Abstract
MYB transcription factors (TFs) have been implicated in various biology processes in model plants. However, functions of the great majority of MYB TFs in wheat (Triticum aestivum L.) have not been characterized. The soil-borne fungal pathogens Bipolaris sorokiniana and Rhizoctonia cerealis are the causal agents of important destructive diseases of wheat. Here, the TaPIMP2 gene, encoding a pathogen-induced MYB protein in wheat, was isolated through comparative transcriptomic analysis, and its defensive role was studied. TaPIMP2 was proved to localize in nuclei. TaPIMP2 responded in a different extent and speed upon infections of B. sorokiniana or R. cerealis. TaPIMP2 displayed different expression patterns after exogenous application of phytohormones, including abscisic acid, ethylene, and salicylic acid. Silencing of TaPIMP2 repressed resistance of wheat cultivar Yangmai 6 to B. sorokiniana, but did not alter resistance of wheat line CI12633 to R. cerealis. TaPIMP2 overexpression significantly improved resistance to B. sorokiniana rather than R. cerealis in transgenic wheat. Moreover, TaPIMP2 positively modulated the expression of pathogenesis-related genes, including PR1a, PR2, PR5, and PR10. Collectively, TaPIMP2 positively contributes to wheat resistance to B. sorokiniana possibly through regulating the expression of defense-related genes, and TaPIMP2 plays distinct roles in defense responses to different fungal infection.
Similar content being viewed by others
Introduction
Plants possess an elaborate and complex immune system to recognize and respond to pathogen infection1. Major progress has been made towards the understanding of plant immune system. Numerous defense-related events, including pathogen perception, signal generation and transmission, and activation of defense products restricting pathogen invasion, have been identified. Perception of pathogens by host plants induces robust and selective transcriptional reprogramming that is central for triggering effective defense response to invading pathogens. Phytohormones, such as salicylic acid (SA), jasmonic acid (JA), ethylene, and abscisic acid (ABA), plays pivotal roles in regulation of defense signaling network2. Accumulating evidence indicates that transcription factors (TFs) play important regulatory roles in plant defense response to biotic stress3, 4.
Myeloblastosis (MYB) TFs belong to one of the largest TF families5, 6. MYB TFs contain one to four highly conserved DNA-binding MYB domains, and can be divided into 4 major subgroups, 1R-MYB (MYB-related), R2R3-MYB, 3R-MYB, and 4R-MYB6. The MYB TFs have been studied in certain plant species, such as Arabidopsis thaliana, maize (Zea mays), rice (Oryza sativa), grapevine (Vitis vinifera), poplar (Populus tremuloides), apple (Malus domestica), and wheat (Triticum aestivum). MYB proteins have been confirmed to play crucial regulation roles in diverse physiological and biological processes, including cell cycle regulation, cell wall biosynthesis, development and reproduction, hormonal signaling, primary and secondary metabolism, and tolerance to abiotic stress7,8,9,10,11,12,13,14. Some MYB TFs participate in plant defense responses to invading pathogens7, 15,16,17,18,19,20. For example, two R2R3-MYB proteins from Arabidopsis, BOS1 (BOTRYTIS-SUSCEPTIBLE1, AtMYB108)7 and AtMYB9615, are required for defense responses to necrotrophic pathogens (Botrytis cinerea and Alternaria brassicicola) and to the bacteria pathogen Pseudomonas syringae DC3000, respectively. Overexpression of TiMYB2R-1, an R2R3-MYB gene in Thinopyrum intermedium, significantly enhanced resistance of the transgenic wheat to take-all disease caused by Gaeumannomyces graminis 16. In barley, an MYB TF HvMYB6 positively regulates basal resistance and MLA-mediated immunity response to Blumeria graminis 17. In wheat, TaPIMP1, the pathogen-induced MYB protein1, positively contributes host resistance to infection of the fungal pathogen Bipolaris sorokiniana and drought stresses18. Silencing of the R2R3-MYB gene TaMYB4 and the MYB1R gene TaLHY increased susceptibility to the biotrophic fungal pathogen Puccinia striiformis f. sp. Tritici (Pst) in wheat19, 20, respectively. However, functional roles of the great majority of MYB TFs have not yet been investigated in plant species.
Wheat is one of most important staple crops and plays a fundamental role in food security worldwide. Wheat production was often negatively affected by the infection of a wide variety of pathogens. Common root rot is one of the most serious wheat diseases worldwide. Bipolaris sorokiniana, a soil-borne fungal pathogen, is the causal agent of common root rot. B. sorokiniana also can cause leaf spot blotch, seedling blight, head blight, and black point of grains in wheat and barley, and other species of cereals and grasses21, 22. Additionally, the sharp eyespot, mainly caused by the necrotrophic fungus Rhizoctonia cerealis, is a destructive disease that seriously reduces wheat grain yield in some regions of the world23. For example, in China, since 2005 to date, wheat plants in at least 8 million ha have been subjected to sharp eyespot disease, resulting in yield losses of 10~30% (http://www.agri.cn/V20/bchqb/201501/t20150121_4344729.htm). The efficient and environmentally-safe means to protect wheat from B. sorokiniana and R. cerealis infection is to develop disease-resistant varieties. The efficiency of breeding depends on the availability of resistant germplasm and on an understanding the wheat defense responses. However, traditional breeding of resistant wheat varieties is difficult since no completely-resistant wheat accessions have been available. Previous studies18,19,20, 24 reported that 4 wheat MYB genes, including TaPIMP1, TaMYB4, TaLHY, and TaRIM1, positively participate in infection of the soil-borne fungal pathogen B. sorokiniana, the biotrophic fungal pathogen Pst, and the necrotrophic pathogen R. cerealis, respectively. Genetic engineering of resistance-related MYB TFs has been proposed to be an important strategy for improving the disease resistance of wheat. Therefore, it is vital to identify important resistance-related MYB genes and to dissect their functions.
In this study, an MYB gene was identified that displayed the differential induction between the resistant CI12633 and susceptible Wenmai 6 upon inoculation with R. cerealis based on the wheat microarray data obtained previously by our lab (GEO accession number GSE69245)25. The transcription of this MYB gene was also up-regulated in B. sorokiniana-resistant wheat Yangmai 6 after infection of B. sorokiniana. In a previous study, the wheat MYB gene TaPIMP1 was characterized, which contributes to host resistance to B. sorokiniana infection and drought tolerance18. Thus, the MYB gene characterized in this study was designated as TaPIMP2. TaPIMP2 exhibited distinct expression patterns in responses to the infections of B. sorakiniana and R. cerealis. The defense functions of TaPIMP2 to these two pathogens were investigated through generation and assessment of TaPIMP2 knockdown and overexpression wheat plants. The alteration of expression level of TaPIMP2 affected the wheat resistance degree to B. sorokiniana infection rather than R. cerealis. These results suggested that TaPIMP2 plays distinct roles in different wheat responses to the infection of B. sorokiniana or R. cerealis.
