Abstract
We have conducted a multicenter study of the diagnostic accuracy of the MTBDRplus 2.0 assay in compared with conventional and molecular reference standard in four tuberculosis (TB)-specialized hospitals of China. A total of 5038 patients were enrolled in this study. The overall sensitivity of the assay for the diagnosis of TB was 92.7% [1723/1858, 95% confidence interval (95% CI): 91.5–93.9]. In smear-positive/culture-positive cases the sensitivity was 97.7% (995/1018, 95% CI: 96.6–98.6), whereas in smear-negative/culture-positive cases it was 86.7% (728/840, 95% CI: 84.2–88.9). The agreement rate between MTBDRplus 2.0 and Xpert MTB/RIF was 97.7% (1015/1039, 95% CI: 96.6–98.5) for smear-positive cases and 97.0% (3682/3794, 95% CI: 96.5–97.6) for smear-negative cases. As compared with phenotypic drug susceptibility testing, the MTBDRplus 2.0 correctly identified 298 of 315 patients (94.6%, 95% CI: 91.5–96.8) with rifampicin-resistance. As noted previously, isoniazid resistance is associated with many different mutations and consequently the sensitivity compared to phenotypic testing was lower (81.0%, 95% CI: 76.8–84.7). In conclusion, this assay is a rapid, accurate test in terms of increased sensitivity for detecting smear-negative TB patients, as well as an alternative for detecting both RIF and INH resistance in persons with presumptive TB, whereas the absence of a mutation in the specimens must be interpreted cautiously.
Similar content being viewed by others
Introduction
Multidrug-resistant tuberculosis (MDR-TB), defined as resistance to at least isoniazid (INH) and rifampicin (RIF), poses as a major threat to global tuberculosis control1,2,3. According to the 2015 World Health Organization (WHO) report, there were an estimated 580,000 MDR /RIF-resistant tuberculosis (RR-TB) cases, and 250,000 deaths from MDR/RR-TB in 20154. China, India and the Russian Federation carried the greatest estimated burden of MDR tuberculosis, accounting for 54% of the global cases4, 5. Despite the high incidence of MDR-TB occurring in high TB-burden countries, only about a quarter of MDR-TB cases were detected and reported in 2014, reflecting the real difficulties in the diagnosis of MDR-TB6.
Bacterial culture-based drug susceptibility testing (DST) is still the standard for detecting MDR-TB7. Due to the slow growth rate of Mycobacterium tuberculosis (MTB), these methods always require several weeks to generate available results8. This creates a diagnostic delay that increases morbidity and mortality for patients, and enhances transmission in the community9. In recent years, an increasing array of new tools for the diagnosis of multidrug resistance has been introduced to overcome the inherent delays due to the use of phenotypic DST methods6. In 2008, the GenoType MTBDRplus (Hain Lifescience, Germany) was approved by the WHO for the detection of MTB and for identifying rifampicin (RIF) and isoniazid (INH) resistance from cultured isolates and from smear-positive sputum samples10, 11. The Genotype MTBDRplus assay provided a rapid option for detecting multidrug resistance from smear-positive samples11, while its turnaround time was still no less than 10 days for smear-negative samples due to the requirement for prior bacterial culture. Another endorsed molecular test platform for the detection of MTB and drug-resistance to RIF is Xpert MTB/RIF (Cepheid, USA)12. This automated molecular assay can diagnose TB and RIF-resistant TB directly from sputum samples in 2 hours, which overcomes many of the current operational difficulties13, 14. Despite exhibiting high accuracy for the diagnosis of TB, an obvious shortcoming of Xpert is that it can only detect RIF resistance rather than multidrug resistance. Considering the limitations of current available kits, there is an urgent need to develop molecular diagnostic tools for the diagnosis of MDR-TB from both smear-positive and smear-negative samples.
