Abstract
Biological ocean data collected from ships find reuse in aggregations of historical data. These data are heavily relied upon to document long term change, validate satellite algorithms for ocean biology and are useful in assessing the performance of autonomous platforms and biogeochemical models. Existing aggregate products have largely been restricted to the surface ocean, omit physical data or have limited biological data. We present the first version of a BIOlogical ocean data reforMATting Effort (BIO-MATE) to begin to fill a gap in subsurface bio-physical data aggregates in a reproducible way. BIO-MATE uses open-source R software that reformats openly sourced published datasets from oceanographic voyages. These reformatted biological and physical data from underway sensors, profiling sensors, pigments analysis and particulate organic carbon analysis are stored in an interoperable BIO-MATE data product for easy access and use. Specific QA/QC protocols can now be easily applied to the BIO-MATE data product to support a variety of surface and subsurface applications.
Similar content being viewed by others
Background & Summary
Marine phytoplankton blooms support ocean food-webs and influence global climate through the biological carbon pump1,2,3. Ocean physics and other environmental drivers control the timing, magnitude and extent of phytoplankton blooms through complex bio-physical relationships4,5,6,7. To study these relationships, integrated data structures that link biological and physical ocean data are needed. Ship-based data are the gold standard for accurate biological oceanographic measurements8. These data are often published separately to physical ocean data, stored across different repositories and in multiple formats. This makes it difficult and time-consuming to aggregate and link biological and physical data. The described data product attempts to make this task easier.
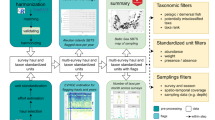
The biological ocean data reformatting effort (BIO-MATE) works to link existing, open-access biological and physical datasets across oceanographic voyages and promote their re-use (Fig. 1). This has been done by developing a BIO-MATE R software package that not only reformats published datasets, but also cross-references between biological and physical data and allows access to citation information (https://github.com/KimBaldry/BIOMATE-Rpackage). The resulting BIO-MATE data product allows users to easily access, manipulate and cite published ship-based datasets of different dimensions for multiple applications.
The BIO-MATE data product can be accessed via the IMAS Data Portal9 and the Australian Ocean Data Network (https://portal.aodn.org.au/). The aggregation includes four data streams: (1) data collected from shipboard underway sensors, (2) profiling sensors mounted on sampling rosettes, (3) lab analysis for phytoplankton pigments and (4) lab analysis for particulate organic carbon (POC). These data streams are cross-referenced by unique expedition codes (EXPOCODE) and profiling station identifications (CTD_ID). An additional data stream contains supporting information for the data product including a list of oceanographic voyages, investigator contact information and data citations for reformatted datasets. We have also included an aggregated data table for biological data. Users are requested to refer to supporting data and cite all data products accessed through BIO-MATE, as well as the BIO-MATE data product itself. We consulted the distribution licenses of all data sources to ensure that with this condition data are re-used lawfully.
The data product has been used to understand how the response of in-situ fluorometers changes in the Southern Ocean, to assess non-photochemical quenching corrections and to investigate the role of ocean physics in mediating subsurface chlorophyll features10. These examples highlight the malleability of this data product to improve our understanding of biological oceanography. Other potential applications include validating satellite observations11,12, developing new ways to validate in-situ bio-optical observations collected by autonomous profiling platforms in the presence of dynamic fronts8,13,14, training ocean state estimations15, informing bio-physical models and using multi-variate analyses to understand bio-physical relationships.
We recognise the massive effort in producing the thousands of data records in this data product. This includes the investigators and data officers who have spent countless hours in ship time, project organisation, grant writing, laboratory analysis, data processing and report writing. Oceanographic data are often collected with regional studies in mind, but their value increases with publication and re-use. We encourage all investigators to publish their data for re-use through data products like BIO-MATE.
Methods
Published datasets in BIO-MATE
The BIO-MATE aggregate data product9 brings together ship-based data that have been collected by a Principal Investigator (PI), published to a publicly accessible database and re-cited in this data descriptor16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349,350,351,352,353,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410,411,412,413,414,415,416,417,418,419,420,421,422,423,424,425,426,427,428,429,430,431,432,433,434,435,436,437,438,439,440,441,442,443,444,445,446,447,448,449,450,451,452,453,454,455,456,457,458,459,460,461,462,463,464,465,466,467,468,469,470,471,472,473,474,475,476,477,478,479,480,481,482,483,484,485,486,487,488,489,490,491,492,493,494,495,496,497,498,499,500,501,502,503,504,505,506 (Fig. 2). The first version of BIO-MATE includes published datasets associated with four types of measurements:
-
1.
sensors in the vessels underway seawater in-take (underway sensor data stream),
-
2.
profiling sensors mounted to sampling rosettes (profiling sensor data stream),
-
3.
pigments measured in the laboratory (pigment data stream), and
-
4.
POC measured in the laboratory (POC data stream).
Data records from the pigment data stream were first identified in data repositories that host biological data. Pigment data records were identified using the search term “chlorophyll” and a latitude bound of 30 to 90 S from PANGAEA506 (https://www.pangaea.de/), SeaBASS500,501 (https://seabass.gsfc.nasa.gov/), the Australian Ocean Data Network (AODN, https://portal.aodn.org.au/), GLODAP466,467 (https://www.glodap.info/), the Palmer Long Term Ecological Record (PAL-LTER, https://pal.lternet.edu/data), the Biological and Chemical Oceanography Data Management Office (BCO-DMO, https://www.bco-dmo.org/data), the CSIRO Marlin Data Trawler (Marlin, https://www.cmar.csiro.au/data/trawler/) and the Australian Antarctic Data Center (AADC, https://data.aad.gov.au/). Data records from the profiling sensors and underway sensors data streams were then identified in these repositories and in the CCHDO (https://cchdo.ucsd.edu/) and MGDS493 (https://www.marine-geo.org/). Although we largely constrained the first version of BIO-MATE aggregate data product to the Southern Ocean, further versions can be expanded to the global ocean.
From available pigment data records, 178 relevant voyages were identified using unique 12-digit expedition codes (EXPOCODES) assigned as follows; National Oceanographic Data Centre (NODC) platform codes followed by voyage 8 digit start dates (YYYYMMDD). NODC platform and country codes are recorded on Git Hub (https://github.com/KimBaldry/BIO-MATE/product_data/supporting_information/codes) and within the BIO-MATE software (https://github.com/KimBaldry/BIOMATE-Rpackage/inst/codes). If the vessel name or voyage start/end dates were absent, this information was found using Google to discover voyage records. This voyage information was used to do a final Google keywords search (i.e. ship name, synonyms for voyages, year, “underway”, “CTD”, “chlorophyll”,”POC”, “cruise report” and “data”) to determine any absent records and to discover accompanying cruise reports.
Semi-automated BIO-MATE workflow for reformatting datasets
A semi-automated workflow and the BIO-MATE R software (https://github.com/KimBaldry/BIOMATE-Rpackage) were used to reformat published datasets, and produce the BIO-MATE data product (Fig. 3). Downloaded data files were split by EXPOCODES if they recorded data within a larger dataset (e.g PAL-LTER data records). Files for the profiling sensor data stream were further split into individual profiles. Processing metadata were manually entered into a table to inform the BIO-MATE R software and a bulk run of the software was performed to reformat data files. The workflow is described in more detail in the following subsections (Fig. 2).
Download of published datasets
Published datasets were manually downloaded from open source repositories and stored locally in accordance with data policies. Some manual reformatting of a small portion of downloaded data had to be performed on old datasets, prior to the application of reformatting scripts, due to formatting irregularities. Downloaded data files, and their amendments used to create the BIO-MATE data product, are not published in BIO-MATE, but are available upon request to the corresponding author.
Splitting large datasets with BIO-MATE software
The BIO-MATE R software requires each file to only contain observations from a single voyage. Further, the profiling sensor data stream requires each file to only contain observations from a single profiling cast, held in a discrete directory for each voyage.
The split_delim_file function splits files using identified variables containing EXPOCODE synonyms and/or profiling station information. This function can be used to split a single, large data file into smaller files as required. For this version of the data product, a number of files had to be split to be ingested into the BIO-MATE core functions. A record of these can be found in Git Hub in the project notebook (https://github.com/KimBaldry/BIO-MATE/blob/main/BIO-MATE.Rmd).
Processing metadata
Information on file formats, dataset information, citation information, location data variables and ocean data variables are needed to reformat published datasets with BIO-MATE software. This information is called processing metadata herein and was manually entered and stored as comma delimited text files. The processing metadata required to run BIO-MATE software is described in the supplement Processing Metadata Table and differs for each data stream. All processing metadata used to construct the BIO-MATE aggregated data product is stored in Git Hub (https://github.com/KimBaldry/BIO-MATE/tree/main/product_data/processing_metadata).
Dataset citation with BibTEX files
Information is included in the BIO-MATE data product, for citing published datasets, laboratory analysis methodologies (for the pigment and POC data streams) and the data repositories through which published data records were accessed. Each citation was recorded as a BibTeX entry, compatible with EndNote, R and LaTeX. Each BibTeX entry has a tag that is referenced in the processing metadata. This tag is used to link citations to their corresponding data records when datasets are ingested in the BIO-MATE software. Citation information is then printed in the header information in reformatted files. Where possible BibTeX entries were sourced from data repositories. If BibTeX entries were not found, they were created manually.
All BibTeX entries are stored on Git Hub (https://github.com/KimBaldry/BIO-MATE/product_data/supporting_information/citations) and in the BIO-MATE software (https://github.com/KimBaldry/BIOMATE-Rpackage/inst/citations). A look-up table is included in the BIO-MATE software to help users find relevant BibTeX entries needed to cite datasets appropriately (https://github.com/KimBaldry/BIOMATE-Rpackage/tree/main/data). A function export_ref supports the export of a smaller BibTeX file based on user selections of EXPOCODES and data streams that they have accessed through the product. This allows references to be easily appended to a bibliography as required.
Reformatting and linking data streams with BIO-MATE R software
The BIO-MATE R software was run to reformat data files to the WHP (CCHDO)-Exchange format (https://exchange-format.readthedocs.io/en/latest/index.html), using the original or split data files, processing metadata and citation information as input. The software arranges reformatted WHPE files into four data streams in local directories that include separate WHPE files, for each EXPOCODE, and for underway sensors, profiling sensor casts, pigment measurements, and POC measurements.
Each data stream has its own reformatting function within the BIO-MATE R software (UWY_to_WHPE, PROF_to_WHPE, PIG_to_WHPE, POC_to_WHPE). The software requires physical (underway sensor and profiling sensor) data streams to be reformatted before biological (pigment and POC) data streams, to accommodate a biological-physical matching algorithm within the PIG_to_WHPE and POC_to_WHPE functions. The algorithm links biological data in the pigment and POC data streams to the physical data in the profiling sensor and underway sensor data streams. Biological data records are given a profiling sensor identification tag (CTD_ID) if matched to physical data in BIO-MATE.
To match biological data to physical data, the algorithm first uses EXPOCODES to find relevant physical data in profiling sensor data streams. It then matches biological and physical data records by comparing station number (STNBR) and cast number (CASTNO) records. If matches are detected using STNBR and CASTNO, the validity of these matches is checked by comparing time and position information, but if position and time were not recorded in biological datasets (4157 pigment and 1948 POC records) it is assumed that the STNNBR and CASTNO records are correct if they match between data streams (e.g. in the JGOFS records). If position or time was recorded, a check on identified matches is performed to see if both the biological and physical data record data either within 24 hours of each other or within 8 km507. This quick check catches cases where STNNBR and CASTNO are used in similar ways within physical and biological sampling, but exact matches do not correspond to the same sampling event.
If matches couldn’t be identified using STNBR and CASTNO between datasets a more rigorous search was performed using a database of time and position information from all profiling sensor data relating to the EXPOCODE. Matches were then found for biological data, if it contains position information, by finding the closest profiling sensor record within 1km in the database. If time information exits, matches are identified as the closest profiling sensor record within 6 hours, otherwise only matching date information is required. These position and time constraints are tighter than if STNNBR and CASTNBR records were matched. Using this process all biological sampling events had matching physical sampling events. Matching has only been implemented with physical profiling sensor data and not to physical underway data. Underway surface data do not require station IDs and are more simply found with EXPOCODE and position and/or time.