Results
Isolation and sequence analyses of TaPIMP2 gene
Through Agilent Wheat GeneChip-based transcriptomic analyses, we identified 1,533 genes (GEO accession number GSE69245) that were expressed in more than 2-fold higher level in R. cerealis-resistant wheat line CI12633 than in R. cerealis-susceptible wheat cultivar Wenmai 6 following 4 and 21 day post inoculation (dpi) with R. cerealis isolate R030125. Among them, a probe with the number A_99_P129195, corresponding to the 3′-end sequence of a wheat MYB cDNA (GenBank accession no. AB252145), was selected to be characterized. Its transcript levels in CI12633 increased 2.854-fold at 4 dpi and 5.204-fold at 21 dpi than that in Wenmai 6. The wheat gene corresponding to the probe A_99_P129195 was named TaPIMP2 (pathogen-induced MYB protein 2 in Triticum aestivum). The full-length cDNA sequence (1234 bp) of TaPIMP2 was obtained through 2 round 3′-RACE PCRs from R. cerealis-infected stem cDNA of CI12633 and deposited in the GenBank database (GenBank accession no. KX683396), as well as can be cloned from B. sorokiniana -infected stem cDNA of the wheat line Yangmai 6. The genomic DNA sequence of TaPIMP2 was also amplified from CI12633. Comparison of the genomic DNA and cDNA sequence revealed that the TaPIMP2 gene structure includes two exons and an intron with 141 nucleotides. The cDNA sequence of TaPIMP2 includes an ORF with 882 nucleotides (from no. 67 to 948 bp). The ORF sequence of TaPIMP2 shares a 44.51% identity with TaRIM1.
The deduced protein TaPIMP2 is consisted of 293 amino acids, and contains two conserved MYB domains (Fig. 1). In previous studies from our laboratory we characterized two MYB genes, TaPIMP1 18 and TaRIM1 24, which were involved into wheat resistance to B. sorokiniana and R. cerealis, respectively. Here, the putative protein sequence of TaPIMP2 was aligned with TaPIMP1 and TaRIM1 (Fig. 1). The result showed that two MYB domains [one (R2) located at amino acids 15–65 and other one (R3) at 68–116] were conserved among these 3 MYB proteins. Except the conserved MYB domains, whole amino acid sequences of these 3 proteins exhibited a low sequence similarity and identity. The whole protein sequence of TaPIMP2 shared 35.0% sequence identity with TaRIM1 (GenBank accession no. KU864997) and 26.9% sequence identity with TaPIMP1 (GenBank accession no. EF587267), respectively. In the reconstructed phylogenetic tree including the 18 known-functional MYB proteins from 8 plant species, TaPIMP2 was clustered into the R2R3-MYB subgroup (Fig. 2). The phylogenetic tree analysis showed that TaPIMP2 was the closest to AtMYB72 from Arabidopsis, while TaPIMP1 and TaRIM1, other defense-related wheat MYB proteins, were clustered into different clades in R2R3-MYB subgroup, respectively. These results indicated that TaPIMP2 is a R2R3 MYB protein and might be involved in wheat defense responses.
A phylogenetic analysis of 18 known-functional MYB members from 8 plant species. TaPIMP2 from wheat was indicated by black box. The Genbank accession number of proteins including this analysis: Ntmyb1: AAB41101; TaRIM1: AMP18876; AtMYB96: AED97610; AtMYB30: Q9SCU7; AtMYB72: NP_176012; GhMYB108: ALL53614; AtMYB108: Q9LDE1; TaPIMP1: ABU93236; OsJAMyb: AAK08983; AtMYB3R3: NP_001078127; ZmMYB3R3: NP_001151448; TaMYB3R1: ADO32617; OsMYB3R2: NP_001044767; StMYB1R-1: NP_001275346; GmMYB176: ABH02865; TaLHY: ADW09013; CCA1: AEC10760.
Expression patterns of TaPIMP2 in wheat challenging with R. cerealis and B. sorokiniana
Hexaploid wheat (AABBDD) contains three sub-genomes. Here, the expression profiles of TaPIMP2 homoeologous members from the 3 differential sub-genomes were investigated. Using nucleotide acid sequence of TaPIMP2 as query to blast wheat survey sequences database (https://urgi.versailles.inra.fr/blast), we obtained three homoeologous members that displayed highly sequence identity with TaPIMP2. The three sequences were TRIAE_CS42_1AL_TGACv1_002296, TRIAE_CS42_1BL_TGACv1_030624, and TRIAE_CS42_1DL_TGACv1_062692. TaPIMP2 displayed the highest sequence identity (99.9%) with TRIAE_CS42_1BL_TGACv1_030624. These results implied that TaPIMP2 have three homoeologous members in hexaploid wheat, which were derived from 1A, 1B, and 1D subgroup, respectively. The three homoeologous members were designated as TaPIMP2-1A, TaPIMP2-1B, andTaPIMP2-1D. The homoeologous member-specific primers were designed for detecting the transcriptions of the homoeologous genes.
The transcriptional patterns of TaPIMP2 homoeologous members in wheat stems defense to R. cerealis or B. sorokiniana were investigated by RT-qPCR. As shown in Fig. 3a and b, after pathogens infection, three homoeologous members exhibited similar expression trends. TaPIMP2-1B and TaPIMP2-1D possessed similar transcriptional levels, while TaPIMP2-1A had a slightly lower transcriptional level than the former two members. Compared with the untreated wheat plants, the transcription of TaPIMP2 in R. cerealis-resistant wheat line CI12633 stems was gradually elevated after inoculated with R. cerealis. The transcriptional level reached to ~4-fold at both 1 dpi (day post inoculation) and 2 dpi, subsequently decreased (at 4 dpi similar to that in untreated plants), and increased to the peak at 7 dpi (~14-fold). In addition, the expression of TaPIMP2 was measured in wheat cultivar Yangmai 6 after inoculation with B. Sorokiniana. Wheat cultivar Yangmai 6 exhibited resistance to infection with B. sorokiniana. Upon B. sorokiniana infection, transcriptional accumulation of TaPIMP2 rapidly and dramatically increased ~60-fold at 1 dpi compared with untreated plants, then gradually decreased at following days (2, 3, and 4 dpi), with ~25-fold at 4 dpi compared with untreated plants (Fig. 3b). This distinct expression profiles of TaPIMP2 in wheat defense responses against the above two different fungal pathogens suggested that TaPIMP2 may be involved in defense responses to infection of B. sorokiniana or R. cerealis in a different extent.