Hain Lifescience has developed a new MTBDRplus 2.0 test with a higher analytical sensitivity when compared with the original MTBDRplus 15, 16, which allows this new version to be performed on both smear-positive and smear- negative clinical specimens. By combining an optimized PCR master mix and a redesigned amplification program the new test version has increased sensitivity. Here, we have conducted a multicenter study on the diagnostic accuracy of the MTBDRplus 2.0 test across four TB-specialized hospitals in China. Our aim was to assess the performance of this assay for detecting TB and MDR-TB patients from persons with presumptive TB as compared with the conventional and molecular reference standards.
Results
Patients
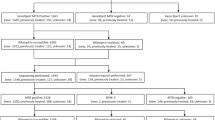
A total of 5038 patients were enrolled in this study. As shown in Table 1, 1078 (21.4%) had smear-positive/culture-positive tuberculosis; 955 (19.0%) had smear-negative/culture-positive tuberculosis; 1899 (37.6%) were defined as clinically diagnosed cases; and 1106 (22.0%) with no clinical evidence of tuberculosis, were classified as non-TB patients. Of the 5038 patients, 265 (5.3%) were excluded from the comparison between liquid culture and MTBDRplus 2.0 for a variety of reasons, including 86 culture contamination, 114 nontuberculous mycobacteria, and 65 smear-positive/culture-negative cases. In addition, all the cases with the valid results detected by both MTBDRplus 2.0 and Xpert MTB/RIF were included in the comparison between these two methods, and there were 4833 cases included in the final analysis.
Performance for case detection
The performance of MTBDRplus 2.0 for the diagnosis of TB is shown in Table 2. The overall sensitivity of the assay was 92.7% (1723/1858, 95% confidence interval [CI], 91.5–93.9). We observed no significant variation in overall sensitivity across the four sites (P = 0.077). In smear-positive/culture-positive cases the sensitivity was 97.7% (995/1018, 95% CI, 96.6–98.6), whereas in smear-negative/culture-positive cases it was 86.7% (728/840, 95% CI, 84.2–88.9), which was significantly lower than that in the smear-positive/culture-positive group (P < 0.01). The estimated specificity for the diagnosis of TB was 94.5% for MTBDRplus 2.0 (2755/2915, 95% CI, 93.6–95.3). Statistical analysis revealed that MTBDRplus 2.0 exhibited excellent agreement with liquid culture for detecting TB cases, with a kappa value of 0.87.
We also compared the agreement rate for detecting TB between MTBDRplus 2.0 and Xpert MTB/RIF. As shown in Table 2, the overall agreement rate between the two assays was 97.2% (4697/4833, 95% CI, 96.7–97.6), with a kappa value of 0.94. When separating participants into smear-positive and smear negative cases, the agreement rate was 97.7% (1015/1039, kappa value: 0.83) for smear-positive cases and 97.0% (3682/3794, kappa value: 0.92) for smear-negative cases. Statistical analysis revealed that there was no significant difference between smear-positive and smear-negative groups (P = 0.267).
Performance for detecting drug resistance
Using the phenotypic liquid culture method, BACTEC MGIT960 DST (BD Diagnostics, USA) 315 samples were identified as RIF resistant. In contrast, MTBDRplus 2.0 identified 17 of these samples as susceptible to RIF, indicating an overall sensitivity of 94.6% (95% CI, 91.5–96.8) for detecting RIF resistance. For 17 of 315 patients for whom results on the MTBDRplus 2.0 were discrepant against phenotypic testing, 16 (94.1%) were confirmed to be genotypically resistant by sequencing of the rpoB gene. Interestingly, the results of RIF susceptibility determined by Xpert MTB/RIF among 16 false negative cases were susceptible. In addition, the overall specificity of MTBDRplus 2.0 for RIF susceptibility was 95.6% (95% CI, 94.4–96.7). We observed that 59 patients identified as RIF-resistant by MTBDRplus 2.0 showed phenotypic susceptibility to RIF (Table 3). Further DNA sequencing analysis revealed that 46 (78.0%) out of 59 cases harbored genotypic mutations within the rpoB gene, including 4 (6.8%) strains with mutation in codon 511, 2 (3.4%) in codon 513, 1 (1.7%) in codon 515, 4 (6.8%) in codon 516, 3 (5.1%) in codon 522, 9 (15.3%) in codon 526, 18 (30.5%) in codon 531 and 5 (8.5%) in codon 533. In contrast, no genotypic mutations were detected in a screen of the rpoB gene from other false positive 13 strains (22.0%).
