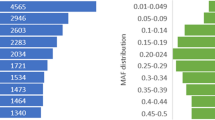
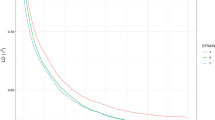
Abstract
The progress of aquaculture heavily depends on the efficient utilization of diverse genetic resources to enhance production efficiency and maximize profitability. Single nucleotide polymorphisms (SNPs) have been widely used in the study of aquaculture genomics, genetics, and breeding research since they are the most prevalent molecular markers on the genome. Currently, a large number of SNP markers from cultured fish species are scattered in individual studies, making querying complicated and data reuse problematic. We compiled relevant SNP data from literature and public databases to create a fish SNP database, FishSNP (http://bioinfo.ihb.ac.cn/fishsnp), and also used a unified analysis pipeline to process raw data that the author of the literature did not perform SNP calling on to obtain SNPs with high reliability. This database presently contains 45,690,243 (45 million) nonredundant SNP data for 13 fish species, with 30,288,958 (30 million) of those being high-quality SNPs. The main function of FishSNP is to search, browse, annotate and download SNPs, which provide researchers various and comprehensive associated information.
Similar content being viewed by others
Background & Summary
In the post-genomics era, the endeavor to improve the efficiency, sustainability, product quality, and profitability of aquaculture production is increasingly rely on the diverse genetic knowledges obtained from genome study of each aquaculture species1. The genome study is frequently based on gene type, gene quantity, and sequence variation. Insertions, deletions, translocations of large and small fragments, polymorphisms of tandem sequence (satellite DNA) repetitions, and single nucleotide polymorphisms (SNPs) are the most common types of sequence variation. SNPs are common in the genome2 and easily detectable by high-throughput sequencing, making them the primary molecular markers in the study of fish genome variation3,4,5. The presence of homologous genes or sequences initially complicates allele identification, however the simplicity with which SNPs can be scored promotes allele discrimination6. SNP markers have been widely employed in fish disease resistance7,8, feed conversion efficiency9,10, growth rate11,12, muscle yield13, reproductive characteristics14,15 and tolerance to environmental stressors16,17 since the emergence of high-throughput sequencing technology. Similarly, SNP marker-related sequencing data increased rapidly. However, there have been various issues with using SNP markers in fish research.
First, the SNP results of fish research are dispersed throughout the literature, and fish SNP data in public databases are still quite limited. There are now two major databases that contain some fish SNP information: the EMBL-EBI European Variation Archive (EVA, https://www.ebi.ac.uk/eva) and the Animal QTLdb (https://www.animalgenome.org/tools/SNPnmids). EVA is devoted to collecting data on variation types in species other than humans, including SNP information in zebrafish and several non-model fish, with salmon and trout data accounting for more than half of all SNPs18,19. EVA primarily collects pertinent SNPs and background information files that authors upload. However, the disclosure of SNP information in studies is often not required, reducing the integrity of EVA data collection. Furthermore, EVA has not confirmed the author’s marker information, and its correctness remains unknown. AnimalQTLdb mostly collects molecular markers from livestock and poultry, however it also has over 60 K rainbow trout SNPs20,21. AnimalQTLdb uses EVA as the standard, compares custom markers to EVA, keeps the same SNP site, and inherits with its ID. There are a few SNP databases that focus on one or two specific fish species, for example, SalmonDB for Atlantic salmon and rainbow trout22, and SNPfisher for zebrafish23. However, for most important economic fish, their SNPs and related information are still scattered in vast mounts of literatures. A fish SNP database that integrates genomic information, annotations, and covers more complete fish species is urgently needed for fish research.