Transcriptional patterns of TaPIMP2 homoeologous member response to fungal infection. Transcription of TaPIMP2 homoeologous member response to infection with Rhizoctonia cerealisin in wheat line CI12633 (a) and infection with Bipolaris sorokiniana in wheat cultivar Yangmai 6 (b) was examined by RT-qPCR. The samples were collected from stem of infected wheat plants. The relative gene expression was quantified using the comparative threshold (2−ΔΔCT) method. The transcriptional level of TaPIMP2-1B from the untreated wheat plants was used as control. The mean and standard deviation were calculated using data from three independent biological replicates. Asterisks indicate statistically significant differences. (Student’s t-test: *P < 0.05).
TaPIMP2 transcriptional responses to exogenous hormones in wheat plants
Transcriptional profiles of TaPIMP2 homoeologous members, TaPIMP2-1A, -1B, and -1D were analyzed by qPCR in Yangmai16 leaves after treatments with exogenous defense-related hormones ABA, 1-aminocyclopropane-1-carboxylic acid (ACC, precursor of ethylene), and SA. As shown in Fig. 4a, TaPIMP2 expression levels were gradually induced by ABA. The accumulation reached a peak at 3 hour after the treatment, increased ~50 fold compared with control plants. After treatment with ACC, transcription of TaPIMP2 was increased to ~3 fold at 0.5 hour, subsequently decreased to 0.5 fold at 1, 3, and 6 hour compared with control plants (Fig. 4b). However, transcription of TaPIMP2 was repressed by exogenous SA, which showed a reduction to 0.2-fold of control plants at 1, 3, and 6 hour after treatment (Fig. 4c). TaPIMP2 homoeologous members shared similar expression tendency, and the accumulation of TaPIMP2-1D was slightly lower than those of TaPIMP2-1B and -1A.
Transcriptional patterns of TaPIMP2 homoeologous members after treatment with hormones. The wheat cultivar Yangmai 16 plants were sprayed with abscisic acid (ABA), 1-aminocyclopropane-1-carboxylic acid (ACC, precursor of ethylene), salycilic acid (SA), or Tween-20 solution. The transcriptional level of TaPIMP2-1B from the wheat plants treated with Tween-20 was used as control. The mean and standard deviation were calculated using data from three independent biological replicates. Asterisks indicate statistically significant differences. (Student’s t-test: *P < 0.05, **P < 0.01).
TaPIMP2 is a nuclear-expression protein
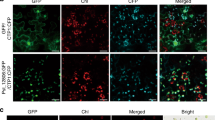
We investigated the subcelluar localization of TaPIMP2-1B in both wheat mesophyll protoplasts and onion epidermal cells. The p35S::TaPIMP2-GFP (green fluorescence protein) or control p35S::GFP construct was separately transiently expressed in wheat mesophyll protoplasts. As shown in Fig. 5a, the fluorescence signal of the TaPIMP2-GFP fusion protein was observed in nucleus of wheat mesophyll protoplasts, whereas the fluorescence of the control GFP was distributed in both cytoplasm and nucleus. As onion epidermal cells are lacked of interfering chlorophyll fluorescence, green fluorescence signals were readily observed. The p35S::TaPIMP2-GFP or control p35S::GFP construct was separately introduced into onion epidermal cells through particle bombardment. Confocal imaging of transient expression in the onion epidermal cells showed that TaPIMP2-GFP localized in the nucleus, whereas the fluorescence of the control GFP was distributed in both cytoplasm and nucleus (Fig. 5b). The above results proved that TaPIMP2 is localized and can be expressed in nucleus in both wheat mesophyll protoplasts and onion epidermal cells.
Subcellular localization of TaPIMP2 in wheat mesophyll protoplasts (a) and onion epidermal cells (b). TaPIMP2-GFP (green fluorescent protein) fusion protein is localized to nucleus. GFP alone is distributed in both cytoplasm and nucleus. The red fluorescence is from chloroplast autofluoresence. Images were captured using the following wavelengths, green fluorescence (excitation, 488 nm; emission, 509 nm) and red fluoresence (excitation, 448 nm; emission, 647 nm). Bars: 50 μm.
TaPIMP2 is required for wheat resistance to B. sorokiniana
As TaPIMP2 transcription was induced after challenge with R. cerealis or B. sorokiniana, its defensive roles in wheat were investigated by barley stripe mosaic virus (BSMV)-mediated virus-induced gene silencing (VIGS). For the silencing purpose, a TaPIMP2-specific fragment (247 bp length, from 729 to 975 nt of cDNA sequence, spanning the 3′ terminal of ORF and 3′ untranslated region) was inserted in an antisense orientation into Nhe I restriction site of the BSMV γ strand to generate the recombinant vector γ:TaPIMP2. BSMV α, β, and γ or γ:GFP or recombinant γ:TaPIMP2 were transcribed and mixed to result in BSMV:GFP or BSMV:TaPIMP2 viruses. The BSMV:GFP or BSMV:TaPIMP2 viruses were inoculated onto leaves of 2 wheat lines (R. cerealis-resistant CI12633 and B. sorokiniana-resistant Yangmai 6). After 7 days of the virus inoculation, the expression of BSMV coat protein (CP) gene was readily detected in new emerged leaves (Fig. 6a), and the typical BSMV symptom was present in these tissues (Fig. 6b), proving that these inoculated plants were successfully infected by either BSMV:GFP or BSMV:TaPIMP2.
Responses of TaPIMP2-silenced wheat plants to infection with Rhizoctonia cerealis or Bipolaris sorokiniana. (a) The barley stripe mosaic virus coat protein (cp) gene was detected by RT-PCR. (b) Phenotypes of wheat leaves infected with BSMV:TaPIMP2 or BSMV:GFP (control). (c) Transcriptional analysis of TaPIMP2 in stems of TaPIMP2-silencing CI12633 wheat and control plants. The expression level of TaPIMP2 in control CI12633 plants was set to 1. (d) The typical sharp eyespot symptoms were displayed in the susceptible control wheat Wenmai 6. (e) Transcriptional analysis of TaPIMP2 in TaPIMP2-silencing Yangmai 6 wheat and control plants. The expression level of TaPIMP2 in control Yangmai 6 plants was set to 1. (f) The typical common root rot symptoms were displayed in the TaPIMP2-silencing Yangmai 6. Values represent the mean and standard deviation from three independent biological replicates (Student’s t-test: *P < 0.05).