Quality assurance
Limited quality assurance has been performed on the BIO-MATE data product and is variable across published datasets. As a supplement we include some insights into the quality of pigment data and chlorophyll fluorescence profiles which has been obtained through visual inspection (Supplementary QA/QC). The initial integrity of these data records lies with the Principal Investigators of the published data record. As a result, reformatted data have varying levels of quality control and post-processing. We have included cruise report citations in our product to aid in further data quality assurance efforts.
This allows a range of users to benefit from the BIO-MATE aggregate product and ensures data quality remains at the standard it was published. The quality assurance required of physical and biological ocean data varies according to application and is up to the user to confirm the data is suitable for their application. Future versions of BIO-MATE could implement quality assurance metrics under community consensus. The data can now be easily ingested by other data synthesis efforts, like GLODAP466,467 and the World Ocean Database (WOD), which implement established QA/QC protocols.
Data Records
The BIO-MATE data product9 is stored on the IMAS data portal (https://data.imas.utas.edu.au) and available on the AODN (https://portal.aodn.org.au/), formatted as four data streams linked through unique EXPOCODES. Supporting data contains a metadata table and BibTeX citation files. The spatial extent of the data records is confined largely to the Southern Ocean and was collected from 1985–2018 (Fig. 4). A summary of the data records in the BIO-MATE aggregate data product is presented in Tables 1–2.
Underway sensor data stream
The underway sensor data stream contains a comma delimited WHP-Exchange file for each voyage ([EXPOCODE]_UWY.csv). The format of this file consists of headers to store metadata, followed by a data table that reports records collected by underway sensors mounted on the vessel (Data Records Table 1).
Profiling sensor data stream
The profiling sensor data stream contains a comma delimited WHP-Exchange file for each unique profiling cast conducted on each voyage ([EXPOCODE][station number][cast number]_ctd1.csv). The file is formatted to store metadata as headers which is followed by the data table that reports records from profiling sensors mounted on a sampling rosette (Data Records Table 2).
Pigment data stream
The pigment data stream contains a comma delimited WHP-Exchange file for each voyage (named [EXPOCODE]PIG[SOURCED_FROM]_[METHOD].csv). The format of this file consists of headers to store supporting information, followed by a data table that records measurements from the laboratory analysis of seawater samples for pigments performed by principal investigators (Data Records Table 3). The laboratory analyses considered are fluorometric determination and high-performance liquid chromatography (HPLC).
Particulate organic carbon data stream
The POC data stream contains a comma delimited WHP-Exchange file for each voyage (named [EXPOCODE]POC[SOURCED_FROM]_[METHOD].csv). The format of this file consists of headers to store supporting information, followed by a data table that records measurements from the lab analysis of seawater samples for particulate organic carbon performed by principal investigators (Data Records Table 4).
Supporting data
Supporting data are included in the BIO-MATE aggregate data product to support the correct citation of data and guide user access to data. This data includes (1) A BibTeX file, that contains information to reference all BIO-MATE data records (2) An index table indicating data availability and citation tags against data records listed by EXPOCODE, data stream, method and source, (3) A records table for all data repositories from which BIO-MATE data was sourced and (4) A records table for all pigment and POC analysis methods used in BIO-MATE data.
Technical Validation
We validated the quality of the BIO-MATE data compilation, by displaying a number of key data distributions and trends. This validation does not confirm the quality of individual data points, in which the authors have placed no additional quality assurance to the published datasets.
The location data associated with the published datasets has been interpreted correctly by the software. This is evident from the success of the biophysical matching algorithm, along with the spatial distribution of the data and recorded sampling depths (Fig. 4). The data are predominantly collected in the month of January between 1991–2010. This is consistent with the fact that ship-based sampling in the Southern Ocean is conducted during Austral summer and displays a lag time in publishing most recent datasets to data repositories. All data are in the ocean, not on land, confirming the absence of spurious location data, and most samples are located in the Southern Ocean which is consistent with our search constraints. Finally information on sampling time of ship-based biological data is as expected, and CTD sampling times (start, bottom and end) are sequential and follow a trend with sampling depth (Fig. 5).
The biological ocean data associated with the published datasets has been interpreted correctly by the software. Overall, fluorometrically derived chlorophyll (FCHLORA), HPLC derived chlorophyll a (Chl a) and HPLC derived total chlorophyll (TCHLA) measurements show a log-normal distribution, as expected. High values (>10 μg/l) are constrained to the coastal zones as expected (Fig. 6).
The (a) distribution of chlorophyll-a derived from high-performance liquid chromatography (Chla), total chlorophyll-a derived from high-performance liquid chromatography (TCHLA) and chlorophyll-a derived from fluorometric determination (FCHLORA) in the BIO-MATE data compilation and (b) the location of high (>10 μg/l) Chla, TCHLA and FCHLA measurements.
There is a linear relationship between chlorophyll-a derived from HPLC methods and chlorophyll derived from fluorometric methods (Fig. 7). Five fluorometric methods to derive chlorophyll have coincident HPLC measurements. Briefly, the ANTXVIII492_and JGOFS491 methods shows good correlation between the fluorometric and HPLC. The PALMER_LTER497 method shows considerable variability, which may be due to the coastal location of most samples and the influence of accessory pigments, but further investigation is needed. Only a small number of coincident HPLC measurements were collected alongside other fluorometric methods (<11), making it difficult to assess their quality.
Our validation plots show that fluorometric determination of chlorophyll tends to overestimate chlorophyll in the Southern Ocean. However, considerable variability is observed as the over-estimation or underestimation of chlorophyll-a by fluorometry is regionally dependant with changing phytoplankton assemblages508,509,510. A recent international intercomparison has also highlighted higher uncertainties, possibly due to low filtration volumes and different extraction and storage methods and suggests new standards for these measurements511. Despite these uncertainties, visual inspection of the profiles show that distributions of chlorophyll with depth are often well captured by fluorometric measurements.
The physical ocean data associated with published datasets has been interpreted correctly by the software. Temperature and salinity ranges fall within expected vales for the ocean and display expected trends with latitude (Fig. 8).
Usage Notes
The community is welcome to contribute to the development of BIO-MATE software and to contribute published data to the aggregation, by following a user guide (Fig. 3).
Contributing to BIO-MATE software development
It is recommended that changes to BIO-MATE software be made through Git Hub. Contributors can fork the existing repository (https://github.com/KimBaldry/BIOMATE-Rpackage) and make changes directly to the source code. Once changes are made, they can be directed back to the BIO-MATE R package repository and released as an updated version of the BIO-MATE software. If the BIO-MATE source code is to be significantly developed, we suggest that the corresponding author is contacted and a hand-over of the software is negotiated. We encourage the addition of new data streams to BIO-MATE, the expansion of BIO-MATE capabilities, the addition of quality assurances and increases in software efficiency.
Contributing data to BIO-MATE
Users can create their own workflows using the BIO-MATE R package to reformat data and information (Fig. 3; https://github.com/KimBaldry/BIO-MATE/blob/main/BIO-MATE.Rmd). Once data have been reformatted, they can be submitted to the corresponding author via Git Hub (https://github.com/KimBaldry/BIOSHARE-submissions) or direct communication. We ask that all data submitted to BIO-MATE are published elsewhere and that users enter an accurate citation for the data they are submitting.
Currently, BIO-MATE only supports data files stored in delimited text formats, with structured headers and columns in a data table, and NetCDF format. The user is required to enter in some metadata to inform the software on input formats (Supplementary Processing Metadata Table).
Recommended use in data analyses
We encourage the use of the data aggregate product as a new integrated database of biological and physical data. Data files from selected voyages can be identified using unique EXPOCODES and CTD_IDs. This makes it easy to use multiple data streams in analysis, by indexing files across these EXPOCODES. Alternatively, the selection tool on the IMAS repository helps users to select voyages using spatial bounds.
Code availability
All data processing was performed in R software (Version 3.6.3). The BIO-MATE R software is freely available (https://github.com/KimBaldry/BIOMATE-Rpackage). The semi-automated workflow and accompanying processing data used to construct the data product, along with the code used to create the data descriptor is freely accessible via Git Hub (https://github.com/KimBaldry/BIO-MATE).
References
Raymond, W. S. The ocean’s role in climate. Oceanography 31, 32–40, https://doi.org/10.5670/oceanog.2018.225 (2018).
Ainley, D. G., Fraser, W. R., Smith, W. O., Hopkins, T. L. & Torres, J. J. The structure of upper level pelagic food webs in the Antarctic: Effect of phytoplankton distribution. J. Mar. Syst. 2, 111–122, https://doi.org/10.1016/0924-7963(91)90017-O (1991).
Basu, S. & Mackey, K. R. M. Phytoplankton as key mediators of the biological carbon pump: Their responses to a changing climate. Sustainability 10, 869, https://doi.org/10.3390/su10030869 (2018).
Carranza, M. M. et al. When mixed layers are not mixed. Storm-driven mixing and bio-optical vertical gradients in mixed layers of the Southern Ocean. J. Geophys. Res. Oceans 123, 7264–7289, https://doi.org/10.1029/2018JC014416 (2018).
Prairie, J. C., Sutherland, K. R., Nickols, K. J. & Kaltenberg, A. M. Biophysical interactions in the plankton: A cross-scale review. Limnol. and Oceanogr. Fluids and Environments 2, 121–145, https://doi.org/10.1215/21573689-1964713 (2012).
Wihsgott, J. U. et al. Observations of vertical mixing in autumn and its effect on the autumn phytoplankton bloom. Prog. Oceanogr. 177, 102059, https://doi.org/10.1016/j.pocean.2019.01.001 (2019).
Brody, S. R. & Lozier, M. S. Characterizing upper-ocean mixing and its effect on the spring phytoplankton bloom with in situ data. ICES J. Mar. Sci. 72, 1961–1970, https://doi.org/10.1093/icesjms/fsv006 (2015).
Mignot, A., D’Ortenzio, F., Taillandier, V., Cossarini, G. & Salon, S. Quantifying observational errors in biogeochemical-argo oxygen, nitrate, and chlorophyll a concentrations. Geophys. Res. Lett. 46, 4330–4337, https://doi.org/10.1029/2018GL080541 (2019).
Baldry, K. BIOMATE data compilation (v1.0). University of Tasmania https://doi.org/10.25959/ETRA-4655 (2023).
Baldry, K., Strutton, P. G., Hill, N. A. & Boyd, P. W. Subsurface chlorophyll maxima reduce the performance of non-photochemical quenching corrections in the Southern Ocean. Front. Mar. Sci. 10, 1302999, https://doi.org/10.3389/fmars.2023.1302999 (2024).
Valente, A. et al. A compilation of global bio-optical in situ data for ocean-colour satellite applications – version two. Earth Syst. Sci. Data 11, 1037–1068, https://doi.org/10.5194/essd-11-1037-2019 (2019).
Johnson, R., Strutton, P. G., Wright, S. W., McMinn, A. & Meiners, K. M. Three improved satellite chlorophyll algorithms for the Southern Ocean. J. of Geophys. Res. Oceans 118, 3694–3703, https://doi.org/10.1002/jgrc.20270 (2013).
Sauzède, R. et al. Vertical distribution of chlorophyll a concentration and phytoplankton community composition from in situ fluorescence profiles: A first database for the global ocean. Earth Syst. Sci. Data 7, 261–273, https://doi.org/10.5194/essd-7-261-2015 (2015).
Roesler, C. et al. Recommendations for obtaining unbiased chlorophyll estimates from in situ chlorophyll fluorometers: A global analysis of WET labs ECO sensors. Limnol. and Oceanogr. Methods 15, 572–585, https://doi.org/10.1002/lom3.10185 (2017).
Verdy, A. & Mazloff, M. R. A data assimilating model for estimating Southern Ocean biogeochemistry. J. of Geophys. Res. Oceans 122, 6968–6988, https://doi.org/10.1002/2016JC012650 (2017).
Rohardt, G. Physical oceanography during Polarstern cruise ANT-III/3. PANGAEA https://doi.org/10.1594/PANGAEA.734977 (2010).
Nöthig, E.-M. & Cornils, A. Nutrients, biochemical parameters and primary production during Polarstern cruise ANT-III/3 (PS06) - Part 2. PANGAEA https://doi.org/10.1594/PANGAEA.897335 (2019).
Hempel, G. Die expedition ANTARKTIS-III mit FS polarstern 1984/85. Reports on Polar Research 25, 1–209, https://doi.org/10.2312/BzP_0025_1985 (1985).
Gordon, A. L. Physical oceanography during Polarstern cruise ANT-V/2. PANGAEA https://doi.org/10.1594/PANGAEA.734981 (2010).