Second, while the issue of SNP marker accuracy is prevalent to some extent, it is an inherent challenge in fish molecular biology research. Most fish genome research is significantly less extensive than that of humans and model animals, and SNP calling is frequently based on rather limited sequencing data. Poor sequencing quality and depth, as well as inappropriate data processing techniques, will inevitably contribute to an increase in false positives in this situation. Some highly similar homologous sequences scattered throughout the genome have only a few single nucleotide differences. These are treated as alleles during SNP calling, resulting in “pseudo-SNP” markers, which is a typical problem when employing SNP markers24. Fish commonly experienced two or more rounds whole-genome duplication events, such as common carp, salmon and trout. The duplication event adds to the genome’s complexity and has crucial consequences for evolutionary studies25,26,27, However, it also copies homologous sequences, resulting in additional “pseudo-SNPs”28.
In general, synthesis analysis of fish data from multiple research on the same fish species promotes mutual confirmation and bias removal. However, comparing SNP detection results from fish samples is infeasible. The fundamental reason for this is that the most commonly utilized sequencing methods in fish are restriction site-associated DNA sequencing (RAD-seq) that are cost-effective but low genome coverage29,30. Most SNP marker sets produced using RAD-seq have inadequate repeatability31. In many circumstances, we may identify various sets of SNP loci from separate investigations, different batches of library builds, and even different samples within the same batch, and there is little or even no overlap between these SNP sets. The upgrading of the reference genome and the development of new versions will make SNP marker interaction verification even more challenging.
To address all of the aforementioned issues, this study collected and categorized a large number of fish SNPs and created the Fish SNP database, which provides trustworthy SNP information for fish research. We acquire SNP data from three sources: (1) fish SNP marker data in public databases; (2) SNP data reported in published literature; and (3) SNP markers obtained by processing original literature data using a unified approach. We noticed that some published studies provided raw sequencing data without SNP information during the collection process, so we used the GATK haplotype process with higher accuracy32,33,34 and general hard filtering parameters for SNP marker filtering35 to obtain SNP information. We used a unified pipeline to analyze data that the original authors did not call for SNPs, and we retrieved SNPs with excellent reliability. To eliminate “pseudo-SNP” markers, we used the Mendelian segregation ratios test in pedigree data and the Hardy Weinberg equilibrium test in random populations to identify real SNPs. Various published study results were combined to generate a uniform SNP ID that was compatible with numerous genome versions. An annotation tool, utilizing SnpEff36,37 to annotate new SNPs, is also accessible to users, facilitating the creation of a complete genome variation profile of fish species. We anticipate that FishSNP will give more comprehensive SNP information for researchers in aquaculture genomics, genetics and breeding.
Methods
Data resources of large scale loach
We performed whole genome sequencing on 20 large scale loach. The large-scale loach adults were collected from the Baishazhou Aquatic Product Market in Wuhan, Hubei Province, China. Ten females and three males were selected for breeding on April 30th, 2020. Their offspring were kept at 20 °C, 25 °C, and 30 °C, three different water temperatures in the lab, and fed enough red worms twice a day. The tails of all these samples were preserved in 95% ethanol, and the DNA was extracted using the CTAB protocol. High-throughput pair-end (150 bp/end) whole genome sequencing (DNBSEQ-T7, Illumina Inc.) was performed on ten female parents (BGI, Wuhan, China), and high-throughput pair-end (150 bp/end) whole genome sequencing (BGISEQ-500, Illumina Inc.) was performed on ten offspring reared at 20 °C (BGI, Wuhan, China). All experiments and animal treatments were carried out according to the principles of Animal Care and Use Committee of Institute of Hydrobiology, Chinese Academy of Sciences.
Other data resources and integration
The study first assessed the value of fish in culture and evolutionary research, and the existing status of fish SNP inclusion in public SNP databases before selecting 13 fish species to collect and organize data. On the one hand, we collected as much SNP-related literature about these fishes as possible and mined SNPs from it as the FishSNP database’s baseline dataset. Simultaneously, we collected VCF files from EVA for the respective species, integrated, and annotated SNP data as an essential component of FishSNP. SNPs from various sources were integrated based on the SNP’s site information or the alignment information of the flanking sequences.
Although some publications include SNP information in the appendix, the formats and reference genomes are often incompatible. The study compared the genome version of the article with the genome version provided in our database for the attachments containing the genome version and SNP site information. If the genome versions are consistent, the SNP attachments were directly converted to VCF files. Otherwise, the SNP’s 150 bp flanking sequences on the original reference genome or the flanking sequences provided in attachments were aligned to the version of genome in FishSNP using bowtie2.0, then the markers’ positions were obtained.