The silencing of TaPIMP2 in stems of BSMV-infected wheat plants was analyzed by RT-qPCR. The conserved region sequence of TaPIMP2-1A, -1B, and -1D was used to design qPCR primers, which were used to detect simultaneously transcriptional levels of three TaPIMP2 homoeologous members. The results showed that TaPIMP2 transcriptional level was significantly decreased to 25~39% in BSMV:TaPIMP2-infected (TaPIMP2-silenced) CI12633 plants compared with BSMV:GFP-infected control plants (Fig. 6c). The R. cerealis isolate R0301 was inoculated on the stems of these BSMV-infected plants to evaluate the resistance role of TaPIMP2 to sharp eyespot. Following inoculation with R. cerealis for 40 d, sharp eyespot symptoms did not obviously appear at inoculation site at both TaPIMP2-silencing and control wheat CI12633 plants, whereas serious sharp eyespot symptoms were presented in the R. cerealis-susceptible wheat cultivar Wenmai 6 (Fig. 6d). B. sorokiniana-resistance wheat cultivar Yangmai 6 plants were used for evaluating defense role of TaPIMP2 to common root rot disease by VIGS. Compared with control plants, transcriptional level of TaPIMP2 decreased to 21.5~32.2% in BSMV:TaPIMP2-infected Yangmai 6 plants (Fig. 6e). After inoculation with B. sorokiniana for 40 d, common root rot disease symptoms were heavily developed in TaPIMP2-silenced Yangmai 6, but BSMV:GFP-infected (control) Yangmai 6 plants exhibited weaker symptoms (Fig. 6f). The above results suggested that TaPIMP2 was required for wheat defense to B. sorokiniana infection in Yangmai 6, but not required for wheat defense, at least in the resistant wheat line CI12633, to R. cerealis.
Overexpression of TaPIMP2 enhances wheat resistance to B. sorokiniana
To further explore the functional role in wheat defense response, TaPIMP2-overexpression wheat plants were generated. As the sequence of the target probe A_99_P129195 in Agilent Wheat GeneChip was corresponding to the homoeologous member TaPIMP2-1B, the TaPIMP2-1B was selected for transformation into wheat. The ORF sequence of TaPIMP2-1B was inserted into a modified monocot transformation vector pAHC25-myc26, resulting in the overexpression vector pUBI::myc-TaPIMP2 (Fig. 7a). In the transformation construct, the expression of the fused c-myc-TaPIMP2 of a c-myc epitope tag and TaPIMP2-1B was driven by a maize ubiquitin promoter (UBI) and terminated by the terminator of the Agrobacterium tumefaciens nopaline synthase gene (Tnos).
Molecular characterization of TaPIMP2-overexpression wheat plants and responses to Bipolaris sorokiniana. (a) Scheme of the TaPIMP2 expression cassette on the transformation vector pUBI::myc-TaPIMP2. Ubi: maize ubiquitin promoter; Tnos: terminator of Agrobacterium tumefaciens nopaline synthase gene. The arrow indicates the amplified regions of transgenic plants by PCR. (b) PCR patterns of the TaPIMP2-transgenic wheat lines using the primers specific to the TaPIMP2-Tnos cassette. M: molecular marker; P: plasmid pUBI::myc-TaPIMP2; Y16: the recipient Yangmai 16. (c) Relative expression levels of TaPIMP2 in overexpressing wheat lines and untransformed Yangmai 16. The relative transcript level in the untransformed Yangmai 16 was set to 1. (d) Western blotting analysis of the TaPIMP2-overexpressing wheat lines and untransformed Yangmai 16 using an anti-myc antibody. (e) The typical common root rot symptoms of TaPIMP2-overexpression and untransformed Yangmai 16. Values represent the mean and standard deviation from three independent biological replicates (Student’s t-test: *P < 0.05).
The pUBI::myc-TaPIMP2 plasmid was bombarded to immature embryos of the spring wheat cultivar Yangmai 16 for generating transgenic wheat plants. The presence of introduced TaPIMP2 transgene was detected by the targeted PCR product (213 bp) using the primer pairs locating in TaPIMP2 and Tnos sequences (Fig. 7b), and the copy number in transgenic wheat plants was estimated by droplet digital PCR (ddPCR). The primers used in ddPCR assay were the same with those used in PCR detection for transgenic plants. The ddPCR has proven to be a reliable and reproducible method in estimation of transgene copy number in wheat27 and tobacco28. TaSTOP1-A was used as the reference with two copies in hexaploid wheat29. The data from ddPCR assay showed that copy numbers of introduced TaPIMP2-Tnos chimera were 8.210, 5.895, 8.145, and 11.206 in transgenic lines OE393, OE394, OE395, and OE396, respectively. Hence, the copy numbers of introduced TaPIMP2 gene were estimated as 8, 6, 8, and 11 in transgenic wheat lines OE393, OE394, OE395, and OE396, respectively.
Four transgenic lines containing TaPIMP2-Tnos (OE393, OE394, OE395, and OE396) were selected for disease resistance evaluation. RT-qPCR analysis showed that the transcript levels of TaPIMP2 in stems from four overexpression transgenic lines were significantly elevated compared with untransformed (wild type, WT) Yangmai 16 (Fig. 7c). Using anti-c-myc as antibody, western blotting assay showed that the introduced c-myc-TaPIMP2 fusion protein was significantly accumulated in all four transgenic wheat lines, whereas no corresponding band was detected in untransformed Yangmai 16 (Fig. 7d). These results indicated that the introduced myc-TaPIMP2 was overexpressed and translated in these 4 transgenic wheat lines.
The defense responses of these 4 TaPIMP2-overexpression lines in T4 generation were evaluated following inoculating with B. sorokiniana or R. cerealis. Following inoculation with B. sorokiniana for 40 d, the disease severity scoring results showed that these four overexpression lines exhibited significantly enhanced resistance to common root rot compared to untransformed Yangmai 16. The average disease indices of the four overexpression lines were 25.22, 27.20, 20.44, and 24.56, respectively, whereas the disease index of untransformed Yangmai 16 was 52.26 (Fig. 7e). Furthermore, the responses of the four TaPIMP2-overexpression wheat lines to R. cerealis in T1-T4 generations were evaluated. The sharp eyespot infection types and disease indices did not exhibit significant differences between the four overexpressing wheat lines and untransformed wheat plants (Table S1). These results indicated that overexpression of TaPIMP2 enhanced wheat resistance to infection of B. sorokiniana rather than R. cerealis.
TaPIMP2 regulates transcription of pathogenesis-related genes in wheat
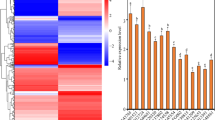
Pathogenesis-related (PR) proteins, including PR1a, PR2, PR5, and PR10, were shown to be involved in wheat resistance to B. sorokiniana or R. cerealis infection in previously publications18, 30. To explore if TaPIMP2 regulates the expression of PR genes, the above-mentioned PR protein-encoding genes were selected for transcriptional quantification analysis by RT-qPCR in TaPIMP2-overexpression wheat lines, TaPIMP2-silenced wheat plants, and their corresponding control wheat plants. RT-qPCR analysis results showed that the transcriptional levels of PR1a, PR2, PR5, and PR10 were significantly increased in TaPIMP2-overexpression wheat lines compared with untransformed Yangmai 16, but significantly decreased in TaPIMP2-silenced wheat plants than that in BSMV:GFP-infected plants (Fig. 8). The results clearly revealed that TaPIMP2 positively regulated the expression of certain defense-related genes in wheat.