The comparison between the MTBDRplus 2.0 test and MGIT DST showed an overall sensitivity of 81.0% (95% CI, 76.8–84.7) for the detection of INH resistance. The sensitivity of MTBDRplus 2.0 for the diagnosis of INH resistance exhibited significant heterogeneity among the four sites in this study, ranging from 74.4% in Xi’an to 94.7% in Anhui (P = 0.004). Among the 78 phenotypically resistant strains showing wild-type bands at the inhA and katG genes, sequencing revealed that 71 strains carried no mutations in these two genes, thus confirming the MTBDRplus results. We also calculated the specificity of MTBDRplus for detecting INH resistance, and it correctly determined susceptibility to INH in 1187 of 1224 cases, with an overall specificity of 97.0% (95% CI, 95.9–97.9) (Table 4). Out of 37 strains with potentially “false positive” results, the presence of nucleotide substitution was observed among 27 strains (72.9%, 27/37), including 22 (59.5%) strains harboring mutations in the promoter region of inhA and 5 (13.5%) in the katG gene.
Discussion
The low detection rate of MDR-TB is one of the major challenges influencing the gap between diagnosis and treatment in China (WHO). There are several reasons responsible for this issue: (1) the implementation of rapid molecular assays for the detection of drug resistance is unsatisfactory in resource-poor settings with a high MDR-TB burden; (2) at present, all the molecular assays approved by the Chinese Food and Drug Administration are only approved for use on smear-positive clinical samples, including the endorsed Xpert MTB/RIF. Hence, more than 50% of MDR-TB cases may be missed due to lack of accessibility to molecular laboratory examinations for smear negative patients17. In this study, we first conducted a multicenter study to assess the diagnostic accuracy of the new version 2 of the GenoType MTBDRplus test for the detection of multidrug resistance among persons with presumptive TB. Our results suggest that the MTBDRplus 2.0 assay provides an accurate option for the diagnosis of MDR-TB among both smear-positive and smear-negative TB cases.
The MTBDRplus 2.0 assay outperformed smear microscopy because it detected a significant proportion of smear-negative TB cases. The proportion of rapidly diagnosed cases relative to smear microscopy significantly improved from 53.6% to 92.7%. According to our observations in this study, nearly one third of culture-positive patients were smear-negative. Considering that the four city hospitals in this study were top level TB specialized hospitals, most of the patients seeking care there were likely associated with more severe symptoms, thereby accounting for more smear-positive patients. Hence, there may be a higher proportion of smear-negative culture-positive patients in the county and prefectural level dispensaries. Consistent with our hypothesis, the national survey conducted in 2010 revealed that smear-negative patients contributed half of the bacteriologically positive tuberculosis burden in China in 201017. Taking into account the high prevalence of smear-negative patients, there is an urgent need to scale up use of a rapid molecular assay, such as MTBDRplus 2.0, to improve the early detection of TB in smear-negative patients in China. In addition, we also found that the sensitivity (97.7%) of MTBDRplus 2.0 for detection of MTB in smear-positive patients was significantly higher than that (86.7%) for the smear-negative group, indicating that the bacterial load correlated strongly with test performance. Similar results were observed by Theron and colleagues, showing that the sensitivity of Xpert MTB/RIF in smear-negative TB was limited by bacterial load9. Sample concentration may serve as an option for improving the performance of the molecular assays. Based on our previous experience, sample concentration could increase the positivity rate of MTBDRplus in smear-positive sputum samples, especially for sputum at the smear scanty level. Further studies are required to evaluate the effectiveness of sample concentration for the detection of MTB in smear-negative clinical samples.