SNPs were called using an in-house pipeline after some other literatures provided sequencing data instead of SNPs. We employed a unified SNP calling procedure to retrieve VCF files from the original sequencing data, and we used the corresponding genome and SnpEff software (version 4.3)36 for functional annotation (Fig. 1). This procedure necessitated the usage of reference genomes, and we prioritized the genome version that offers structural annotation information on NCBI, the most recently published genome version, and the genome version used by EVA in that order.
Each SNP found in literature was given a unique ID in FishSNP. From left to right, the FishSNP ID consists of three fields. The first two characters “FS” represent the database’s identifier, the next three digits form the serial number of a species ranging from 000 to 999, which is displayed in the “Help” section of the FishSNP website, and the remaining numbers represent the serial number of an SNP. Users can also search all SNPs integrated from the EVA database using EVA’s official ID.
SNP calling and validation process
Prefetch (2.9.3-1) was used to download the sequencing data, and vdb-validate (2.9.6) was used to evaluate the data integrity, then the sequence data was converted into the original fastq file by fastq-dump (2.9.6). Fastq quality filter (parameter “-q 20 - p 70 -z -Q 33”) was used to clean the original data, and the paired-end data was coupled together using an in-house script. Clean data were aligned to the genome using bowtie2 (2.3.5) with default parameters38, followed by a series of procedures from the GATK package (4.1.1.0) (https://github.com/broadinstitute/gatk/releases), including SortSam, Add Or Replace Read Groups, Mark Duplicates, Fix Mate Information, and Haplotype Caller, all of which run with default parameters. Each sample resulted in a gvcf file; Merge the gvcf files of multiple samples (CombineGVCFs, GenotypeGVCFs) and perform hard filtering (Variant Filtration, the parameters are QD < 2.0 || FS > 60.0 || MQ < 35.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 || SOR > 3.0)35,39. Due to the double-enzyme digestion sequencing principle in data of RAD-seq method, the procedure to mark duplicates (MarkDuplicates) will cause SNPs to be mistakenly deleted by the subsequent process. So MarkDuplicates was applied only to the whole genome re-sequencing data.
We have developed in-house scripts for conducting population tests based on Mendelian segregation ratio and Hardy-Weinberg equilibrium. The script used for the Mendelian segregation ratio test employs a significance cutoff of 0.01 for the P value obtained from the chi-square test. Additionally, we utilized plink1.940 (https://www.cog-genomics.org/plink2) to perform the Hardy-Weinberg equilibrium test, applying the parameter “–hwe 0.000001” to filter error SNPs. Subsequently, quality classification was carried out based on P values. All these scripts have been made publicly available in FishSNP’s download module, or directly at, http://bioinfo.ihb.ac.cn/software/FishSNP and https://github.com/hliihbcas/FishSNP.
Data Records
All raw data and SNP data of large scale loach were deposited into the GSA database41,42,43 and EVA44. All The SNPs collected in this study are summaried by species in Table 1, and the statistics of articles and populations are listed in Supplementary Tables S1 and S2. The datasets have been archived at Figshare45. We have organized the data by creating a dedicated folder for each species, and within each species folder, SNP data are stored according to the respective genome version. The “Unmapped” data consists of information gathered from literature sources that cannot be mapped to any currently available genome version. And we also provide symbols to mark the data resources such as EVA. All the literature sources utilized in this research are enumerated in Supplementary Tables S3 and S4. In Supplementary Table S3, we have referenced these papers for accessibility and presented concise descriptions for each of them. In Supplementary Table S4, comprehensive details are presented, including information such as population size, test types, bioproject references, and sequencing methods.