Schnack-Schiel, S. B. & Dieckmann, G. S. Major pigment concentrations measured on water bottle samples during Polarstern cruise ANT-V/2. PANGAEA https://doi.org/10.1594/PANGAEA.217354 (2004).
Schnack-Schiel, S. B. The Winter-Expedition of RV Polarstern to the Antarctic ANT V/1-3. Reports on Polar Research 32, 1–259 (1987).
Hempel, I. & Veth, C. Physical oceanography during Polarstern cruise ANT-VII/3. PANGAEA https://doi.org/10.1594/PANGAEA.92067 (2003).
Hempel, I., Veth, C., Smetacek, V., van Beusekom, J. & Jacques, G. Biogeochemical and oceanographic data measured on water bottle samples during Polarstern cruise ANT-VII/3. PANGAEA https://doi.org/10.1594/PANGAEA.92066 (2003).
Smetacek, V., Hempel, I. & Schalk, P. H. The expedition ANTARKTIS-VII/3 EPOSLEG2 of RV polarstern in 1988/89. Reports on Polar Research 65, 1–199 (1989).
Smetacek, V., de Baar, H. J. W., Bathmann, U., Lochte, K. & Rutgers van der Loeff, M. M. Physical oceanography during Polarstern cruise ANT-X/6. PANGAEA https://doi.org/10.1594/PANGAEA.759261 (1997).
Smetacek, V., de Baar, H. J. W., Bathmann, U., Lochte, K. & Rutgers van der Loeff, M. M. Pigments measured on water bottle samples during Polarstern cruise ANT-X/6. PANGAEA https://doi.org/10.1594/PANGAEA.761047 (1997).
Smetacek, V., de Baar, H. J. W., Bathmann, U., Lochte, K. & Rutgers van der Loeff, M. M. Nutrients and pigments measured on water bottle samples during Polarstern cruise ANT-X/6. PANGAEA https://doi.org/10.1594/PANGAEA.761042 (1997).
Bathmann, U., Smetacek, V., Baar, H. J., de, Fahrbach, E. & Krause, G. Die expeditionen ANTARKTIS x/6-8 des forschungsschiffes polarstern 1992/93. Reports on Polar Research 135, 1–235, https://doi.org/10.2312/BzP_0135_1994 (1994).
Strass, V. H. Physical oceanography during Polarstern cruise ANT-XVIII/2. PANGAEA https://doi.org/10.1594/PANGAEA.742583 (2010).
Riebesell, U. Particulate organic carbon and nitrogen concentrations measured on water bottles samples during Polarstern cruise ANT-XVIII/2. PANGAEA https://doi.org/10.1594/PANGAEA.727657 (2009).
Bathmann, U., El Naggar, S. E. D. & Smetacek, V. The expeditions ANTARKTIS XVIII/1-2 of the research vessel Polarstern in 2000. Reports on Polar Research 400, 1–232, https://doi.org/10.2312/BzPM_0400_2001 (2001).
Smetacek, V. & Riebesell, U. Pigments measured on water bottle samples during Polarstern cruise ANT-XVIII/2. PANGAEA https://doi.org/10.1594/PANGAEA.126394 (2003).
Peeken, I. & Nachtigall, K. Phytoplankton pigments measured on water bottle samples during Polarstern cruise ANT-XVIII/2. PANGAEA https://doi.org/10.1594/PANGAEA.869823 (2014).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect1. PANGAEA https://doi.org/10.1594/PANGAEA.126293 (2003).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect2. PANGAEA https://doi.org/10.1594/PANGAEA.126294 (2003).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect3. PANGAEA https://doi.org/10.1594/PANGAEA.126295 (2003).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect4. PANGAEA https://doi.org/10.1594/PANGAEA.126296 (2003).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect5. PANGAEA https://doi.org/10.1594/PANGAEA.126297 (2003).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect6. PANGAEA https://doi.org/10.1594/PANGAEA.126298 (2003).
Smetacek, V. & Riebesell, U. Chlorophyll and pheopigments in surface water along transect PS58_transect7. PANGAEA https://doi.org/10.1594/PANGAEA.126299 (2003).
Strass, V. H. Physical oceanography during Polarstern cruise ANT-XXI/3 (EIFEX). PANGAEA https://doi.org/10.1594/PANGAEA.733410 (2010).
Smetacek, V. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXI/3 (EIFEX). PANGAEA https://doi.org/10.1594/PANGAEA.666229 (2007).
Bathmann, U., Smetacek, V. & Helmke, E. The expeditions ANTARKTIS XXI/3-4-5 of the research vessel polarstern in 2004. Reports on Polar Research 500, 1–302, https://doi.org/10.2312/BzPM_0500_2005 (2005).
Peeken, I. & Hoffmann, L. Phytoplankton pigments measured on water bottle samples during Polarstern cruise ANT-XXI/3 (EIFEX). PANGAEA https://doi.org/10.1594/PANGAEA.869824 (2014).
Peeken, I. & Hoffmann, L. Phytoplankton pigments in surface water during Polarstern cruise ANT-XXI/3 (EIFEX). PANGAEA https://doi.org/10.1594/PANGAEA.869825 (2014).
Strass, V. H. Physical oceanography during Polarstern cruise ANT-XXIV/2 (SCACE). PANGAEA https://doi.org/10.1594/PANGAEA.733665 (2010).
Bathmann, U. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXIV/2. PANGAEA https://doi.org/10.1594/PANGAEA.727464 (2009).
Bathmann, U. The expedition of the research vessel Polarstern to the Antarctic in 2007/2008 ANT-XXIV/2. Reports on Polar Research 604, 1–205, https://doi.org/10.2312/BzPM_0604_2010 (2010).
Peeken, I., Bracher, A. & Murawski, S. Phytoplankton pigments measured on water bottle samples during Polarstern cruise ANT-XXIV/2. PANGAEA https://doi.org/10.1594/PANGAEA.873199 (2017).
Schmitt, B. & Rohardt, G. Physical oceanography during Polarstern cruise ANT-XXIV/4. PANGAEA https://doi.org/10.1594/PANGAEA.742855 (2010).
Macke, A. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXIV/4. PANGAEA https://doi.org/10.1594/PANGAEA.727466 (2009).
Macke, A. The expedition of the research vessel Polarstern to the Antarctic in 2008 ANT XXIV/4. Reports on Polar Research 591, 1–64, https://doi.org/10.2312/BzPM_0591_2009 (2009).
Bracher, A. Phytoplankton pigment concentrations during Polarstern cruise ANT-XXIV/4. PANGAEA https://doi.org/10.1594/PANGAEA.848584 (2015).
Schröder, M. & Wisotzki, A. Physical oceanography during Polarstern cruise ANT-XXVI/3. PANGAEA https://doi.org/10.1594/PANGAEA.754248 (2010).
Gohl, K. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXVI/3. PANGAEA https://doi.org/10.1594/PANGAEA.742729 (2010).
Gohl, K. The expedition of the research vessel Polarstern to the Amundsen sea, antarctica, in 2010 ANT XXVI/3. Reports on Polar Research 617, 1–173, https://doi.org/10.2312/BzPM_0617_2010 (2010).
Peeken, I. & Nachtigall, K. Phytoplankton pigments measured on water bottle samples during Polarstern cruise ANT-XXVI/3. PANGAEA https://doi.org/10.1594/PANGAEA.869826 (2014).
Peeken, I. & Nachtigall, K. Pigments in surface water during Polarstern cruise ANT-XXVI/3. PANGAEA https://doi.org/10.1594/PANGAEA.869827 (2014).
Rohardt, G. & Bracher, A. Physical oceanography during Polarstern cruise ANT-XXVI/4. PANGAEA https://doi.org/10.1594/PANGAEA.758127 (2011).
Körtzinger, A. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXVI/4. PANGAEA https://doi.org/10.1594/PANGAEA.753224 (2010).
Körtzinger, A. The expedition of the research vessel Polarstern to the Antarctic in 2010 ANT XXVI/4. Reports on Polar Research 631, 1–73, https://doi.org/10.2312/BzPM_0631_2011 (2011).
Bracher, A. Phytoplankton pigment concentrations during Polarstern cruise ANT-XXVI/4. PANGAEA https://doi.org/10.1594/PANGAEA.848585 (2015).
Strass, V. H. et al. Physical oceanography during Polarstern cruise ANT-XXVIII/3. PANGAEA https://doi.org/10.1594/PANGAEA.840334 (2016).
Wolf-Gladrow, D. A. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXVIII/3. PANGAEA https://doi.org/10.1594/PANGAEA.780004 (2012).
Wolf-Gladrow, D. The expedition of the research vessel Polarstern to the Antarctic in 2012 ANT XXVIII/3. Reports on Polar Research 661, 1–191, https://doi.org/10.2312/BzPM_0661_2013 (2013).
Bracher, A. Phytoplankton pigment concentrations during Polarstern cruise ANT-XXVIII/3. PANGAEA https://doi.org/10.1594/PANGAEA.848588 (2014).
Roca-Martı́, M. et al. Particulate Thorium-234 (Th-234), organic carbon (POC) and organic nitrogen (PON) concentrations using in-situ pumps (particles >53 μm) during Polarstern cruise ANT-XXVIII/3. PANGAEA https://doi.org/10.1594/PANGAEA.848818 (2015).
Meyer, B. & Rohardt, G. Physical oceanography during Polarstern cruise ANT-XXIX/7. PANGAEA https://doi.org/10.1594/PANGAEA.836537 (2014).
Meyer, B. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track ANT-XXIX/7. PANGAEA https://doi.org/10.1594/PANGAEA.823259 (2013).
Meyer, B. & Auerswald, L. The expedition of the research vessel Polarstern to the Antarctic in 2013 ANT XXIX/7. Reports on Polar Research 674, 1–130, https://doi.org/10.2312/BzPM_0674_2014 (2014).
Meyer, B. et al. Chorophyll a and particulate organic carbon measured on water bottle samples during Polarstern cruise ANT-XXIX/7. PANGAEA https://doi.org/10.1594/PANGAEA.864704 (2017).
Rohardt, G. & Boebel, O. Physical oceanography during Polarstern cruise PS103 (ANT-XXXII/2). PANGAEA https://doi.org/10.1594/PANGAEA.881076 (2017).
Boebel, O. The expedition PS103 of the research vessel Polarstern to the Weddell sea in 2016/2017. Reports on Polar Research 710, 1–160, https://doi.org/10.2312/BzPM_0710_2017 (2017).
Boebel, O. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track PS103 (ANT-XXXII/2). PANGAEA https://doi.org/10.1594/PANGAEA.889513 (2018).
Bracher, A. Phytoplankton pigment concentrations in the Southern Ocean during RV Polarstern cruise PS103 in Dec 2016 to Jan 2017. PANGAEA https://doi.org/10.1594/PANGAEA.898941 (2019).
Rohardt, G. & Boebel, O. Physical oceanography during Polarstern cruise PS117. PANGAEA https://doi.org/10.1594/PANGAEA.910663 (2020).
Boebel, O. & Rohardt, G. Continuous thermosalinograph oceanography along Polarstern cruise track PS117. PANGAEA https://doi.org/10.1594/PANGAEA.905562 (2019).
Boebel, O. The expedition PS117 of the research vessel Polarstern to the Weddell sea in 2018/2019. Reports on Polar Research 732, 1–205 (2019).
Trull, T. RV Investigator voyage summary IN2015_V01. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=309&start=61#IN2015_V01 (2015).
CSIRO. RV Investigator voyage IN2015_V01 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/1dcbfe47-d8e3-4c36-e053-08114f8cc252 (2015).
CSIRO. RV Investigator voyage IN2015_V01 underway UWY data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/1160f9f9-c2b1-027f-e053-08114f8c709e (2015).
Coffin, M. RV Investigator voyage summary IN2016_V01. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=309&start=51#IN2016_V01 (2016).
CSIRO. RV Investigator voyage IN2016_V01 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/6ba5a0ce-b87a-4bda-b16c-3527279c3bca (2016).
CSIRO. RV Investigator voyage IN2016_V01 underway UWY data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/291d1744-409c-2a7e-e053-08114f8ca4d4 (2016).
Clementson, L. RV Investigator IN2016_V01, pigment and ocean colour data. CSIRO Marine National Facility https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Trull, T. RV Investigator voyage summary IN2016_V02. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=309&start=51#IN2016_V02 (2016).
CSIRO. RV Investigator voyage IN2016_V02 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/d57331f4-3213-4817-b47e-a2ad0292148a (2016).
