demattei, M. V., Bigot, Y. & Federici, B. A. Evidence for the evolution of ascoviruses from iridoviruses. Journal of general virology. 84(11), 2999–3009 (2003).
Asgari, S., Davis, J., Wood, D., Wilson, P. & McGrath, A. Sequence and organization of the Heliothis virescens ascovirus genome. Journal of general virology. 88(4), 1120–1132 (2007).
Cheng, X. W., Carner, G. R. & Brown, T. M. Circular configuration of the genome of ascoviruses. Journal of general virology. 80(6), 1537–1540 (1999).
Huang, G. H. et al. Phylogenetic position and replication kinetics of Heliothis virescens ascovirus 3h (HvAV-3h) isolated from Spodoptera exigua. PloS ONE. 7(7), e40225 (2012).
Huang, G. H., Hou, D. H., Wang, M., Cheng, X. W. & Hu, Z. Genome analysis of Heliothis virescens ascovirus 3h isolated from China. Virologica Sinica 32(2), 147–154 (2017).
Bideshi, D. K., Tan, Y., Bigot, Y. & Federici, B. A. A viral caspase contributes to modified apoptosis for virus transmission. Genes & Development. 19(12), 1416–1421 (2005).
Li, S. J. et al. A comparison of growth and development of three major agricultural insect pests infected with Heliothis virescens ascovirus 3h (HvAV-3h). PloS ONE. 8(12), e85704 (2013).
Hu, J. et al. Characterization and Growing Development of Spodoptera exigua (Lepidoptera: Noctuidae) larvae Infected by Heliothis virescens ascovirus 3h (HvAV-3h). Journal of Economic Entomology. 109(5), 2020–2026 (2016).
Valasek, M. A. & Repa, J. J. The power of real-time PCR. Advances in physiology education. 29(3), 151–159 (2005).
Fleige, S. & Pfaffl, M. W. RNA integrity and the effect on the real-time qRT-PCR performance. Molecular aspects of medicine. 27(2), 126–139 (2006).
Huggett, J., Dheda, K., Bustin, S. & Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes and Immunity. 6(4), 279–284 (2005).
Garcia-Crespo, D., Juste, R. A. & Hurtado, A. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Veterinary Research. 1(1), 1 (2005).
Dheda, K. et al. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 37, 112–119 (2004).
Jain, M., Nijhawan, A., Tyagi, A. K. & Khurana, J. P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications. 345(2), 646–651 (2006).
Radonić, A. et al. Reference gene selection for quantitative real-time PCR analysis in virus infected cells: SARS corona virus, Yellow fever virus, Human Herpesvirus-6, Camelpox virus and Cytomegalovirus infections. Virology Journal. 2(1), 1 (2005).
Salem, T. Z., Allam, W. R. & Thiem, S. M. Verifying the stability of selected genes for normalization in q-PCR experiments of Spodoptera frugiperda cells during AcMNPV infection. PloS ONE. 9(10), e108516 (2014).
Xue, J. L., Salem, T. Z., Turney, C. M. & Cheng, X. W. Strategy of the use of 28S rRNA as a housekeeping gene in real-time quantitative PCR analysis of gene transcription in insect cells infected by viruses. Journal of Virological Methods. 163(2), 210–215 (2010).
Niu, J., Cappelle, K., de Miranda, J. R., Smagghe, G. & Meeus, I. Analysis of reference gene stability after Israeli acute paralysis virus infection in bumblebees Bombus terrestris. Journal of Invertebrate Pathology. 115, 76–79 (2014).
Radonić, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications. 313(4), 856–862 (2004).
Zhang, S., Zhang, Y. J., Su, H. H., Gao, X. W. & Guo, Y. Y. Identification and expression pattern of putative odorant-binding proteins and chemosensory proteins in antennae of the Microplitis mediator (Hymenoptera: Braconidae). Chemical Senses. 34(6), 503–512 (2009).
Deveraux, Q. L. & Reed, J. C. IAP family proteins—suppressors of apoptosis. Genes & development 13(3), 239–252 (1999).
Crook, N. E., Clem, R. J. & Miller, L. K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. Journal of virology 67(4), 2168–2174 (1993).
Clem, R. J. & Miller, L. K. Control of programmed cell death by the baculovirus genes p35 and iap. Molecular and Cellular Biology 14(8), 5212–5222 (1994).