Technical Validation
Population tests for Mendelian segregation ratio or Hardy-Weinberg equilibrium rely on the completeness of population information, typically including pedigree details for family-based tests and individual population information for Hardy-Weinberg equilibrium tests. Collecting such information poses significant challenges due to variations in the level of commitment from authors/uploaders and the lack of uniform requirements for data uploading across different journals or public databases. To ensure the clarity regarding the completeness of the collected data, we have presented a comprehensive list of the acquired data (Supplementary Table S4), and provided annotations indicating the availability of different aspects of these datasets in the columns labeled “Description”, “Attachment detail” and “Project detail”.
If a literature provides sequencing data rather than SNPs, the database will only include those datasets that meet the requirements for population tests. So far, the SNP markers directly described in the literatures (or their attachments) do not disclose the genotype of every sample, therefore redoing a population test is unfeasible. It can only be classified as tested or untested based on whether or not the population test was performed as specified in the article. For SNPs with known genotypes of samples, the chi-square test was used on the SNP markers mined by the in-house pipeline to see if they met the Mendelian segregation ratio in pedigree data or the Hardy-Weinberg equilibrium in random group data. Markers with high reliability (P > 0.01) are labeled “Pass” in the database, while “Deviation” (P ≤ 0.01) warrants careful consideration in data analysis, and “Untested” denotes cases where population testing was infeasible due to insufficient essential information (Table 1). If an SNP is found in multiple data sets, the geometric mean of its various P values appears in the SNP’s basic information panel on the database website, whereas the information for a single detection appears in the “Populations” section.
Although EVA does not provide quality control information for all 16,484,194 EVA-derived SNPs, we checked all markers with P values and found 5,264,880 SNPs that were shared with EVA, among which those with better reliability (P > 0.01) occupy 80.35%, a proportion obviously lower than that of the remaining markers (93.98%), suggesting that the quality of SNPs derived from literature and our pipeline is higher than that of EVA.
Usage Notes
We created the FishSNP database to enhance the intuitive and efficient utilization of SNP data. FishSNP encompasses four primary functions for retrieving SNP information: search, browse, annotate, and download (Fig. 2). Users can search and visualize SNP data in various formats, and new SNP data can be subject to be annotated and uploaded to the database. Detailed instructions on how to use these features are available on the “Help” page of the website.
The datasets, which include essential information for each SNP, as well as the P value from the population test, are provided in a VCF format, with detailed explanations of the test results at the beginning of each file. In addition, we have mapped the SNP data to various available genome versions and made them accessible through separate folders under each species, allowing users to select and utilize the data according to their specific needs.
Code availability
The process and script files can be downloaded at http://bioinfo.ihb.ac.cn/software/FishSNP or https://github.com/hliihbcas/FishSNP.
References
Abdelrahman, H. et al. Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research. Bmc Genomics 18, https://doi.org/10.1186/s12864-017-3557-1 (2017).
Sachidanandam, R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933, https://doi.org/10.1038/35057149 (2001).
Helyar, S. J. et al. Application of SNPs for population genetics of nonmodel organisms: new opportunities and challenges. Mol Ecol Resour 11, 123–136, https://doi.org/10.1111/j.1755-0998.2010.02943.x (2011).
Flanagan, S. P. & Jones, A. G. The future of parentage analysis: From microsatellites to SNPs and beyond. Mol Ecol 28, 544–567, https://doi.org/10.1111/mec.14988 (2019).
Sun, Y.-L. et al. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 433, 19–27, https://doi.org/10.1016/j.aquaculture.2014.05.035 (2014).
Vignal, A., Milan, D., SanCristobal, M. & Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genetics selection evolution 34, 275–305 (2002).
Hillestad, B., Makvandi-Nejad, S., Krasnov, A. & Moghadam, H. K. Identification of genetic loci associated with higher resistance to pancreas disease (PD) in Atlantic salmon (Salmo salar L.). BMC Genomics 21, 388, https://doi.org/10.1186/s12864-020-06788-4 (2020).
Jin, R. M. et al. Characterization of mandarin fish (Siniperca chuatsi) IL-6 and IL-6 signal transducer and the association between their SNPs and resistance to ISKNV disease. Fish Shellfish Immunol 113, 139–147, https://doi.org/10.1016/j.fsi.2021.04.003 (2021).