CSIRO. RV Investigator voyage IN2016_V02 underway UWY data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/2cf37561-1c43-0d22-e053-08114f8c7cd2 (2016).
Clementson, L. RV Investigator IN2016_V02, pigment and ocean colour data. CSIRO Marine National Facility https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Sloyan, B. RV Investigator voyage summary IN2016_V03. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=309&start=51#IN2016_V02 (2016).
CSIRO. RV Investigator voyage IN2016_V03 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/d57331f4-3213-4817-b47e-a2ad0292148a (2016).
CSIRO. RV Investigator voyage IN2016_V03 underway UWY data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/2cf37561-1c43-0d22-e053-08114f8c7cd2 (2016).
Sloyan, B. M. et al. Dissolved inorganic carbon (DIC), total alkalinity, pH on total scale, and other hydrographic and chemical data obtained during the SOCCOM float deployment expedition onboard r/v investigator cruise IN2016_V03 along the GO-SHIP section P15S (EXPOCODE 096U20160426) from 2016-04-26 to 2016-06-30 (NCEI accession 0168141). National Oceanic and Atmospheric Administration, Department of Commerce https://doi.org/v10.7289/v5p26wdd (2017).
Rintoul, S. RV Investigator voyage summary IN2018_V01. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=309&start=51#IN2016_V02 (2018).
CSIRO. RV Investigator voyage IN2018_V01 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/d57331f4-3213-4817-b47e-a2ad0292148a (2018).
CSIRO. RV Investigator voyage IN2018_V01 underway UWY data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/2cf37561-1c43-0d22-e053-08114f8c7cd2 (2018).
Clementson, L. RV Investigator IN2016_V03, pigment and ocean colour data. CSIRO Marine National Facility https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
Rosenberg, M. Aurora Australis Southern Ocean oceanographic (CTD) data, cruise au9006 (AAMBER2). Australian Antarctic Data Centre https://doi.org/10.4225/15/58afb50245d5c (2002).
Wright, S. Chlorophyll a data collected on voyage 3 of the Aurora Australis in the 2002-2002 season - CLIVAR (Climate Variability) voyage. Australian Antarctic Data Centre https://doi.org/10.4225/15/5212D592E3ED9 (2013).
Reeve, J. Aurora Australis voyage 6 (AAMBER2) 1990-91 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199091060 (1999).
Vaudrey, D. J. CTD processing notes for AU9101. Australian Antarctic Data Centre https://doi.org/10.4225/15/5497925FD0BC0 (2001).
Rosenberg, M. & Ronai, B. Database of CTD data collected in the Southern Ocean by the AAD, ACE CRC and part of the Southern Ocean Atlas data set., Ver. 1. Australian Antarctic Data Centre https://doi.org/10.4225/15/5497925FD0BC0 (2001).
Wright, S. Chlorophyll a data collected on voyage 3 of the Aurora Australis in the 2002-2002 season - CLIVAR (Climate Variability) voyage. Australian Antarctic Data Centre https://doi.org/10.4225/15/520B0A83F2C4A (2013).
Reeve, J. Aurora Australis voyage 1 and 1.1 (WOCE91) 1991-92 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199192010 (1999).
Wright, S. Chlorophyll a data collected on voyage 4 of the Aurora Australis in the 1991-1992 season. Australian Antarctic Data Centre https://doi.org/10.4225/15/520B19F725597 (2013).
Reeve, J. Aurora Australis voyage 4 1991-92 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199192040 (1999).
Hosie, G. & Watts, D. Aurora Australis Southern Ocean oceanographic (CTD) data, cruise 1992/93 V7 (KROCK). Australian Antarctic Data Centre https://doi.org/10.26179/5bd7d5ef98cae (2018).
Wright, S. Aurora Australis voyage 7 (KROCK) 1992-93 chlorophyll a data. Australian Antarctic Data Centre https://doi.org/10.4225/15/520C5139985CB (2013).
Reeve, J. Aurora Australis voyage 7 (KROCK) 1992-93 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199293070 (1999).
Mark Rosenberg, R. E. & Rintoul, S. Aurora Australis marine science cruise AU9309/AU9391 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/58b397ccc65ad (2001).
Rosenberg, M. Aurora Australis WOCE Southern Ocean oceanographic data, cruise au9309/9391. Australian Antarctic Data Centre https://doi.org/10.4225/15/58b397ccc65ad (2001).
Wright, S. Aurora Australis voyage 9 (WOES) 1992-93 chlorophyll a data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199293090 (2013).
Reeve, J. Aurora Australis voyages 9 and 9.1 (WOES and WORSE) 1992-93 underway data. Australian Antarctic Data Centre, https://data.aad.gov.au/metadata/records/199293090 (1999).
Wright, S. The role of antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/ASAC_40 (2013).
Reeve, J. Aurora Australis voyage 4 1993-94 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199394040 (1999).
Rosenberg, M. Aurora Australis Marine Science Cruise AU9407 - Oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/5493AEC974038 (2001).
Rosenberg, M. Aurora Australis WOCE Southern Ocean oceanographic data, cruise au9407. Australian Antarctic Data Centre https://doi.org/10.4225/15/5493AEC974038 (2001).
Wright, S. Role of Antarctic marine protists in trophodynamics and global change and impact of UV-B on these organisms - Voyage 7 of the Aurora Australis, 1993-1994 - SHAM. Australian Antarctic Data Centre https://doi.org/10.4225/15/580eafa998da7 (2004).
Reeve, J. Aurora Australis voyage 7 (SHAM) 1993-94 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199394070 (1999).
Rosenberg, M., Eriksen, R., Bell, S. & Rintoul, S. Aurora Australis marine science cruise AU9404 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/58b3a6b793939 (2001).
Rosenberg, M. Aurora Australis WOCE Southern Ocean oceanographic data, cruise au9404. Australian Antarctic Data Centre https://doi.org/10.4225/15/58b3a6b793939 (2001).
Reeve, J. Aurora Australis voyage 4 (WOCET) 1994-95 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199495040 (1999).
Rosenberg, M. et al. Aurora Australis marine science cruises AU9501, AU9604 and AU9601 - oceanographic field measurements and analysis, inter-cruise comparisons and data quality notes. Australian Antarctic Data Centre https://doi.org/10.4225/15/5493B203ADEAE (2001).
Rosenberg, M. Aurora Australis WOCE Southern Ocean oceanographic data, cruise au9501. Australian Antarctic Data Centre https://doi.org/10.4225/15/5493B203ADEAE (2001).
Wright, S. Role of Antarctic marine protists in trophodynamics and global change and impact of UV-B on these organisms - V1 of the Aurora Australis. Australian Antarctic Data Centre https://doi.org/10.4225/15/580d988844bb9 (2004).
Reeve, J. Aurora Australis voyage 1 (ABSTAIN) 1995-96 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199596010 (1999).
Rosenberg, M. et al. Aurora Australis marine science cruises AU9501, AU9604 and AU9601 - oceanographic field measurements and analysis, inter-cruise comparisons and data quality notes. Australian Antarctic Data Centre https://doi.org/10.4225/15/54978B0D737F5 (2008).
Steve Rintoul, M. R. & Bindoff, N. Aurora Australis Southern Ocean oceanographic data, cruise au9604 - BROKE. Australian Antarctic Data Centre https://doi.org/10.4225/15/54978B0D737F5 (2008).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - BROKE Samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/580d8b6967f23 (2010).
Reeve, J. Aurora Australis voyage 4 (BROKE) 1995-96 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199596040/ (1999).
Rintoul, S., Rosenberg, M. & Trull, T. Aurora Australis SAZ Southern Ocean oceanographic data, cruise au97_9806. Australian Antarctic Data Centre https://doi.org/10.4225/15/58b4f13eeefef (2001).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - voyage 6 1997/1998 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/580058219609f (2011).
Reeve, J. Aurora Australis voyage 6 (SNARK) 1997-98 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199798060 (1999).
Rosenberg, M. et al. Mertz Polynya Experiment, marine science cruises AU9807, AU9801, AU9905, AU9901 and TA0051 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.26179/5c9c01b329429 (2004).
Rosenberg, M., Bindoff, N. & Williams, G. Mertz Polynya Experiment, Aurora Australis science cruises au9807 and au9901, and Tangaroa science cruise ta0051 - ship-based CTD, ADCP, LADCP and mooring data. Australian Antarctic Data Centre https://doi.org/10.26179/5c9c01b329429 (2004).
Wright, S. & van den Enden, D. Biology of the Mertz Glacier Polynya - protist data. Australian Antarctic Data Centre https://doi.org/10.4225/15/54979A67E82D9 (2012).
Reeve, J. Aurora Australis voyage 1 (IDIOTS) 1999-00 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/199900010 (1999).
Rosenberg, M. Aurora Australis Southern Ocean oceanographic data, voyage 6, 2000-2001 - KACTAS. Australian Antarctic Data Centre https://doi.org/10.26179/5e7411e49faf2 (2020).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - KACTAS samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5212CDBC6EC44 (2013).
Reeve, J. Aurora Australis Voyage 6 2000-2001 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200001060 (2001).
Rosenberg, M., Rintoul, S., Bray, S., Moy, C. & Johnston, N. Aurora Australis marine science cruise AU0103, CLIVAR-SR3 transect: oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/58a5360c6c0db (2004).
Rintoul, S. & Rosenberg, M. Aurora Australis marine science cruise au0103 (CLIVAR_SR3) - CTD and ADCP data. Australian Antarctic Data Centre https://doi.org/10.4225/15/58a5360c6c0db (2004).
Reeve, J. Aurora Australis voyage 3 2001-2002 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200102030 (2001).
Rosenberg, M. Aurora australis Southern Ocean oceanographic data, voyage 7, 2001-2002 - LOSS. Australian Antarctic Data Centre https://doi.org/10.26179/5e8d6b44ae94b (2020).
Rosenberg, M. Kerguelen Deep Western Boundary Current Experiment and CLIVAR I9 Transect, marine science cruises AU0304 and AU0403 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/58ad0bd50bd58 (2004).
Rintoul, S., Rosenberg, M., Bindoff, N., Church, J. & Yasushi, F. Kerguelen Plateau Deep Western Boundary Current Experiment (DWBC), Aurora Australis marine science cruise au0304 - ship-based CTD, ADCP and LADCP data. Australian Antarctic Data Centre https://doi.org/10.4225/15/58ad0bd50bd58 (2004).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Voyage 4 2002/2003 samples (KAOS). Australian Antarctic Data Centre https://doi.org/10.4225/15/521412D6095CA (2013).
Reeve, J. Aurora Australis voyage 4 2002-2003 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200203040 (2003).
Rosenberg, M. CTD Data report for Aurora Australis marine science cruise AU0404, V4 2003/2004 (“HIPPIES” Heard Is. Fishing Voyage). Australian Antarctic Data Centre https://doi.org/10.4225/15/58afb50245d5c (2004).
Rosenberg, M. & Ronai, B. Database of CTD data collected in the Southern Ocean by the AAD, ACE CRC and part of the Southern Ocean atlas data set., Ver. 1. Australian Antarctic Data Centre https://doi.org/10.4225/15/58afb50245d5c (2005).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - HIPPIES samples, Ver. 1. Australian Antarctic Data Centre https://doi.org/10.4225/15/52845DCD3AD19 (2013).
Reeve, J. Aurora Australis voyage 4 2003-2004 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200304040 (2004).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Voyage 1 2004/2005 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/52855D1CCA2B8 (2013).
Wiley, P. Aurora Australis voyage 1 2004-2005 underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200405010 (2005).
Rosenberg, M., Rintoul, S., Fukamachi, M. & Church, J. CLIVAR I9 transect and Kerguelen Plateau deep western boundary current experiment, aurora australis marine science cruise au0403 - ship-based CTD and ADCP data. Australian Antarctic Data Centre https://doi.org/10.4225/15/58ad124987ff6 (2006).
Rosenberg, M. BROKE West Survey, marine science cruise AU0603 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.26179/5ceb6d79c35a4 (2019).
Rosenberg, M. & Gorton, R. BROKE West Survey, marine science cruise AU0603 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.26179/5ceb6d79c35a4 (2019).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Voyage 3, BROKE-West, Aurora Australis 2005/2006 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/528C426A13819 (2010).
Wiley, P. Aurora Australis voyage 3 2005-2006 (BROKE-West) underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200506030 (2006).
Rosenberg, M. SAZ-SENSE, Marine science cruise AU0703 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/57B13F22B2F87 (2010).