Expression of defense-associated genes in TaPIMP2-overexpressing wheat lines and TaPIMP2-silenced wheat plants by RT-qPCR. The transcript levels of these genes in TaPIMP2-overexpressing wheat lines are relative to those in untransformed Yangmai 16, whereas the levels in TaPIMP2-silenced wheat plants are relative to those in the control plants infected with BSMV:GFP. Values represent the average standard error of three independent biological replicates (Student’s t-test: *P < 0.05).
Discussion
In this study, TaPIMP2, a pathogen-induced MYB gene, was cloned from wheat based on comparative transcriptome. The deduced protein TaPIMP2 belongs to R2R3 MYB subgroup. Comparison of whole protein sequences showed that TaPIMP2 has no obvious sequence similarity to both TaPIMP1 and TaRIM1 except the MYB domains (Fig. 1). Previous studies showed that some MYB genes being involved in defense responses were induced after challenge with the pathogens. For example, a wheat R2R3-MYB gene TaPIMP1 is induced after B. sorokiniana infection and positively regulates host defense response to the pathogen infection18. Both TaMYB4 and TaLHY were induced after infection of Pst 19, 20. TaRIM1 was induced upon infection with R. cerealis 24. Here, RT-qPCR analysis results indicated that infection of R. cerealis and B. sorokiniana triggered the transcriptional accumulation of TaPIMP2 in wheat lines (Fig. 3a and b). However, the induced transcription patterns were different upon infection between the two pathogens. After infection of R. cerealis, TaPIMP2 transcriptional level gradually increased at 1, 2, and 4 dpi, then sharply elevated and reached the peak (~14-fold) at 7 dpi (Fig. 3a). Upon challenging with B. sorokiniana, TaPIMP2 transcript was rapidly and dramatically accumulated to ~60-fold at 1 dpi than that of untreated plants (Fig. 3b). The above results suggest that TaPIMP2 showed a more sharp and rapid response to infection of B. sorokiniana than to R. cerealis infection.
Previous studies revealed that mutant/silencing of certain MYB genes (AtMYB108, TaPIMP1, TaMYB4, TaLHY, and TaRIM1) impaired resistance or enhanced susceptibility to different diseases, while overexpression of the MYB genes, such as TaPIMP1 and TaRIM1, improved significantly resistance of transgenic plants to infection of pathogens7, 18,19,20, 24. In the present study, we dissected the functional roles of TaPIMP2 to B. sorokiniana and R. cerealis through generation and assessment of TaPIMP2 silencing and overexpression wheat plants. The results indicated that silencing of TaPIMP2 significantly impaired wheat Yangmai 6 resistance to B. sorokiniana, but silencing of TaPIMP2 did not obviously impair the resistance to R. cerealis in sharp eyespot-resistant wheat line CI12633 (Fig. 6). This discrepancy in responses to R. cerealis and B. sorokiniana in TaPIMP2-silenced wheat plants may be ascribed to that TaPIMP2 could be involved in different defense pathways in CI12633 and Yangmai 6. Another possible reason may be that TaPIMP2 is functionally redundant with other genes in CI12633 resistance response to R. cerealis. For example, in Arabidopsis, two MYB proteins MYB33 and MYB65 participate in anther development. However, neither single knockout mutant, myb33 or myb65, display disturbed anther development31. Moreover, overexpression of TaPIMP2 significantly increased resistance of transgenic wheat Yangmai 16 to common root rot caused by B. sorokiniana, but did not significantly alter resistance degree of the transgenic wheat lines to R. cerealis infection (Table S1). These results suggested that TaPIMP2 positively contributes to resistance response to B. sorokiniana, but is not sufficient for improving disease resistance in Yangmai 16 to R. cerealis. Special MYB proteins may have multiple roles in different biotic and abiotic stresses7, 8, 15, 18. It is very interesting to further explore the role of TaPIMP2 in other areas including signal pathway.
Phytohormone, including SA, JA, ethylene, and ABA, as well as other small phytohormones, have been evidenced to play pivotal roles in immunity network2, 32,33,34. Usually, SA is associated with biotrophic pathogen resistance, whereas JA and ethylene are associated with necrotrophic pathogen resistance responses2. ABA plays a important role in regulation of defense responses to abiotic and biotic stresses2, 33. A number of TF families, e. g. ethylene-responsive factor (ERF) and MYB TFs, play important roles in the transmission of pathogen-derived defense signals. For instance, the Arabidopsis BOS1 gene controls both JA- and ABA-inducible genes, consequently participates in defense responses to necrotrophic pathogens and abiotic stresses7. AtMYB96-mediated ABA signaling promotes drought tolerance and resistance to the pathogen Pseudomonas syringae pv. tomato DC3000 infection by inducing SA biosynthesis8, 15. In Arabidopsis, cooperation of JA and ethylene in activation of defense against necrotrophic pathogens can be explained by the activation of the nodes ERF1 or ORA59, two members of the AP2/ERF transcription factor subfamily35, 36. Previously, our lab studies revealed that the wheat MYB TF TaPIMP1, being up-regulated by exogenous ABA and SA positively regulates host resistance to B. sorokiniana and drought through ABA and SA signaling pathways18; and a wheat ERF TF TaPIE1 being induced by ethylene positively contributes to resistance to R. cerealis, and freezing stresses mainly through the ethylene signaling pathway30. In this study, the transcription of TaPIMP2 was rapidly and strongly induced by ABA, while was transiently induced by ACC, the precursor of ethylene (Fig. 4). These data suggested that TaPIMP2 might be modulated in a distinct extent through ABA and ethylene signal pathways in defense responses to B. sorokiniana and R. cerealis. The underlying mechanism needs to be explored in future.