Luo, L. et al. Selection of growth-related genes and dominant genotypes in transgenic Yellow River carp Cyprinus carpio L. Funct Integr Genomics 18, 425–437, https://doi.org/10.1007/s10142-018-0597-9 (2018).
Barría, A., Benzie, J. A. H., Houston, R. D., De Koning, D. J. & de Verdal, H. Genomic Selection and Genome-wide Association Study for Feed-Efficiency Traits in a Farmed Nile Tilapia (Oreochromis niloticus) Population. Front Genet 12, 737906, https://doi.org/10.3389/fgene.2021.737906 (2021).
Robledo, D., Rubiolo, J. A., Cabaleiro, S., Martínez, P. & Bouza, C. Differential gene expression and SNP association between fast- and slow-growing turbot (Scophthalmus maximus). Sci Rep 7, 12105, https://doi.org/10.1038/s41598-017-12459-4 (2017).
Salem, M. et al. RNA-Seq identifies SNP markers for growth traits in rainbow trout. PLoS One 7, e36264, https://doi.org/10.1371/journal.pone.0036264 (2012).
Al-Tobasei, R. et al. Identification of SNPs associated with muscle yield and quality traits using allelic-imbalance analyses of pooled RNA-Seq samples in rainbow trout. BMC Genomics 18, 582, https://doi.org/10.1186/s12864-017-3992-z (2017).
Mohamed, A. R. et al. Polygenic and sex specific architecture for two maturation traits in farmed Atlantic salmon. BMC Genomics 20, 139, https://doi.org/10.1186/s12864-019-5525-4 (2019).
Maekawa, M. et al. Sex-Associated SNP Confirmation of Sex-Reversed Male Farmed Japanese Flounder Paralichthys olivaceus. Mar Biotechnol (NY) 25, 718–728, https://doi.org/10.1007/s10126-023-10235-2 (2023).
Kess, T. et al. Genomic basis of deep-water adaptation in Arctic Charr (Salvelinus alpinus) morphs. Mol Ecol 30, 4415–4432, https://doi.org/10.1111/mec.16033 (2021).
Zhao, S. S., Su, X. L., Yang, H. Q., Zheng, G. D. & Zou, S. M. Functional exploration of SNP mutations in HIF2αb gene correlated with hypoxia tolerance in blunt snout bream (Megalobrama amblycephala). Fish Physiol Biochem 49, 239–251, https://doi.org/10.1007/s10695-023-01173-w (2023).
Cezard, T. et al. The European Variation Archive: a FAIR resource of genomic variation for all species. Nucleic Acids Res 50, D1216–D1220, https://doi.org/10.1093/nar/gkab960 (2022).
Cook, C. E. et al. The European Bioinformatics Institute in 2016: Data growth and integration. Nucleic Acids Res 44, D20–26, https://doi.org/10.1093/nar/gkv1352 (2016).
Hu, Z. L., Park, C. A. & Reecy, J. M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res 47, D701–D710, https://doi.org/10.1093/nar/gky1084 (2019).
Hu, Z. L., Park, C. A. & Reecy, J. M. Bringing the Animal QTLdb and CorrDB into the future: meeting new challenges and providing updated services. Nucleic Acids Res 50, D956–D961, https://doi.org/10.1093/nar/gkab1116 (2022).
Di Génova, A. et al. SalmonDB: a bioinformatics resource for Salmo salar and Oncorhynchus mykiss. Database (Oxford) 2011, bar050, https://doi.org/10.1093/database/bar050 (2011).
Butler, M. G. et al. SNPfisher: tools for probing genetic variation in laboratory-reared zebrafish. Development 142, 1542–1552, https://doi.org/10.1242/dev.118786 (2015).
Castaño Sánchez, C., Palti, Y. & Rexroad, C. SNP analysis with duplicated fish genomes: differentiation of SNPs, paralogous sequence variants, amd multisite variants. Next generation sequencing and whole genome selection in aquaculture, 133–150 (2011).