Mark Rosenberg, W. R. H. & Griffiths, B. CTD data collected during the SAZ-SENSE voyage, January-February 2007. Australian Antarctic Data Centre https://doi.org/10.4225/15/57B13F22B2F87 (2010).
Howard, W., Wright, S. & Griffiths, B. Samples collected for HPLC pigments, microscopy and flow cytometry for the SAZ-SENSE project. Australian Antarctic Data Centre https://doi.org/10.4225/15/556E63D646382 (2014).
Jono Reeve, W. R. H. & Griffiths, B. Aurora Australis voyage V3 2006/07 (SAZ-SENSE) track and underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/200607030 (2009).
Rosenberg, M. & Rintoul, S. Aurora Australis marine science cruises AU0803 and AU0806 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/58ad18180df56 (2019).
Rosenberg, M. & Rintoul, S. Southern Ocean CTD data collected from the CASO cruise of the Aurora Australis. Australian Antarctic Data Centre https://doi.org/10.4225/15/58ad18180df56 (2019).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Voyage 6, Aurora Australis 2007/2008 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5292DD6705559 (2013).
Rintoul, S. R., van Wijk, E. M. Aurora australis southern ocean oceanographic data, cruise au1121 2010/11 VMS. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/au1121 (2014).
Steve Rintoul, M. R. Aurora australis marine science cruise AU1203 (V3 2011/2012), CLIVAR I9S transect and Adelie Land - ship-based CTD data. Australian Antarctic Data Centre https://doi.org/10.4225/15/527B046ACEEC4 (2013).
Rosenberg, M. & Eriksen, R. Aurora Australis marine science cruise AU1603 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/5adec86550a1a (2019).
Bestley, S. & Rosenberg, M. Aurora Australis marine science cruise AU1603 - oceanographic field measurements and analysis. Australian Antarctic Data Centre https://doi.org/10.4225/15/5adec86550a1a (2019).
Symmons, L. Aurora Australis voyage 3 2015/16 track and underway data. Australian Antarctic Data Centre https://data.aad.gov.au/metadata/records/201516030 (2016).
CSIRO. Franklin voyage FR 10/97 CTD data. CSIRO Marine National Facility https://www.marine.csiro.au/data/trawler/dataset.cfm?survey=FR199710&data_type=ctd (1999).
Clementson, L. Franklin cruise 10/97 HPLC pigment and ocean colour data. CSIRO Marine National Facility https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
Parslow, J. Cruise report SS 01/95: January 14 - February 2, 1995. CSIRO Marine National Facility https://doi.org/10.4225/08/586fdceb6d29a (1995).
CSIRO. Southern Surveyor voyage SS 01/95 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search;jsessionid=fwib9h30l5g11n86sk9ouv655#/metadata/7780aa58-6434-453b-9bf6-b8ba0bb93533 (2014).
Clementson, L. Southern Surveyor SS01/95 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
Tillbrook, B. SS 11/95 Survey details and related metadata, reports, events and data. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=8&start=161#SS199511 (1995).
CSIRO. Southern Surveyor voyage SS 11/95 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/005cb0d3-c96e-4124-8852-19fa741b633a (1998).
Clementson, L. Southern Surveyor voyage SS 11/95 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 11/95 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/8121b9d8-cefb-4fec-a6a3-1659bfab00dc (2001).
Williams, A. & Bax, N. SS 02/96 Survey details and related metadata, reports, events and data. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_list.cfm?q=&source_id=8&start=161#SS199511 (1996).
CSIRO. Southern Surveyor voyage SS 02/96 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/3463b550-9abc-4816-8d39-9dfd13a700ed (2001).
Clementson, L. Southern Surveyor voyage SS 02/96 HPLC pigment and ocean colour data. CSIRO Marine National Facility https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 02/96 underway Data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/cfc98f1a-498e-43d1-a732-f4c6b1a8eb38 (2001).
Young, J. SS 03/96 survey details and related metadata, reports, events and data. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS199603 (1996).
CSIRO. Southern Surveyor voyage SS 03/96 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/900e2c7e-b0d2-4740-a458-8503a5340de7 (1998).
Clementson, L. Southern Surveyor voyage SS 03/96 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
Williams, A. & Liron, C. Cruise report SS 06/96 November 20 - December 19, 1996. https://doi.org/10.4225/08/586fdc688f7a9 (1996).
CSIRO. Southern Surveyor voyage SS 06/96 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/abc685f6-528a-4c18-b2b0-38f197e010c5 (1996).
CSIRO. Southern Surveyor voyage SS 06/96 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/4578cb46-47e4-4d2e-abd0-0fe631e96f2b (1996).
Clementson, L. Southern Surveyor voyage SS 06/96 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
Koslow, T. & Kloser, R. Cruise report SS 01/99: January 10 - February 4, 1999. https://doi.org/10.4225/08/58712d755cf79 (1996).
Clementson, L. Southern Surveyor voyage SS 01/96 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 02/99 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/c7052a7f-705d-4fb5-95b1-142888b30430 (2004).
Clementson, L. Southern Surveyor voyage SS 02/99 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 02/99 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/a69eeaef-844c-45d4-a3ee-09a7d771665d (2004).
Koslow, T. Voyage summary SS07/2003. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS200307 (2003).
CSIRO. Southern Surveyor voyage SS 07/2003 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/97576cdc-fe47-4f0a-81b2-8595b73006e9 (2007).
Clementson, L. SRFME ocean colour data - offShore, Southern Surveyor SS 07/2003. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
Koslow, T. Voyage summary SS01/2004. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS200401 (2004).
CSIRO. Southern Surveyor voyage SS 01/2004 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/dc42f374-b762-4bea-956a-f44a6cda956e (2000).
Clementson, L. SRFME ocean colour data - offShore, Southern Surveyor SS 01/2004. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 01/2004 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/f82466ce-fbd7-4986-8008-a828c8747189 (2011).
Kloser, R. Voyage summary SS 07/2004. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS200407 (2004).
CSIRO. Southern Surveyor voyage SS 07/2004 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/5811196d-970a-4415-9df6-db9e75ecc58e (2007).
Clementson, L. Southern Surveyor voyage SS 07/2004 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 07/2004 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/7c6f72fb-88e7-4cc5-a83b-e8ae9b98ea00 (2007).
Alan Williams, R. K. & Bax, N. Voyage summary SS07/2005. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS200401 (2004).
CSIRO. Southern Surveyor voyage SS 07/2005 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/d93be039-6534-4f53-a7c9-b0c9cc991699 (2008).
Clementson, L. Southern Surveyor voyage SS 07/2005 HPLC pigment and ocean colour Data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 07/2005 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/8ead389a-489f-4ad1-ad08-9a4675b3712b (2008).
Williams, A. & Kloser, R. Voyage summary SS07/2005. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS200510 (2004).
CSIRO. Southern Surveyor voyage SS 10/2005 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/0c369d29-6063-4cce-9d58-61ec8c6b4f0f (2006).
Clementson, L. Southern Surveyor voyage SS 10/2005, pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 10/2005 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/52a34094-58c0-4dd0-987e-b821739db500 (2007).
Trull, T. SS 03/2006. Voyage details and related metadata, reports, events and data. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS200603 (2006).
Brodie, P. Southern Surveyor voyage SS 03/2006 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/e09df8fb-ec43-4495-a64e-9bd5bd3a0f4d (2011).
Clementson, L. Southern Surveyor voyage SS 03/2006, pigment and ocean colour data. https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS 03/2006 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/46c96e26-0064-406c-a1ec-b3bec4103940 (2011).
Williams, A. & Kloser, R. Voyage summary SS2010_v06. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS2010_V06 (2010).
CSIRO. Southern Surveyor voyage SS2010_V06 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/bc1b3741-e742-5039-e044-00144f7bc0f4 (2012).
Clementson, L. Southern Surveyor voyage SS2010_v06 HPLC pigment data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS2010_V06 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/bc1b3741-e7ef-5039-e044-00144f7bc0f4 (2011).
Doblin, M. Voyage summary SS2010_V09. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS2010_V09 (2010).
CSIRO. Southern Surveyor voyage SS2010_V09 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/bc1b3741-e7d9-5039-e044-00144f7bc0f4 (2012).
Clementson, L. Southern Surveyor voyage SS2010_V09 HPLC pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS2010_V09 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/bc1b3741-e78a-5039-e044-00144f7bc0f4 (2010).
Phillips, H. VOYAGE SUMMARY SS2012_V04. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS2012_V04 (2010).
CSIRO. Southern Surveyor voyage SS2012_V04 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/ccb13735-44d1-3ec9-e043-08114f8c29d9 (2012).
Clementson, L. Southern Surveyor voyage SS2012_V04 pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS2012_V04 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/dc065655-d9f8-4782-e043-08114f8c8963 (2012).
Wilcox, C. Voyage summary SS2012_T06. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS2012_T06 (2012).
CSIRO. Southern Surveyor voyage SS2012_T06 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/d864efad-75a4-0745-e043-08114f8ca335 (2013).
Clementson, L. Southern Surveyor voyage SS2012_T06 pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS2012_T06 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/dc075527-6b13-26ae-e043-08114f8c5f4c (2012).
Trull, T. SS2013_V04. Voyage details and related metadata, reports, events and data. CSIRO Marine National Facility https://www.cmar.csiro.au/data/trawler/survey_details.cfm?survey=SS2013_V04 (2013).
Kippo, H. Southern Surveyor voyage SS2013_V04 CTD data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/ed3ab483-5373-2bf5-e043-08114f8cf4f8 (2013).
Clementson, L. Southern Surveyor voyage SS2013_v04, pigment and ocean colour data. Australian Ocean Data Network https://portal.aodn.org.au/search?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2020).
CSIRO. Southern Surveyor voyage SS2013_V04 underway data. CSIRO Marine National Facility https://marlin.csiro.au/geonetwork/srv/eng/catalog.search#/metadata/f4fed110-7324-268e-e043-08114f8c160 (2011).
Garcı́a, M. A. et al. Physical oceanography during Hespérides cruise Fruela95. PANGAEA https://doi.org/10.1594/PANGAEA.825643 (2002).
Ricardo Anadón, M. E. pH, alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations using CTD, bottle and other instruments from the HESPERIDES in the south Atlantic Ocean from 1995-12-03 to 1996-01-05 (NCEI accession 0113549). NCEI https://doi.org/10.3334/cdiac/otg.carina_29he19951203 (2013).
Garcı́a, M. A. et al. Physical oceanography during Hespérides cruise Fruela96. PANGAEA https://doi.org/10.1594/PANGAEA.825644 (2002).
Ricardo Anadón, M. E. pH, alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations using CTD, bottle and other instruments from the HESPERIDES in the south Atlantic Ocean from 1996-01-17 to 1996-02-05 (NCEI accession 0113755). https://doi.org/110.3334/CDIAC/otg.CARINA_29HE19960117 (2013).
Johnson, G. C. Documentation from cruise 31DSCG94_1. CCHDO https://cchdo.ucsd.edu/cruise/31DSCG94_1 (2015).
Johnson, G. C. CTD data from cruise 31DSCG94_1. CCHDO https://cchdo.ucsd.edu/cruise/31DSCG94_1 (2015).
Feely, R. A., M. R., Bullister, J. L. Dissolved inorganic carbon, pH, alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations using CTD, bottle and other instruments from NOAA ship Discoverer in the Gulf of Alaska and north Pacific Ocean from 1991-03-07 to 1991-04-07 (NCEI accession 0115175). NCEI https://doi.org/10.3334/cdiac/otg.31dicgc91_2 (2013).
Smith, W. Directory of data in /jgofs/southern/nbp96_4A/. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp96_4A/ (2005).
Morrison, J. Final version CTD, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp96_4A/ (2005).
Anderson, B. shipboard underway data. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp96_4A/ (2005).
Smith, W. O. Fluorometric chlorophyll-a & phaeopigments. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp96_4A/ (2005).
Bidigare, R. R. Pigments, HPLC method, sampled from bottle and TM casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp96_4A/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp96_4A/ (2005).
Marra, J. Directory of data in /jgofs/southern/nbp97_1/. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_1/ (2005).
Morrison, J. Final version CTD, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_1/ (2005).
Anderson, B. Shipboard underway data. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_1/ (2005).
Smith, W. O. Fluorometric chlorophyll-a & phaeopigments. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_1/ (2005).
Bidigare, R. R. Pigments, HPLC method, sampled from bottle and TM casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_1/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_1/ (2005).
Ducklow, H. Directory of data in /jgofs/southern/nbp97_3/. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_3/ (2005).