Various PR proteins played important roles in plant defense responses to different pathogen infection37,38,39. In this study, we analyzed the transcriptional levels of four selected PR genes (including PR1a, PR2, PR5, and PR10) in TaPIMP2-overexpression wheat lines, TaPIMP2-silencing wheat plants, and their corresponding control wheat plants (Fig. 8). The results revealed that TaPIMP2 positively modulated the expression of the tested PR genes. Increasing evidence indicates that transcription factors may modulate the expression of defense-related genes through binding to corresponding cis-acting elements. For example, the wheat MYB TF TaPIMP1 can activate the expression of 5 defense-related genes by binding to the MYB-binding sequence (MBS) cis-elements in their promoters18. An MYB TF BjMYB1 from Brassica juncea could active the expression of a chitinase gene BjCHI1 through binding to the W-box element WbI-4 in its promoter40. The upland cotton MYB TF GhMYB108 could bind to the MBS cis-element41. Previous studies showed that the promoter regions of PR1a and PR10 contain ACI and MBS cis-elements18, 24. Our analyses on the promoter regions of PR2 and PR5 revealed that 2 MYB-binding sequences (ACI and MBS) and 1 W-box cis-element were present in these promoters (Table S2). These data suggested that TaPIMP2 might bind to MBS cis-elements in these promoters of the tested PR genes and active directly or indirectly their expression, leading to enhanced resistance in overexpression transgenic wheat.
In conclusion, TaPIMP2, a pathogen-induce MYB protein-encoding gene in wheat, was isolated and its defense function against R. cerealis or B. sorokiniana was characterized. Transcriptional level of TaPIMP2 was rapidly and strongly increased after treatments with B. sorokiniana and ABA, while was moderately induced by R. cerealis and transiently by ethylene, or depressed by SA, respectively. TaPIMP2 positively regulates wheat resistance defense to common root rot caused by B. sorokiniana through modulating transcription of certain PR genes. Our study provided new insight into functional diversity of MYB TFs in plant defense responses to infection of different pathogens.
Materials and Methods
Plant and fungal materials, and treatments
Four wheat lines/cultivars, CI12633, Wenmai 6, Yangmai 6, and Yangmai 16 were used in this study. Among them, CI12633 is resistant to R. cerealis infection, whereas Wenmai 6 is highly susceptible. Yangmai 6 is resistant to infection of B. sorokiniana. Yangmai 16 is moderately susceptible to both R. cerealis and B. sorokiniana infection, and used as transgenic recipient. The pathogenic fungus R. cerealis isolate R0301 was provided by Profs. Huaigu Chen and Shibin Cai from Jiangsu Academy of Agricultural Sciences, China. B. sorokiniana isolate Hn was provided by Prof. Hongjie Li from Institute of Crop Science, Chinese Academy of Agricultural Sciences.
Wheat plants were grown in a 14 h light/10 h dark (22 °C/10 °C) regime. The wheat plants inoculation with R. cerealis was followed the method of Chen et al.23. Briefly, the soaked wheat kernels were autoclaved for 20 min at 121 °C and then inoculated with fungal discs cut at the edge of freshly cultured R. cerealis on patato dextrose agar plate. The inoculated kernels were incubated for 14 days at 25 °C. The R. cerealis-colonized wheat kernels were placed on the soil surface in contact with the stem base of wheat plants. The inoculation of B. sorokiniana was conducted followed by Zhang et al. procedures18. Briefly, the B. sorokiniana-colonized wheat kernels were placed on the soil surface in contact with the stem base of wheat plants.
The wheat cultivar Yangmai 16 plants at the three-leaf stage were sprayed with 1.0 mM ABA, 50 μM ACC, 1.0 mM SA, and 0.1% Tween-20 solution. Wheat plants treated with Tween-20 solution were used as mock control. The wheat leaves were sampled for detecting the transcription of TaPIMP2.
RNA extraction and cDNA synthesis
Total RNA was extracted using TRIZOL reagent (Invitrogen) and subjected to DNase I (TaKaRa) digestion and purification. The first-strand cDNA was synthesized using 2-μg purified RNA, AMV reverse transcriptase and oligo(dT15) primers (TaKaRa) according to the product instruction.
Cloning of full-length sequence of TaPIMP2 and sequences analyses
Full-length cDNA sequence of TaPIMP2 was obtained from CI12633 through 3′-RACE core set kit (TaKaRa) and RT-PCR. The full-length of cDNA and genomic DNA of TaPIMP2 were obtained from CI12633 using the primers TaPIMP2-fl-F and TaPIMP2-fl-R. The purified PCR products were cloned into the pMD-18T vector (TaKaRa) and sequenced. The cDNA and genomic DNA was analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Pfam software (http://pfam.xfam.org/) was used for prediction of the conserved domains and motifs. A phylogenetic tree was constructed by the neighbor-joining method using MEGA 5.0 software. The amino acid sequence alignment of MYB proteins was analyzed by Clustal X software.
Subcellular localization of TaPIMP2 in wheat and onion cells
The coding region of TaPIMP2 lacking the stop codon was amplified using the primers TaPIMP2-GFP_F and TaPIMP2-GFP_R containing restriction sites Sal I and BamH I, respectively. The PCR products was digested with restriction enzymes Sal I and BamH I, then subcloned in-frame into the 5-terminus of the green fluorescent protein (GFP) coding region in the p35S::GFP vector (kindly provided by Dr. DaowenWang, Chinese Academy of Sciences), resulting in the TaPIMP2-GFP fusion construct p35S::TaPIMP2-GFP.
The p35S::TaPIMP2-GFP or p35S::GFP vector construct was separately transformed into onion epidermal cell by particle bombardment. For wheat protoplast assay, the TaPIMP2-GFP fusion or GFP alone construct was separately introduced into wheat protoplasts via the PEG-mediated transfection method following Yoo et al.42. The transformed onion epidermal cells or wheat protoplasts cells were incubated at 25 °C for 15 h. The GFP signals were observed and photographed using a Confocal Laser Scanning Microscopy (Zeiss LSM 700, Germany) with a Fluar 10X/0.50 M27 objective lens and SP640 filter.
Real-time quantitative RT-PCR (RT-qPCR) analyses of TaPIMP2 and defense-related genes
For RT-qPCR assay, the stems of wheat plants were sampled for examining the transcriptional level of target genes in TaPIMP2-silencing plants and –overexpression wheat plants. RT-qPCR was performed using SYBR Green I Master Mix (TaKaRa) using ABI PRISM 7500 detective system according to the manufacturer’s instruction. The primers of RT-qPCR for TaPIMP2 homoeologous members and 4 defense-related genes were listed in the Table 1. RT-qPCR reaction was set up with the following thermal profile: 95 °C for 5 min, 41 cycles of 95 °C for 15 s and 60 °C for 31 s. The relative transcript levels of tested genes were calculated using the 2−ΔΔCT method43, where the wheat actin gene (Genbank accession no. BE425627) was used as reference.
Functional analysis of TaPIMP2 through VIGS
To generate BSMV:TaPIMP2 construct, TaPIMP2-specific cDNA fragment was amplified by primers TaPIMP2-NheIF and TaPIMP2-NheIF containing Nhe I restriction site from cDNA of CI12633. PCR products were digested with Nhe I, and then ligated into the BSMV-γ vector, resulting in recombinant vector BSMV-γ-TaPIMP2.