Guyomard, R., Boussaha, M., Krieg, F., Hervet, C. & Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet 13, 15, https://doi.org/10.1186/1471-2156-13-15 (2012).
Danzmann, R. G. et al. Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon). BMC Genomics 9, 557, https://doi.org/10.1186/1471-2164-9-557 (2008).
Dehal, P. & Boore, J. L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3, e314, https://doi.org/10.1371/journal.pbio.0030314 (2005).
Christensen, K. A. et al. Identification of single nucleotide polymorphisms from the transcriptome of an organism with a whole genome duplication. BMC Bioinformatics 14, 325, https://doi.org/10.1186/1471-2105-14-325 (2013).
Robledo, D., Palaiokostas, C., Bargelloni, L., Martinez, P. & Houston, R. Applications of genotyping by sequencing in aquaculture breeding and genetics. Reviews in Aquaculture 10, 670–682, https://doi.org/10.1111/raq.12193 (2018).
Liu, T., Li, R., Xiao, H. & Chen, S. Research progress of RAD-seq in fish genomics. Journal of Yunnan University. Natural Science 40, 1283–1289 (2018).
Davey, J. W. et al. Special features of RAD Sequencing data: implications for genotyping. Mol Ecol 22, 3151–3164, https://doi.org/10.1111/mec.12084 (2013).
Peng, R., Jones, D. C., Liu, F. & Zhang, B. From Sequencing to Genome Editing for Cotton Improvement. Trends in Biotechnology https://doi.org/10.1016/j.tibtech.2020.09.001 (2020).
Liu, X., Han, S., Wang, Z., Gelernter, J. & Yang, B.-Z. Variant Callers for Next-Generation Sequencing Data: A Comparison Study. Plos One 8, https://doi.org/10.1371/journal.pone.0075619 (2013).
Pirooznia, M. et al. Validation and assessment of variant calling pipelines for next-generation sequencing. Human Genomics 8, https://doi.org/10.1186/1479-7364-8-14 (2014).
De Summa, S. et al. GATK hard filtering: tunable parameters to improve variant calling for next generation sequencing targeted gene panel data. BMC Bioinformatics 18, 119, https://doi.org/10.1186/s12859-017-1537-8 (2017).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92, https://doi.org/10.4161/fly.19695 (2012).
Cingolani, P. in Variant Calling: Methods and Protocols (eds Charlotte Ng & Salvatore Piscuoglio) 289–314 (Springer US, 2022).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, https://doi.org/10.1038/nmeth.1923 (2012).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303, https://doi.org/10.1101/gr.107524.110 (2010).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, https://doi.org/10.1186/s13742-015-0047-8 (2015).
Chen, T. et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics, Proteomics & Bioinformatics 19, 578–583 (2021).
Database resources of the national genomics data center, china national center for bioinformation in 2022. Nucleic Acids Research 50, D27-D38 (2022).
Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA011033) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
European Variation Archive https://identifiers.org/ena.embl:PRJEB65007 (2023).
Zhang, L. et al. FishSNP: a high quality cross-species SNP database of fishes, figshare, https://doi.org/10.6084/m9.figshare.c.6793827.v1 (2024).
Acknowledgements
We thank Ms Zhixian Qiao from the Analysis and Testing Center at Institute of Hydrobiology for her technical supports. The computational work in this study was supported by the Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences, China. This work was supported by the grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding (XDA24010206), the National Key R&D Program of China (2021YFD1200804, 2018YFD0901201) and the National Natural Science Foundation of China (31801055).
Author information
Authors and Affiliations
Contributions
Lei Zhang performed the research, processed the data and wrote the paper. Heng Li organized the script files, and designed the database and web pages. Keyi Ren, Yingyin Cheng and Wanting Zhang contributed the new analysis method. Xiao-Qin Xia, Mijuan Shi and Yaping Wang designed the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, L., Li, H., Shi, M. et al. FishSNP: a high quality cross-species SNP database of fishes. Sci Data 11, 286 (2024). https://doi.org/10.1038/s41597-024-03111-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03111-8