Morrison, J. Final version CTD, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_3/ (2005).
Anderson, B. Shipboard underway data. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_3/ (2005).
Smith, W. O. Fluorometric chlorophyll-a & phaeopigments. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_3/ (2005).
Bidigare, R. R. Pigments, HPLC method, sampled from bottle and TM casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_3/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_3/ (2005).
Smith, W. Directory of data in /jgofs/southern/nbp97_8/. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_8/ (2005).
Morrison, J. Final version CTD, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_8/ (2005).
Anderson, B. Shipboard underway data. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_8/ (2005).
Smith, W. O. Fluorometric chlorophyll-a & phaeopigments. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_8/ (2005).
Bidigare, R. R. Pigments, HPLC method, sampled from bottle and TM casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_8/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp97_8/ (2005).
Dunbar, R. NBP9709. R2R https://doi.org/10.7284/905430 (1997).
Honjo, S. Directory of data in /jgofs/southern/nbp98_2/. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp98_2/ (2005).
Program, USA R/V Nathaniel B. Palmer cruise history. USJGOFS https://www.usap.gov/usapgov/vesselscienceandoperations/ (2020).
Morrison, J. Final version CTD, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp98_2/ (2005).
Anderson, B. Shipboard underway data. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp98_2/ (2005).
Smith, W. O. Fluorometric chlorophyll-a & phaeopigments. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/nbp98_2/ (2005).
Smith, W. & Asper, V. NBP0301B field data. R2R https://doi.org/10.7284/905485 (2003).
Smith, W. NBP0305A field data. R2R https://doi.org/10.7284/905494 (2003).
Smith, W. Underway hydrographic, weather and ship-state data (JGOFS) from Nathaniel B. Palmer expedition NBP0305A (2003). MDGS https://doi.org/10.1594/IEDA/315904 (2010).
Arnold, G. et al. NBP0501 field data. R2R https://doi.org/10.7284/905506 (2005).
Arnold, G. Underway hydrographic, weather and ship-state data (JGOFS) from Nathaniel B. Palmer expedition NBP0501. MDGS https://doi.org/10.1594/IEDA/315941 (2005).
Smith, W. NBP0601A field data. R2R https://doi.org/10.7284/905516 (2006).
Smith, W. Uncalibrated hydrographic data acquired with a CTD in the Ross Sea during the Nathaniel B. Palmer expedition NBP0601A. MDGS https://doi.org/10.1594/IEDA/305330 (2008).
Smith, W. Underway hydrographic, weather and ship-state data (JGOFS) from Nathaniel B. Palmer expedition NBP0601A. MDGS https://doi.org/10.1594/IEDA/305246 (2008).
Gregory, M. B. Calibrated hydrographic data acquired with a CTD from the Scotia Sea, Antarctica acquired during the Nathaniel B. Palmer expedition NBP0606. MDGS https://doi.org/10.1594/IEDA/321960 (2015).
Gregory, M. B. Biological dataset collected from bottle casts from the R/V Lawrence M. Gould and the R/V Nathaniel B. Palmer in the Southern Drake Passage and Scotia Sea in support of National Science Foundation projects OPP 03-30443 and ANT 04-44134 from 15 February 2004 to 09 August 2006 (NCEI Accession 0049902). NCEI https://accession.nodc.noaa.gov/0049902 (2011).
Gregory, M. B. Measurements taken in the blue water zone (BWZ) under NSF funding near Antarctica and Drakes Passage in 2004 to 2006. SEABASS https://10.0.19.203/SeaBASS/NSF-BWZ/DATA001 (2016).
Repository, R. D. NBP1102. R2R https://doi.org/10.7284/904911 (2014).
Swift, J. CTD data from 320620110219. CCHDO https://cchdo.ucsd.edu/cruise/320620110219 (2017).
Swift, J. Underway hydrographic, weather and ship-state data (JGOFS) from Nathaniel B. Palmer expedition NBP1102. MDGS https://doi.org/10.1594/IEDA/317638 (2011).
Repository, R. D. NBP1403. R2R https://doi.org/10.7284/904922 (2014).
Talley, L. CTD data from 320620140320. CCHDO https://doi.org/10.7942/320620140320 (2017).
Chaves, J., Freeman, S., Mannino, A. & Novak, M. Underway hydrographic, weather and ship-state data (JGOFS) from Nathaniel B. Palmer expedition NBP1403. SEABASS https://doi.org/10.5067/SeaBASS/CLIVAR/DATA001 (2015).
Repository, R. D. NBP1511. R2R https://doi.org/10.7284/906667 (2016).
Kelly, M. Calibrated hydrographic data acquired with a CTD from the Southern Ocean acquired during the Nathaniel B. Palmer expedition NBP1511. MDGS https://doi.org/10.1594/IEDA/322377 (2018).
Kelly, M. Underway hydrographic, weather and ship-state data (JGOFS) from the Southern Ocean acquired during the Nathaniel B. Palmer expedition NBP1511. MDGS https://doi.org/10.1594/IEDA/322366 (2018).
Tatiana, R. NBP1701. R2R https://doi.org/10.7284/907210 (2017).
Tatiana, R. Calibrated hydrographic data from the Southern Ocean acquired during R/V Nathaniel B. Palmer expedition NBP1701. MDGS https://doi.org/10.1594/IEDA/324280 (2019).
Tatiana, R. Processed underway hydrographic and weather and ship-state data (JGOFS) from the Southern Ocean acquired during R/V Nathaniel B. Palmer expedition NBP1701. MDGS https://doi.org/10.1594/IEDA/324278 (2019).
Repository, R. D. NBP1704. R2R https://doi.org/10.7284/907629 (2017).
Ackley, S. Calibrated hydrographic data from the Southern Ocean acquired during R/V Nathaniel B. Palmer expedition NBP1704. MDGS https://doi.org/10.1594/IEDA/324287 (2019).
Ackley, S. Raw underway hydrographic and weather and ship-state data (JGOFS) from the Southern Ocean focus site acquired during R/V Nathaniel B. Palmer expedition NBP1704. MDGS https://doi.org/10.1594/IEDA/324284 (2019).
Repository, R. D. NBP1704. R2R https://doi.org/10.7284/907630 (2017).
Mecking, S. Uncalibrated hydrographic data from the Pacific Ocean (southern) acquired during R/V Nathaniel B. Palmer expedition NBP1706. MDGS https://doi.org/10.1594/IEDA/324302 (2018).
Mecking, S. Raw underway hydrographic and weather and ship-state data (JGOFS) from the Pacific Ocean (southern) acquired during R/V Nathaniel B. Palmer expedition NBP1706. MDGS https://doi.org/10.1594/IEDA/324297 (2018).
Repository, R. D. NBP1802. R2R https://doi.org/10.7284/907943 (2018).
Macdonald, A. & Briggs, E. CTD data from 320620140320. CCHDO https://doi.org/10.7942/C2F08X (2020).
Macdonald, A. Raw underway hydrographic and weather and ship-state data (JGOFS) from the Pacific Ocean acquired during R/V Nathaniel B. Palmer expedition NBP1802. MDGS https://doi.org/10.1594/IEDA/324549 (2018).
Orsi, A. & Rosso, I. Documentation from cruise 325020190403. CCHDO https://doi.org/10.7942/C29660 (2020).
Orsi, A. & Rosso, I. CTD data from cruise 325020190403. CCHDO https://doi.org/10.7942/C29660 (2020).
Ross, R. M. LMG99-01 weekly sit rep Jan 2-10. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (1999).
Ross, R. M. LMGould: SciWeekly 25-31 Jan 99. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (1999).
Ross, R. M. LMGould: SciWeekly 1-8 Feb 99. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (1999).
Ross, R. M. Science weekly report LMG00-01. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2000).
Ross, R. M. LMG weekly science report 9-16 Jan. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2000).
Ross, R. M. LM Gould weekly science report 17-22 Jan. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2000).
Ross, R. M. LMG01-01 4 -13 Jan 2001 Palmer LTER cruise. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2001).
Ross, R. M. LMG01-01 14 -20 Jan 2001 Palmer LTER cruise. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2001).
Ross, R. M. LMG01-01 21-27 January Palmer LTER cruise. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2001).
Fraser, B., Vernet, M., Quetin, L. B., Ross, R. M. & Ducklow, H. LMG 0701 situation report 07 – 13 January, 2007. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2007).
Fraser, B., Vernet, M., Quetin, L. B., Ross, R. M. & Ducklow, H. LMG 0701 situation report 14 – 21 January, 2007. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2007).
Fraser, B., Vernet, M., Quetin, L. B., Ross, R. M. & Ducklow, H. LMG 0701 situation report 21 – 28 January, 2007. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2007).
Fraser, W. R., Schofield, O., Steinberg, D., Ducklow, H. & Hollibaugh, J. T. Weekly update 1: LMG cruise 11-01 palmer LTER. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2011).
Fraser, W. R., Schofield, O., Steinberg, D., Ducklow, H. & Hollibaugh, J. T. Weekly update 2: LMG cruise 11-01 palmer LTER. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2011).
Fraser, W. R., Schofield, O., Steinberg, D., Ducklow, H. & Hollibaugh, J. T. Weekly update 3: LMG cruise 11-01 palmer LTER. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2011).
Fraser, W. R., Schofield, O., Steinberg, D., Ducklow, H. & Hollibaugh, J. T. Weekly update 4: LMG cruise 11-01 palmer LTER. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2011).
Fraser, W. R. et al. LMG 12-01: 29 dec. 2011 – 07 february 2012 LTER cruise 20 weekly science report i. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2012).
Fraser, W. R. et al. LMG 12-01: 29 dec. 2011 – 07 february 2012 LTER cruise 20 weekly science report 2. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2012).
Fraser, W. R. et al. LMG 12-01: 29 dec. 2011 – 07 february 2012 LTER cruise 20 weekly science report 3. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2012).
Fraser, W. R. et al. LMG 12-01: 29 dec. 2011 – 07 february 2012 LTER cruise 20 weekly science report 4. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2012).
Ducklow, H. et al. LMG 13-01: 30 dec. 2012 – 07 february 2013 LTER cruise 21 weekly science report i. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2013).
Ducklow, H. et al. LMG 13-01: 30 dec. 2012 – 07 february 2013 LTER cruise 21 weekly science report 2. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2013).
Ducklow, H. et al. LMG 13-01: 30 dec. 2012 – 07 february 2013 LTER cruise 21 weekly science report 3. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2013).
Ducklow, H. et al. LMG 13-01: 30 dec. 2012 – 07 february 2013 LTER cruise 21 weekly science report 4. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2013).
Schofield, O. et al. LMG 14-01: 31 dec. 2013 – 05 february 2013 LTER cruise 22 weekly science report 1. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2014).
Schofield, O. et al. LMG 14-01: 31 dec. 2013 – 05 february 2013 LTER cruise 22 weekly science report 2. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2014).
Schofield, O. et al. LMG 14-01: 31 dec. 2013 – 05 february 2013 LTER cruise 22 weekly science report 3. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2014).
Schofield, O. et al. LMG 14-01: 31 dec. 2013 – 05 february 2013 LTER cruise 22 weekly science report 4. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2014).
Steinberg, D. et al. LMG 15-01: 29 dec. 2014 – 09 february 2015, PAL LTER cruise 23 weekly science report 1. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2015).
Steinberg, D. et al. LMG 15-01: 29 dec. 2014 – 09 february 2015, PAL LTER cruise 23 weekly science report 2. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2015).
Steinberg, D. et al. LMG 15-01: 29 dec. 2014 – 09 february 2015, PAL LTER cruise 23 weekly science report 3. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2015).
Steinberg, D. et al. LMG 15-01: 29 dec. 2014 – 09 february 2015, PAL LTER cruise 23 weekly science report 4. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2015).
Fraser, W. R. et al. LMG 16-01: 03 jan – 17 feb. 2016 LTER cruise 24 weekly science report 1. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2016).
Fraser, W. R. et al. LMG 16-01: 03 jan – 17 feb. 2016 LTER cruise 24 weekly science report 2. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2016).
Fraser, W. R. et al. LMG 16-01: 03 jan – 17 feb. 2016 LTER cruise 24 weekly science report 3. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2016).
Fraser, W. R. et al. LMG 16-01: 03 jan – 17 feb. 2016 LTER cruise 24 final weekly science report. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2016).
Schofield, O., Ducklow, H., Steinberg, D., Fraser, W. R. & Friedlaender, A. Science report for Week 1 of LTER 1701. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2017).