Secondly, the MTBDRplus 2.0 assay demonstrated a good performance across all four sites for the detection of resistance to RIF, which is consistent with numerouspublications15, 18, 19. In contrast, the sensitivity of the assay for detection of resistance to INH was moderate (81.0%). A recent meta-analysis showed that the sensitivity of MTBDRplus was 88.7% for the diagnosis of INH resistance for smear-positive TB patients, which was higher than in our findings18. However, several evaluations of new diagnostic tools in China revealed that the sensitivity for detecting resistance to INH was 80.3% for Genechip8 and 80.2% for MTBDRplus20, respectively, similar to our observations. In addition, a molecular epidemiological study by Pang and other colleagues found that the combination mutations in the katG gene and the promoter of inhA gene only identify less than 75% of INH-resistant isolates in the MDR population in China, whereas 5.1% of MDR isolates only harbored point mutations in the oxyR-ahpC region21. Thus, this lower sensitivity for INH resistance was likely influenced by the high prevalence of oxyR-ahpC region mutations in China, which are not detected by the MTBDRplus 2.0 assay. Similarly, our results exhibited diverse sensitivities across the four sites, which may be also attributed to the heterogeneity of INH-resistant isolates circulating in different local regions in China.
There were several obvious limitations of the MTBDRplus 2.0 assay. First, despite novel modifications introduced into the DNA extraction procedure, the MTBDRplus 2.0 assay also requires extensive manual work, and a relatively long turn-around time when compared with Xpert MTB/RIF. Second, the common mutations in the oxyR-ahpC region need to be included in the MTBDRplus 2.0 assay to improve the sensitivity for INH resistance, which would then be more suitable for use in China. Third, about 15% of smear negative culture positive TB patients could not be detected by the MTBDRplus 2.0 assay, which highlights the urgent need to optimize the procedures to improve the detection sensitivity. Nevertheless, MTBDRplus 2.0 assay provides an ideal solution to simultaneously rule out both RIF and INH resistance from clinical sputum samples. With the support of the Global Fund, 125 prefectural laboratories have been equipped with MTBDR detection equipments. Fortunately, the MTBDR assay platform is also suitable for use with the MTBDRplus 2.0 assay. Hence, it is easy to integrate this novel tool with the existing platform, which will provide a promising solution at the prefectural level.
To the best of our knowledge, this study is the first multicenter evaluation of the MTBDRplus 2.0 assay in a high TB prevalence setting. Our data reveal that this assay is an accurate rapid test in terms of increased sensitivity for detecting smear-negative TB patients. In addition, it established a diagnosis with excellent rule-out value for detecting both RIF and INH resistance from persons with presumptive TB, especially for regions with a high prevalence of mono RIF resistance, as occurs in China. Considering that a small portion of drug-resistant clinical specimens harbor no detectable mutations, especially for INH-resistant cases, the results of the molecular assay must be interpreted cautiously.
Materials and Methods
Study Population
From April 2014 through April 2015, we conducted this study at four TB specialized hospitals: Guangdong Chest Hospital, Shanghai Pulmonary Hospital, Xi’an Chest Hospital, and Anhui Chest Hospital. All patients with symptoms suggestive of pulmonary tuberculosis were consecutively enrolled in this evaluation. We collected sputum specimens of 1.5 mL each from patients. All patients gave written informed consent before they were included in this study. Ethical approval was granted by the Ethics Committee of the Chinese Center for Disease Control and Prevention. The methods were performed in accordance with the approved guidelines.
TB case definitions
A person with presumptive TB patient was defined as someone presenting at the clinic with a cough of 2 weeks or more. Each person with presumptive TB was classified in one of three diagnostic categories: (1) confirmed TB patients: clinical TB symptoms with at least one sputum sample culture-positive for M. tuberculosis. The confirmed patients were further divided into smear-positive culture-positive patients and smear-negative culture-positive patients according to sputum smear results. (2) clinically diagnosed TB patients: clinical TB symptoms without any sputum sample culture-positive for M. tuberculosis plus a clinical-radiologic picture highly suggestive of TB (3) non-TB patients: no evidence of TB based on clinical examination.