The virus BSMV:TaPIMP2 and control virus BSMV:GFP were used to inoculate the CI12633 or Yangmai 6 following Wang et al.44. The stems of BSMV-infected wheat plants were sampled for detection of expression of TaPIMP2. At 20 day after BSMV infection, the fungus R. cerealis or B. sorokiniana was used to inoculate the BSMV infected plants following the methods mentioned above. Disease symptom was observed at 40 day after inoculation with R. cerealis 25 or B. sorokiniana 18.
TaPIMP2-overexpressing transformation vector construction and transformation into wheat
The TaPIMP2 ORF sequence with the Spe I and Sac I restriction sites was amplified and then sub-cloned into the Spe I and Sac I sites of a modified monocot transformation vector pAHC25::myc26. According to the protocol described by Zhu et al.25, the plasmid DNA of resulting overexpression transformation vector pUBI::myc-TaPIMP2 was transformed into immature embryos of the wheat cultivar Yangmai 16 by biolistic bombardment.
PCR and western blotting analyses on TaPIMP2-overexpressing transgenic wheat
The presence of the introduced TaPIMP2 in the transformed wheat plants was monitored by PCR using the primers TaPIMP2-TRANS_F and Tnos-R that specific to TaPIMP2-Tnos cassette. The nontransformed Yangmai 16 and pUBI::myc-TaPIMP2 plasmid DNA were used as negative control and positive control, respectively. The PCR reaction was performed in a total volume of 20 μl containing 1 × Taq buffer, 1.5 mM Mg2+, 0.05 mM dNTP each; 0.4 mM each primer, 1 Unit Taq polymerase (TaKaRa), and 50 ng template DNA, with an initial denaturation at 94 °C 3 min, followed by 35 cycles of 94 °C 45 s, 54 °C 45 s and 72 °C 45 s, and a final extension at 72 °C 10 min. The desired PCR product (213 bp) specific to the introduced TaPIMP2-Tnos cassette was resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
The c-myc-TaPIMP2 fusion protein in the overexpressing transgenic wheat lines was detected by western blotting analysis. Total proteins were extracted from ~0.5 g inoculated stems and sheaths. Total soluble proteins (~10 μg) for each line were separated on 12% SDS-PAGE and transferred to polyvinyl difluoride membranes (Amersham). The blotting membranes were incubated with 900-fold diluted Anti-c-myc Mouse Monoclonal Antibody (Transgen Biotech) at 4 °C overnight, then incubated with 1000-fold diluted Goat Anti-Mouse IgG (H + L), HPR conjugated secondary antibody (Transgen Biotech) at 22 °C for 1 h. The c-myc-TaPIMP2 proteins were visualized using the Pro-light HRP Chemiluminescent Kit (Tiangen Biotech).
The copy number estimation of introduced TaPIMP2-1B gene in transgenic wheat lines
DNA for ddPCR assay was digested by Sal I (TaKaRa). The digested DNA concentration was diluted to 40 ng/μl. For ddPCR reaction, each 20 μl PCR reaction contained 300 ng of DNA template, 100 nmol of each forward primer TaPIMP2-TRANS-Rand reverse primers Tnos-R, and 10 μl EvaGreen Supermix (Bio-Rad). The TaSTOP1-A (Genbank No: KF034796) was used as reference with two copies in hexaploid wheat. The primer TaSTOP1A-F and −R were targeted the reference. Droplets were generated by a QX200 droplet generator (Bio-Rad). A T100 thermal cycler (Bio-Rad) was used to run PCR using the following program: 1 cycle at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 30 s, annealing and extension at 60 °C for 1 min. Droplets were read using a QX200 droplet reader and analyzed with Quantasoft software (Bio-Rad).
Assessment on responses of transgenic and recipient wheat plants to B. sorokiniana and R. cerealis
Thirty plants for each line of TaPIMP2-overexpressors in T4 generations and untransformed Yangmai 16 wheat were evaluated responses to R. cerealis or B. sorokiniana. R. cerealis disease evaluation were followed the method of Zhu et al.25. Disease infection type (IT) of sharp eyespot was scored based on the 0–5 disease scale: 0: no symptoms observed 1: lesions appeared on the sheaths rather than stems; 2: lesions covered less than 1/2 of infected stem perimeter; 3: lesions covered 1/2–3/4 of infected stem perimeter; 4: lesions covered more than 3/4; 5: dead plant. Disease index
where X0–X5 indicated plants with IT: 0–5. B. sorokiniana inoculation and disease evaluation were followed protocols of Zhang et al.18. Disease infection type (IT) of common root rot was categorized from 0 to 4 based on the brown lesion square on the plant stem base. 0: no necrotic lision; 1: lesions covered less than 1/4 of infected stem perimeter; 2: lesions covered 1/4–1/2; 3: lesion covered 1/2–2/3; 4: lesions covered 2/3 to complete stem base. The disease index
where X0–X4 indicated plants with IT: 0–431.
References
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329, doi:10.1104/pp.104.900215 (2006).
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S. & Van Wees, S. C. Networking by small-molecular hormones in plant immunity. Nat. Chem. Biol. 5, 308–316, doi:10.1038/nchembio.164 (2009).
Li, B., Meng, X., Shan, L. & He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host & Microbe 19, 641–650, doi:10.1016/j.chom.2016.04.011 (2016).
Cui, H., Tsuda, K. & Parker, J. E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511, doi:10.1146/annurev-arplant-050213-040012 (2015).
Riechmann, J. L. et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 290, 2105–2110, doi:10.1126/science.290.5499.2105 (2000).
Dubos, C. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581, doi:10.1016/j.tplants.2010.06.005 (2010).
Mengiste, T., Chen, X., Salmeron, J. & Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 15, 2551–2565, doi:10.1105/tpc.014167 (2003).
Seo, P. J. et al. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289, doi:10.1104/pp.109.144220 (2009).
Shim, J. S. et al. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73, 483–495, doi:10.1111/tpj.12051 (2009).
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F. & Hao, Y. J. MdMYB9 and MdMYB11 are involved in the regulatin of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662, doi:10.1093/pcp/pcu205 (2015).
Zhou, J., Lee, C., Zhong, R. & Ye, Z. H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 21, 248–266, doi:10.1105/tpc.108.063321 (2009).
Czemmel, S. et al. The grapevine R2R3-type transcription factor VvMBYF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530, doi:10.1104/pp.109.142059 (2009).
Chagne, D. et al. An ancient duplication of apple MYB transcription factors is responsible for novle red fruit-flesh phenotypes. Plant Physiol. 161, 225–239, doi:10.1104/pp.112.206771 (2013).
Jung, C. et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635, doi:10.1104/pp.107.110981 (2008).
Seo, P. J. & Park, C. M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483, doi:10.1111/j.1469-8137.2010.03183.x (2010).