Martinson, D. et al. Science report for week 2 of LTER 1701. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2017).
Martinson, D. et al. Science report for week 3 of LTER 1701. PAL-LTER https://pal.lternet.edu/publications/station-cruise-reports (2017).
Ducklow, H. Raw underway hydrographic and weather and ship-state data (JGOFS) from the Antarctic Peninsula acquired during R/V Laurence M. Gould expedition LMG1701 (2016). MDGS https://doi.org/10.1594/IEDA/324167 (2019).
Bullister, J. L. & Johnson, G. C. Documentation from cruise 33RO20071215. CCHDO https://cchdo.ucsd.edu/cruise/33RO20071215 (2016).
Bullister, J. L. & Johnson, G. C. Ctd data from cruise 33RO20071215. CCHDO https://cchdo.ucsd.edu/cruise/33RO20071215 (2016).
Wanninkhof, R. Underway physical oceanography and carbon dioxide measurements during Ronald H. Brown cruise 33RO20071215. CCHDO https://doi.org/10.1594/PANGAEA.892094 (2018).
Feely, R. A. et al. Partial pressure (or fugacity) of carbon dioxide, dissolved inorganic carbon, pH, alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations using alkalinity titrator, CTD and other instruments from NOAA ship Ronald H. Brown in the north Pacific Ocean, south Pacific Ocean and southern oceans from 2007-12-15 to 2008-02-23 (NCEI accession 0109903). NCEI https://doi.org/10.3334/cdiac/otg.clivar_p18_2007 (2013).
Sonnerup, R., Carter, B., Purkey, S. & Bourbonnais, A. Documentation from cruise 33RO20161119. CCHDO https://cchdo.ucsd.edu/cruise/33RO20161119 (2020).
Sonnerup, R., Carter, B., Purkey, S. & Bourbonnais, A. CTD data from cruise 33RO20161119. CCHDO https://cchdo.ucsd.edu/cruise/33RO20161119 (2020).
Morison, J. Final CTD data, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_6/ (2005).
Goericke, R. Pigments, HPLC method, sampled from bottle casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_6/ (2005).
Smith, W. O. Fluorometric chlorophyll-a. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_6/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_6/ (2005).
Morison, J. Final CTD data, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_7/ (2005).
Goericke, R. Pigments, HPLC method, sampled from bottle casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_7 (2005).
Smith, W. O. Fluorometric chlorophyll-a. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_7/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_7/ (2005).
Morison, J. Final CTD data, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_8/ (2005).
Morison, J. Processed CTD data from TM casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_8/ (2005).
Goericke, R. Pigments, HPLC method, sampled from bottle casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_8/ (2005).
Smith, W. O. Fluorometric chlorophyll-a. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_8/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_8/ (2005).
Morison, J. Final CTD data, including beam attenuation optics. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_9/ (2005).
Goericke, R. Pigments, HPLC method, sampled from bottle casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_9/ (2005).
Smith, W. O. Fluorometric chlorophyll-a. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_9/ (2005).
Smith, W. O. Particulate organic carbon, nitrogen from CTD casts. USJGOFS http://usjgofs.whoi.edu/jg/dir/jgofs/southern/rr-kiwi_9/ (2005).
Buesseler, K. O., K. H. C. & Siegel, D. USCGC Polar Star cruises COOK19MV, DRFT08RR, PS02_2002 from the Southern Ocean, south of New Zealand in 2002 (SOFeX project). BCO-DMO http://lod.bco-dmo.org/id/dataset/2851 (2007).
Bidigare, R. R. & Landry, M. R. Pigments from HPLC analysis of bottle samples from SOFeX cruisse PS02_2002, COOK19MV, DRFT08RR from the Southern Ocean, south of New Zealand, Southern Ocean in 2002 (SOFeX project). BCO-DMO http://lod.bco-dmo.org/id/dataset/2851 (2007).
Richard Barber, F. C. & Coale, K. H. Fluorometric chlorophyll from Niskin and TM bottle samples from R/V Melville, R/V Roger Revelle cruises COOK19MV, DRFT08RR from the Southern Ocean, south of New Zealand in 2002 (SOFeX project). BCO-DMO http://lod.bco-dmo.org/id/dataset/2933 (2008).
Altabet, M. A. & Timothy, D. Particulate organic matter (POM) from R/V Melville, R/V Roger Revelle cruises COOK19MV, DRFT08RR from the Southern Ocean, south of New Zealand in 2002 (SOFeX project). BCO-DMO http://lod.bco-dmo.org/id/dataset/2926 (2007).
Sloyan, B. M. & Swift, J. Documentation from cruise 33RR200501. CCHDO https://cchdo.ucsd.edu/cruise/33RR200501 (2015).
Sloyan, B. M. & Swift, J. Ctd data from cruise 33RR200501. CCHDO https://cchdo.ucsd.edu/cruise/33RR200501 (2017).
Swift, J. Documentation from cruise 33RR20070204. CCHDO https://cchdo.ucsd.edu/cruise/33RR20070204 (2017).
Swift, J. CTD data from cruise 33RR20070204. CCHDO https://cchdo.ucsd.edu/cruise/33RR20070204 (2017).
Speer, K. G. & Dittmar, T. Documentation from cruise 33RR20080204. CCHDO https://cchdo.ucsd.edu/cruise/33RR20080204 (2009).
Speer, K. G. Ctd data from cruise 33RR20080204. CCHDO https://cchdo.ucsd.edu/cruise/33RR20080204 (2017).
Wanninkhof, R. Underway physical oceanography and carbon dioxide measurements during Roger Revelle cruise 33RR20080204. PANGAEA https://doi.org/10.1594/PANGAEA.851516 (2016).
Macdonald, A. Documentation from cruise 33RR20160208. CCHDO https://doi.org/10.7942/C2H59N (2016).
Macdonald, A. Ctd data from cruise 33RR20160208. CCHDO https://doi.org/10.7942/C2H59N (2016).
Wanninkhof, R. & Pierrot, D. Underway physical oceanography and carbon dioxide measurements during Roger A. Revelle cruise 33RR20160208. PANGAEA https://doi.org/10.1594/PANGAEA.878374 (2017).
Macdonald, A. M. et al. Discrete profile measurements of dissolved inorganic carbon, total alkalinity, pH and other hydrographic and chemical data obtained during the R/V Roger Revelle repeat hydrography and SOCCOM cruise in the Indian Ocean and Southern Ocean: GO-SHIP section I08S (EXPOCODE 33RR20160208), from 2016-02-08 to 2016-03-16 (NCEI accession 0171457). NCEI https://doi.org/10.7289/v5hm56qr (2018).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2002/2003 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5214439081269 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 2 of the Astrolabe - 2002/2003 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521449B6B8AE0 (2013).
McMinn, A. Fast repetition rate fluorometry (FRRF) data collected on the RV l’astrolabe. IMAS https://metadata.imas.utas.edu.au/geonetwork/srv/eng/catalog.search#/metadata/fc64fc7b-29b0-4607-9527-899dfa991d68 (2011).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 4 of the Astrolabe - 2002/2003 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5214565920010 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2003/2004 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/52145994425BF (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 1 of the Astrolabe - 2003/2004 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/52145FA66AF8A (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 2 of the Astrolabe - 2003/2004 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5215A350CF51B (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 4 of the Astrolabe - 2003/2004 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5215A7F3124D6 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2004/2005 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5215AF880C5E4 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2005/2006 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5216E3A703643 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 2 of the Astrolabe - 2005/2006 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521AB4A7693BD (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 4 of the Astrolabe - 2005/2006 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521ABAA06322A (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2006/2007 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521BE0CB374A3 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 2 of the Astrolabe - 2006/2007 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521BE3FA33859 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2007/2008 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521BECBAC1F75 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 2 of the Astrolabe - 2007/2008 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521BF2BAF13CA (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 4 of the Astrolabe - 2007/2008 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521BFF78485E6 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2008/2009 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521C080AE09CE (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 2 of the Astrolabe - 2008/2009 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/521C0DF59E573 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 4 of the Astrolabe - 2008/2009 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5244E3F79E38B (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 0 of the Astrolabe - 2009/2010 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/5244EC8C7BC02 (2013).
Wright, S. The role of Antarctic marine protists in trophodynamics and global change and the impact of UV-B on these organisms - Rotation 4 of the Astrolabe - 2009/2010 samples. Australian Antarctic Data Centre https://doi.org/10.4225/15/52450EBCAC268 (2013).
Fiala, M. ANTARES 2-MD78/ANTARFIX 4 cruise, RV Marion Dufresne. SISMER https://doi.org/10.17600/94200020 (1994).
Fiala, M. & French, C. T. D. data from ANTARES 2-MD78/ANTARFIX 4 cruise. SISMER https://doi.org/10.17600/94200020 (1994).
Mayzaud, P. ANTARES 3 cruise, RV Marion Dufresne. SISMER https://doi.org/10.17600/95200100 (1995).
Mayzaud, P. & French, C. T. D. data from ANTARES 3 cruise. SISMER https://doi.org/10.17600/95200100 (1995).
Marty, J.-C. Pigment concentrations at bottle station ANTARES-III_0210. PANGAEA https://doi.org/10.1594/PANGAEA.136858 (2004).
Marty, J.-C. Pigment concentrations at bottle station ANTARES-III_0310. PANGAEA https://doi.org/10.1594/PANGAEA.136859 (2004).
Marty, J.-C. Pigment concentrations at bottle station ANTARES-III_0460. PANGAEA https://doi.org/10.1594/PANGAEA.136860 (2004).
Marty, J.-C. Pigment concentrations at bottle station ANTARES-III_0490. PANGAEA https://doi.org/10.1594/PANGAEA.136861 (2004).
Marty, J.-C. Pigment concentrations at bottle station ANTARES-III_0510. PANGAEA https://doi.org/10.1594/PANGAEA.136862 (2004).
Marty, J.-C. Pigment concentrations at bottle station ANTARES-III_0590. PANGAEA https://doi.org/10.1594/PANGAEA.136863 (2004).
Descolas-Gros, C. & Edwige, C. Hydrochemistry measured on water bottle samples during Marion Dufresne cruise ANTARES-III (MD102). PANGAEA https://doi.org/10.1594/PANGAEA.803187 (2012).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-01 (EXPOCODE 35MF19980121) in the Indian Ocean from 1998-01-21 to 1998-02-19 (NCEI accession 0113570). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF19980121 (2013).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-02 (EXPOCODE 35MF19980818) in the Indian Ocean from 1998-08-18 to 1998-09-09 (NCEI accession 0113573). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF19980818 (2013).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-03 (EXPOCODE 35MF19981205) in the Indian Ocean from 1998-12-05 to 1998-12-27 (NCEI accession 0113574). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF19981205 (2013).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-04 (EXPOCODE 35MF20000117) in the Indian Ocean from 2000-01-17 to 2000-01-27 (NCEI accession 0113575). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF20000117 (2013).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-05 (EXPOCODE 35MF20010103) in the Indian Ocean from 2000-07-19 to 2000-08-16 (NCEI accession 0113576). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF20000719 (2013).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-06 (EXPOCODE 35MF20010103) in the Indian Ocean from 2001-01-03 to 2001-01-26 (NCEI accession 0113577). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF20010103 (2013).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-08 (EXPOCODE 35MF20020104) in the Indian Ocean from 2002-01-04 to 2002-02-01 (NCEI accession 0113578). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_35MF20020104 (2013).
Metzl, N. VT 62/CARAUS - OISO 11 cruise, RV Marion Dufresne. SISMER https://doi.org/10.17600/4200010 (2004).
Metzl, N. French CTD data from VT 62/CARAUS - OISO 11 cruise. SISMER https://doi.org/10.17600/4200010 (2004).
Claire Lo Monaco, N. M. Dissolved inorganic carbon (DIC), total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations during the R/V Marion Dufresne cruise OISO-11 (EXPOCODE 35MF20040103) in the indian ocean from 2004-01-03 to 2004-02-09 (NCEI accession 0113572). NCEI https://doi.org/10.3334/cdiac/otg.carina_35mf20040103 (2013).
Peeken, I., Bluhm, K. & Zöllner, E. Phytoplankton pigments measured from underway and water bottle samples during Marion Dufresne cruise MD158 (OOMPH MD07 leg 1). PANGAEA https://doi.org/10.1594/PANGAEA.873203 (2017).
Picheral, M. et al. Vertical profiles of environmental parameters measured from physical, optical and imaging sensors during Tara Oceans expedition 2009-2013. PANGAEA https://doi.org/10.1594/PANGAEA.836321 (2014).