Laboratory methods
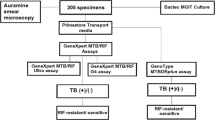
Direct smears from each sputum specimen were examined using Auramine O staining for acid fast bacilli (AFB) (National guidelines for TB laboratories, China Center for Disease Control and Prevention)22. The specimens were further digested with N-acetyl-L-cysteine and sodium hydroxide (NALC-NaOH) for 15 minutes. PBS buffer was added up to a total volume of 45 mL, and the suspension was centrifuged for 15 min at 3,000 × g. Following centrifugation, the supernatant was discarded and the sediment was suspended in 1.5 mL of PBS buffer. A0.5 mL portion of suspension was inoculated into a BACTEC MGIT tube (BD Microbiology Systems, USA); a second 0.5 mL of suspension was used to perform the MTBDRplus 2.0 test according to the manufacturer’s instructions; the final 0.5 mL of suspension was mixed with 1.5 mL of Xpert sample reagent for a 15-minute period at room temperature, then 2 mL of the inactivated material was transferred to a cartridge for GeneXpert MTB/RIF analysis (Cepheid, USA).
Positive cultures from specimens were preliminarily identified as M. tuberculosis complex with the MPT64 antigen kit (Genesis, Hangzhou, China)23. Indirect drug-susceptibility testing with the MGIT 960 SIRT was performed to detect the susceptibility of M. tuberculosis isolates to RIF and INH as per manufacturer’s instructions24. All the participating laboratory workers received standardized training, and were further approved by the National Reference Laboratory of Tuberculosis, China. The four clinical laboratories have participated in DST proficiency tests of the National TB Reference Laboratory since 2007.
DNA Sequencing
Crude genomic DNA was extracted from the positive cultures as previously described21. Briefly, colonies were scraped from the surface of Lowenstein-Jensen medium and heated for 1 h at 95 °C in 500 μl Tris-EDTA (TE) buffer. Followed by centrifugation at 13,000 rpm for 2 min, the supernatant was used as a template for amplification and was sequenced for the corresponding gene fragments (rpoB for RIF resistance; katG and inhA promoter for INH resistance) by the Sanger method with the designed internal primers21. The PCR mixture was prepared in a final volume of 50 μL containing 25 μL 2×PCR Mixture (Genestar, Beijing, China), 2 μL of DNA template and 0.2 μM of each primer set. The PCR was carried out under the following conditions: initial denaturation at 94 °C for 5 min, and then 35 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. The PCR products were sent to Tsingke Biotech Company for DNA sequencing service (Beijing, China). The sequencing results were analyzed by alignment with the corresponding reference strain (M. tuberculosis H37Rv; GenBank accession no. AL123456).
Data analysis
The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated to assess the performance of MTBDRplus 2.0 when using liquid culture and conventional phenotypic DST as reference standards, respectively. All of the data were evaluated by the National Reference Laboratory of Tuberculosis, then enrolled in SPSS15.0 as a data base and analyzed by the software. The kappa statistic was used to gauge the strength of agreement between MTBDRplus 2.0 assay and other methods. Values of the kappa coefficient higher than 0.75 indicated excellent agreement; values between 0.4 and 0.75 fair to good agreement; values lower than 0.4 poor agreement. In addition, the chi square test was performed to compare the performance of MTBDRplus 2.0 assay between different sample groups.
References
Zumla, A. et al. Drug-resistant tuberculosis–current dilemmas, unanswered questions, challenges, and priority needs. J Infect Dis 205(Suppl 2), S228–240 (2012).
Van Rie, A. & Warren, R. M. MDR tuberculosis control: time to change the dogma? Lancet Respir Med 3, 907–909 (2015).
Zhao, Y. et al. National survey of drug-resistant tuberculosis in China. N Engl J Med 366, 2161–2170 (2012).
World Health Organization. Global tuberculosis report 2016. WHO/HTM/TB/2016.13 (2016).
Salfinger, M. Molecular laboratory testing for tuberculosis: innovators, early adopters, or laggards? Clin Infect Dis 58, 977–979 (2014).
Pai, M. & Schito, M. Tuberculosis diagnostics in 2015: landscape, priorities, needs, and prospects. J Infect Dis 211(Suppl 2), S21–28 (2015).