Jiang, X., Luo, L. & Hu, Y. Influence of larval diets on development, fecundity and flight capacity of the beetarmyworm, Spodoptera exigua. Acta Entomologica Sinica. 42(3), 270–276 (1998).
Li, S. J. et al. Development of Microplitis similis (Hymenoptera: Braconidae) on two candidate host species, Spodoptera litura and Spodoptera exigua (Lepidoptera: Noctuidae). Florida Entomologist 98(2), 736–741 (2015).
Li S. J. et al. Imperfection works: Survival, transmission and persistence in the system of Heliothis virescens ascovirus 3h (HvAV-3h), Microplitis similis and Spodoptera exigua. Scientific Reports (2016).
Reed, L. J. & Muench, H. A. simple method of estimating fifty per cent endpoints. American journal of epidemiology. 27(3), 493–497 (1938).
Tillman, P. G., Styer, E. L. & Hamm, J. J. Transmission of ascovirus from Heliothis virescens (Lepidoptera: Noctuidae) by three parasitoids and effects of virus on survival of parasitoid Cardiochiles nigriceps (Hymenoptera: Braconidae). Environmental Entomology. 33(3), 633–643 (2004).
Ma, K. S. et al. Identification and Validation of Reference Genes for the Normalization of Gene Expression Data in qRT-PCR Analysis in Aphis gossypii (Hemiptera: Aphididae). Journal of Insect Science. 16(1), 17 (2016).
Zhu, X. et al. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). PloS ONE. 9(1), e84730 (2014).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research. 29(9), e45–e45 (2001).
Radonić, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications. 313(4), 856–862 (2004).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 3(7), 1–12 (2002).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 64(15), 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnology Letters. 26(6), 509–515 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology. 7(1), 1 (2006).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25(4), 402–408 (2001).
Bustin, S. A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology. 25(2), 169–193 (2000).
Teng, X., Zhang, Z., He, G., Yang, L. & Li, F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. Journal of Insect Science. 12(1), 60 (2012).
Brito, D. V. et al. Molecular characterization of the gene feminizer in the stingless bee Melipona interrupta (Hymenoptera: Apidae) reveals association to sex and caste development. Insect Biochemistry and Molecular Biology. 66, 24–30 (2015).
Ma, L. et al. Molecular characterization and expression profiles of olfactory receptor genes in the parasitic wasp, Microplitis mediator (Hymenoptera: Braconidae). Journal of Insect Physiology. 60, 118–126 (2014).
Chen, H. L., Zhang, H. Y., Throne, J. E. & Zhu, K. Y. Transcript analysis and expression profiling of three heat shock protein 70 genes in the ectoparasitoid Habrobracon hebetor (Hymenoptera: Braconidae). Insect Science. 21(4), 415–428 (2014).
Asgari, S. Replication of Heliothis virescens ascovirus in insect cell lines. Archives of Virology. 151(9), 1689–1699 (2006).
Zhang, H. et al. A cell strain cloned from Spodoptera exigua cell line (IOZCAS-Spex-II) highly susceptible to S. exigua nucleopolyhedrovirus infection. In Vitro Cellular & Developmental Biology-Animal. 45(5–6), 201–204 (2009).
Hussain, M. & Asgari, S. Inhibition of apoptosis by Heliothis virescens ascovirus (HvAV-3e): characterization of orf28 with structural similarity to inhibitor of apoptosis proteins. Apoptosis. 13(12), 1417–1426 (2008).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31371995), the Hunan Provincial Natural Science Foundation for Distinguished Young Scholars of China (14JJ1023), and the National Key Technology R&D Program of China (No. 2012BAD27B02). We thank Dr. Huan Zhang for providing the IOZCAS-Spex-II-A cell line (State Key Laboratory of Integrated Management of Pest Insects and Rodents).
Author information
Authors and Affiliations
Contributions
H.W., and G.H.H. designed the experiment; Z.S.C., and N.N.H. collected data; H.W. and G.H.H. analyzed the data and wrote the manuscript, Z.S.C., H.W., G.H.H., N.N.H. and J.H.L. read, corrected and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, ZS., Han, NN., Li, JH. et al. Selection of reference genes for expression analysis using RT-qPCR in the dissemination system of Heliothis virescens ascovirus 3 h (HvAV-3h). Sci Rep 7, 7045 (2017). https://doi.org/10.1038/s41598-017-07684-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07684-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.