Liu, X. et al. Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J. Exp. Bot. 64, 2243–2253, doi:10.1093/jxb/ert084 (2013).
Chang, C. et al. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell. 25, 1158–1173, doi:10.1105/tpc.113.109942 (2013).
Zhang, Z. et al. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 196, 1155–1170, doi:10.1111/j.1469-8137.2012.04353.x (2012).
Al-Attala, M. N., Wang, X., Abou-Attia, M. A., Duan, X. & Kang, Z. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol. Biol. 84, 589–603, doi:10.1007/s11103-013-0156-7 (2014).
Zhang, Z. et al. TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat. PLoS ONE. 10(5), e0127723, doi:10.1371/journal.pone.0127723 (2015).
Kumar, J. et al. Bipolaris sorokiniana, a cereal pathogen of global concern: cytological and molecular approaches towards better control. Mol. Plant Pathol. 3, 185–195, doi:10.1046/j.1364-3703.2002.00120.x (2002).
Kumar, U., Joshi, A. K., Kumar, S., Chand, R. & Roder, M. S. Mapping of resistance to spot blotch disease caused by Biopolaris sorokiniana in spring wheat. Theor. Appl. Genet. 118, 783–792, doi:10.1007/s00122-008-0938-5 (2009).
Chen, J. et al. Mapping of QTL conferring resistance to sharp eyespot (Rhizoctonia cerealis) in bread wheat at the adult plant growth stage. Theor. Appl. Genet. 126, 2865–2878, doi:10.1007/s00122-013-2178-6 (2013).
Shan, T. et al. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 1, 6, 28777, doi:10.1038/srep28777 (2016).
Zhu, X. et al. The wheat AGC ninase TaAGC1 is a positive contributor to host resistance to the necrotrophic pathogen Rhizoctonia cerealis. J. Exp. Bot. 66, 6591–6603, doi:10.1093/jxb/erv367 (2015).
Christensen, A. H. & Quail, P. H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218, doi:10.1007/BF01969712 (1996).
Collier, R. et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. doi:10.1111/tpj.13517 (2017).
Głowacka, K. et al. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 39, 908–917, doi:10.1111/pce.v39.4 (2016).
Garcia-Oliveira, A. L. et al. Molecular characterization of TaSTOP1 homoeologues and their response to aluminuim and proton (H(+)) toxicity in bread wheat (Triticum aestivum L.). BMC Plant Boil. 13, 134, doi:10.1186/1471-2229-13-134 (2013).
Zhu, X. et al. The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164, 1499–1514, doi:10.1104/pp.113.229575 (2014).
Millar, A. A. & Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell, 17, 705–721, doi:10.1105/tpc.104.027920 (2005).
Thomma, B. P. H. J. et al. Separate jasmonate-dependent and salicylate-dependent defense response pathways in Arabdopsis are essential for resistance to distinct microbial pathogens. Proc. Nat. Acad. Sci. USA, 95, 15107–15111, doi:10.1073/pnas.95.25.15107 (1998).
Adie, B. A. et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell, 19, 1665–1681, doi:10.1105/tpc.106.048041 (2007).
Pieterse, C. M. J., Van der Dose, D., Zamioudis, C., Leon-Reyes, A. & Van Wees, S. C. M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521, doi:10.1146/annurev-cellbio-092910-154055 (2012).
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J. & Solano, R. Ethylene response factor1 integrates signals from ethylene and jasmonate patheays in plant defense. Plant Cell, 15, 165–178, doi:10.1105/tpc.007468 (2003).
Pre, M. et al. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357, doi:10.1104/pp.108.117523 (2008).
Balasubramanian, V., Vashisht, D., Cletus, J. & Sakthivel, N. Plant β-1,3-Glucanases: their biological functions and trangenic expression against phytopathogenic fungi. Biotechnol. Lett. 34, 1983–1990, doi:10.1007/s10529-012-1012-6 (2012).
Agarwal, P. & Agarwal, P. K. Pathogenesis related- 10 proteins are small, structurally similar but with diverse role in stress signaling. Mol. Biol. Rep. 41, 599–611, doi:10.1007/s11033-013-2897-4 (2014).
Ameya, M. et al. Priming of wheat with the green leaf volatile Z-3-hexenyl acetate enhances defense against Fusarium graminearum but boosts deoxynivalenol production. Plant Physiol. 167, 1671–1684, doi:10.1104/pp.15.00107 (2015).
Gao, Y. et al. BjMYB1, a transcription factor implicated in plant defense through activating BjCHI1 chitinase expression by binding to a W-box-like element. J. Exp. Bot. 67, 4647–4658, doi:10.1093/jxb/erw240 (2016).
Cheng, H. Q. et al. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 67, 1935–1950, doi:10.1093/jxb/erw016 (2016).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile celle system for transient gene expression analysis. Nature. 2, 1565–1572 (2007).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25, 402–408, doi:10.1006/meth.2001.1262 (2001).
Wang, G.-F. et al. Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 191, 418–431, doi:10.1111/j.1469-8137.2011.03715.x (2011).
Acknowledgements
The authors are very grateful to Profs Daowen Wang (Chinese Academy of Sciences, China), Huaigu Chen and Shibin Cai (Jiangsu Academy of Agricultural Sciences, China) for their kind providence of the original vector p35S::GFP, and R. cerealis strain R0301. This study was funded by the programs of the Chinese Key Research & Development Plan Project (2016YFD0101004) and a National “Key Sci-Tech” Project (2016ZX08002001-4).
Author information
Authors and Affiliations
Contributions
Z.Z. designed the research. Z.Z. and X.W. wrote the paper. X.W., T.S., Y.H. and X.L. performed the cloning, sequencing, subcellular localization, VIGS and functional assays. H.X. conducted transformation.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wei, X., Shan, T., Hong, Y. et al. TaPIMP2, a pathogen-induced MYB protein in wheat, contributes to host resistance to common root rot caused by Bipolaris sorokiniana . Sci Rep 7, 1754 (2017). https://doi.org/10.1038/s41598-017-01918-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-01918-7
This article is cited by
-
MYB transcription factors—master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses
Plant Cell Reports (2022)
-
Characterization of histological changes at the tillering stage (Z21) in resistant and susceptible wheat plants infected by Tilletia controversa Kühn
BMC Plant Biology (2021)
-
Characterization of the wheat cultivars against Tilletia controversa Kühn, causal agent of wheat dwarf bunt
Scientific Reports (2020)
-
Methyljasmonate and salicylic acid contribute to the control of Tilletia controversa Kühn, causal agent of wheat dwarf bunt
Scientific Reports (2020)
-
Recent developments and applications of genetic transformation and genome editing technologies in wheat
Theoretical and Applied Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.