Campbell, P. J. et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 55, 2032–2041 (2011).
Pang, Y. et al. Multicenter evaluation of Genechip for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol 51, 1707–1713 (2013).
Theron, G. et al. Evaluation of the Xpert MTB/RIF assay for the diagnosis of pulmonary tuberculosis in a high HIV prevalence setting. Am J Respir Crit Care Med 184, 132–140 (2011).
World Health Orgnazition. Molecular line probe patients at risk of multi-drug resistant tuberculosis. Expert group report (2008).
Bai, Y., Wang, Y., Shao, C., Hao, Y. & Jin, Y. GenoType MTBDRplus Assay for Rapid Detection of Multidrug Resistance in Mycobacterium tuberculosis: A Meta-Analysis. PLoS One 11, e0150321 (2016).
Qin, Z. Z. et al. How is Xpert MTB/RIF being implemented in 22 high tuberculosis burden countries? Eur Respir J 45, 549–554 (2015).
Weyer, K. et al. Rapid molecular TB diagnosis: evidence, policy making and global implementation of Xpert MTB/RIF. Eur Respir J 42, 252–271 (2013).
Boehme, C. C. et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363, 1005–1015 (2010).
Crudu, V. et al. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J Clin Microbiol 50, 1264–1269 (2012).
Seifert, M. et al. A performance evaluation of MTBDRplus version 2 for the diagnosis of multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 20, 631–637 (2016).
Wang, L. et al. Tuberculosis prevalence in China, 1990–2010; a longitudinal analysis of national survey data. Lancet 383, 2057–2064 (2014).
Barnard, M. et al. The diagnostic performance of the GenoType MTBDRplus version 2 line probe assay is equivalent to that of the Xpert MTB/RIF assay. J Clin Microbiol 50, 3712–3716 (2012).
Ling, D. I., Zwerling, A. A. & Pai, M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J 32, 1165–1174 (2008).
Li, Q. et al. Multicenter Evaluation of the Molecular Line Probe Assay for Multidrug Resistant Mycobacterium Tuberculosis Detection in China. Biomed Environ Sci 28, 464–467 (2015).
Zhang, Z. et al. Genotyping and molecular characteristics of multidrug-resistant Mycobacterium tuberculosis isolates from China. J Infect 70, 335–345 (2015).
Xia, H. et al. Multicentre evaluation of Ziehl-Neelsen and light-emitting diode fluorescence microscopy in China. Int J Tuberc Lung Dis 17, 107–112 (2013).
Yu, M. C. et al. Evaluation of the rapid MGIT TBc identification test for culture confirmation of Mycobacterium tuberculosis complex strain detection. J Clin Microbiol 49, 802–807 (2011).
Zhang, Z., Wang, Y., Pang, Y. & Liu, C. Comparison of different drug susceptibility test methods to detect rifampin heteroresistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58, 5632–5635 (2014).
Acknowledgements
This work was supported by the National Key Project (2014ZX10003002). We thank the staff working at the four hospitals for patient enrollment and laboratory tests. We thank the staff from National Tuberculosis Reference Laboratory for their technical help. We also would like to thank Dr. Pamela Chedore for her help in editing the language of this manuscript.
Author information
Authors and Affiliations
Contributions
Designed the studies: Y.T., Q.L., Q.W., H.S., J.C., Y.P., and Y.Z. Undertook the experimental work: Y.T., Q.L., Q.W., H.S., J.C., X.C., Y.Y., X.B., C.W., Y.L., and X.W. Analysed the data: Y.T., Q.L., Q.W., H.S., J.C., Y.P., and Y.Z. Contributed to figure and manuscript preparation: Y.T., Q.L., Y.P., and Y.Z.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tan, Y., Li, Q., Wang, Q. et al. Evaluation of the MTBDRplus 2.0 assay for the detection of multidrug resistance among persons with presumptive pulmonary TB in China. Sci Rep 7, 3364 (2017). https://doi.org/10.1038/s41598-017-03473-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03473-7
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.