Claustre, H. HPLC pigments from TARA oceans expedition analysed at LOV. PANGAEA https://doi.org/10.1594/PANGAEA.836321 (2013).
Masao Ishii, M. T. Dissolved inorganic carbon, temperature, salinity and other variables collected from discrete sample and profile observations using CTD, bottle and other instruments from the Hakuho Maru in the Indian Ocean, north Pacific Ocean and south pacific ocean from 2001-12-08 to 2002-01-19 (NCEI accession 0112347). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_49HH20011127 (2013).
Fukasawa, M. Documentation from cruise 49NZ20030803. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20030803 (2015).
Fukasawa, M. CTD data from cruise 49NZ20030803. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20030803 (2015).
Clemenston, L. Data collected on leg 1 of the BEAGLE cruises during August/September 2003. CCHDO https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Watanabe, S. Documentation from cruise 49NZ20030909. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20030909 (2015).
Watanabe, S. CTD data from cruise 49NZ20030909. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20030909 (2015).
Clemenston, L. Data collected on leg 2 of the BEAGLE cruises during September/October 2003. CCHDO https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Clemenston, L. Data collected on leg 3 of the BEAGLE cruises during October 2003. CCHDO https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Yoshikawa, Y. Documentation from cruise 49NZ20031106. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20031106 (2015).
Yoshikawa, Y. CTD data from cruise 49NZ20031106. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20031106 (2015).
Clemenston, L. Data collected on leg 4 of the BEAGLE cruises during November/December 2003. CCHDO https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Fukasawa, M. Documentation from cruise 49NZ20031209. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20031209 (2015).
Fukasawa, M. CTD data from cruise 49NZ20031209. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20031209 (2015).
Clemenston, L. Data collected on leg 5 of the BEAGLE cruises between December 2003 and January 2004. CCHDO https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Clemenston, L. Data collected on leg 6 of the BEAGLE cruises during January and February 2004. CCHDO https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2011).
Katsumata, K. Documentation from cruise 49NZ20121128. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20121128 (2015).
Katsumata, K. CTD data from cruise 49NZ20121128. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20121128 (2015).
Katsumata, K. et al. Dissolved inorganic carbon (DIC), pH on total scale, total alkalinity, temperature, salinity and other variables collected from discrete samples and profile observations during the R/V Mirai cruise GO-SHIP_P14S_S04_2012; MR12-05 leg 2 (EXPOCODE 49NZ20121128) in the Indian, south Pacific and Southern Oceans from 2012-11-28 to 2013-01-04 (NCEI accession 0143950). NCEI https://doi.org/10.3334/cdiac/otg.goship_p14s_s04_2012 (2016).
Uchida, H. Documentation from cruise 49NZ20130106. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20130106 (2015).
Uchida, H. CTD data from cruise 49NZ20130106. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20130106 (2015).
Uchida, H., Murata, A., Aoyama, M., Kumamoto, Y. & Aoki, S. Dissolved inorganic carbon (DIC), pH on total scale, total alkalinity, temperature, salinity and other variables collected from discrete sample and profile observations obtained during the R/V Mirai cruise GOSHIP_S04I_2013 (EXPOCODE 49NZ20130106) in the Indian Ocean and southern oceans from 2013-01-06 to 2013-02-15 (NCEI accession 0156925). NCEI https://doi.org/10.3334/CDIAC/OTG.GOSHIP_S04I_2013 (2016).
Uchida, H. Documentation from cruise 49NZ20170208. CCHDO https://cchdo.ucsd.edu/cruise/49NZ2017020 (2018).
Uchida, H. CTD data from cruise 49NZ20170208. CCHDO https://cchdo.ucsd.edu/cruise/49NZ20170208 (2018).
Uchida, H., Murata, A., Kumamoto, Y., Aoyama, M. & Sasaki, K. Dissolved inorganic carbon (DIC), total alkalinity, chlorofluorocarbons (CFC-11, CFC-12, CFC113), temperature, salinity, oxygen, nutrients and other measurements collected from profile and discrete sample observations during the R/V Mirai cruise MR16-09, GO-SHIP_P17E (EXPOCODE 49NZ20170208) in the south pacific ocean from 2017-02-08 to 2017-03-05 (NCEI accession 0186207). NCEI https://doi.org/10.25921/zmae-wa37 (2019).
Masao Ishii, R. M., Naoki Sato. Dissolved inorganic carbon, pH, temperature, salinity and other variables collected from discrete sample and profile observations using CTD, bottle and other instruments from the SHIRASE in the Indian Ocean, south Pacific Ocean and Tasman Sea from 1992-12-03 to 1993-03-19 (NCEI accession 0113597). NCEI https://doi.org/10.3334/CDIAC/otg.CARINA_49ZS19921203 (2013).
Tsuneo Odate, M. Ishii; Dissolved inorganic carbon, temperature, salinity and other variables collected from discrete sample and profile observations using CTD, bottle and other instruments from the TANGAROA in the Indian Ocean from 2003-02-17 to 2003-03-12 (NCEI accession 0113599). NCEI https://doi.org/10.3334/cdiac/otg.carina_61tg20030217 (2013).
Owens, N. J. P. Documentation from cruise 74JC002_1. CCHDO https://cchdo.ucsd.edu/cruise/74JC002_1 (2015).
Owens, N. J. P. CTD data from cruise 74JC002_1. CCHDO https://cchdo.ucsd.edu/cruise/74JC002_1 (2015).
Firing, Y. Documentation from cruise 74JC20151217. CCHDO https://cchdo.ucsd.edu/cruise/74JC20151217 (2016).
Firing, Y. CTD data from cruise 74JC20151217. CCHDO https://cchdo.ucsd.edu/cruise/74JC20151217 (2017).
de Villiers, S., Siswana, K. & Vena, K. Hydrography and biogeochemistry during S.A. Agulhas II cruise Agu011, in the SW Indian Ocean, April 2014. PANGAEA https://doi.org/10.1594/PANGAEA.848875 (2015).
Olsen, A. et al. The global ocean data analysis project version 2 (GLODAPv2) – an internally consistent data product for the world ocean. Earth Syst. Sci. Data 8, 297–323, https://doi.org/10.5194/essd-8-297-2016 (2016).
Key, R. M. et al. Global ocean data analysis project, version 2 (GLODAPv2). NCEI https://doi.org/10.3334/CDIAC/OTG.NDP093_GLODAPv2 (2015).
System, I. M. O. IMOS - SRS - ocean colour - bio optical database of australian waters. AODN https://catalogue-imos.aodn.org.au/geonetwork/srv/en/metadata.show?uuid=97b9fe73-ee44-437f-b2ae-5b8613f81042 (2019).
Peloquin, J. M. et al. The MAREDAT global database of high performance liquid chromatography marine pigment measurements - Gridded data product (NetCDF) - Contribution to the MAREDAT World ocean atlas of plankton functional types. PANGAEA https://doi.org/10.1594/PANGAEA.793246 (2013).
Peloquin, J. M. et al. The MAREDAT global database of high performance liquid chromatography marine pigment measurements. Earth Syst. Sci. Data 5, 109–123, https://doi.org/10.5194/essd-5-109-2013 (2013).
Taylor, B. B. et al. Bio-optical provinces in the eastern Atlantic Ocean and their biogeographical relevance. Biogeosci. 8, 3609–3629, https://doi.org/10.5194/bg-8-3609-2011 (2011).
Hoffmann, L. J., Peeken, I., Lochte, K., Assmy, P. & Veldhuis, M. Different reactions of Southern Ocean phytoplankton size classes to iron fertilization. Limnol. Oceanogr. 51, 1217–1229, https://doi.org/10.4319/lo.2006.51.3.1217 (2006).
Yentsch, C. S. & Menzel, D. W. A method for the determination of phytoplankton chlorophyll and phaeophytin by fluorescence. Deep Sea Res. and Oceanog. Abstracts 10, 221–231, https://doi.org/10.1016/0011-7471(63)90358-9 (1963).
Ras, J., Claustre, H. & Uitz, J. Spatial variability of phytoplankton pigment distributions in the subtropical south Pacific Ocean: Comparison between in situ and predicted data. Biogeosci. 5, 353–369, https://doi.org/10.5194/bg-5-353-2008 (2008).
Welschmeyer, N. A. Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol. and Oceanogr. 39, 1985–1992, https://doi.org/10.4319/lo.1994.39.8.1985 (1994).
Neveux, J. & Panouse, M. Spectrofluorometric determination of chlorophylls and pheophytin. Archiv fiir Hydrobiologie 109, 567–581 (1987).
Lorenzen, C. J. Determination of chlorophyll and pheo-pigments: spectrophotometric equations. Limnol. and Oceanogr. 12, 343–346, https://doi.org/10.4319/lo.1967.12.2.0343 (1967).
Bidigare, R. R., Van Heukelem, L. & Trees, C. C. Analysis of algal pigments by high-performance liquid chromatography. Algal culturing techniques 327–345 (2005).
Mueller, J. L. et al. Ocean optics protocols for satellite ocean color sensor validation, revision 4. Volume III: Radiometric measurements and data analysis protocols. Goddard Space Flight Space Centre 1–63 https://doi.org/10.25607/OBP-62 (2003).
Van Heukelem, L. & Thomas, C. S. Computer-assisted high-performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments. J. Chromatogr. A 910, 31–49, https://doi.org/10.1016/S0378-4347(00)00603-4 (2001).
Hooker, S. et al. The fifth SeaWiFS HPLC analysis round-robin experiment (SeaHARRE-5). NASA 1–108 (2012).
Álvarez, E., Thoms, S., Bracher, A., Liu, Y. & Völker, C. Modeling photoprotection at global scale: The relative role of nonphotosynthetic pigments, physiological state, and species composition. Global Biogeochem. Cycles 33, 904–926, https://doi.org/10.1029/2018GB006101 (2019).
Wright, S. W. et al. Phytoplankton community structure and stocks in the Southern Ocean (30–80°e) determined by CHEMTAX analysis of HPLC pigment signatures. Deep Sea Res. 2 Top. Stud. Oceanogr. 57, 758–778, https://doi.org/10.1016/j.dsr2.2009.06.015 (2010).
Smith, W. O. et al. Interannual variations in nutrients, net community production, and biogeochemical cycles in the ross sea. Deep Sea Res. 2 Top. Stud. Oceanogr. 53, 815–833, https://doi.org/10.1016/j.dsr2.2006.02.014 (2006).
Wright, S. W. et al. Improved HPLC method for the analysis of chlorophylls and carotenoids from marine phytoplankton. Mar. Ecol. Prog. Ser. 77, 183–196, http://www.jstor.org/stable/24826571 (1991).
Peloquin, J. et al. The MAREDAT global database of high performance liquid chromatography marine pigment measurements. Earth Syst. Sci. Data 5, 109–123, https://doi.org/10.5194/essd-5-109-2013 (2013).
Villiers, S., de, Siswana, K. & Vena, K. In situ measurement of the biogeochemical properties of southern ocean mesoscale eddies in the southwest Indian Ocean, April 2014. Earth Syst. Sci. Data 7, 415–422, https://doi.org/10.5194/essd-7-415-2015 (2015).
Holm-Hansen, O. & Riemann, B. Chlorophyll a determination: Improvements in methodology. Oikos 30, 438–447, http://www.jstor.org/stable/3543338 (1978).
Ishii Masao, M. F., T Suzuki. Report on the surface phytoplankton pigments measured during the JARE-34 cruise to syowa station, antarctica, november 1992 to march 1993. JARE data reports. Marine biology https://doi.org/10.15094/00003303 (1996).
Jeffrey, S. W. & Humphrey, G. F. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochemie und Physiologie der Pflanzen 167, 191–194, https://doi.org/10.1016/S0015-3796(17)30778-3 (1975).
Intergovernmental Oceanographic Commission. Protocols for the joint global ocean flux study (JGOFS) core measurements. Intergovernmental Oceanographic Commission Manuals and Guides 29, 1–170, https://doi.org/10.25607/OBP-1409 (1994).
Smetacek, V., Bathmann, U. & El Naggar, S. E. D. The expeditions ANTARKTIS XVIII/1-2 of the research vessel. Reports on Polar and Marine Research 400 (2001).
Carbotte, S. M. et al. Antarctic multibeam bathymetry and geophysical data synthesis: An On-Line Digital Data Resource for Marine Geoscience Research in the Southern Ocean. USGS 2, 1–4 (2007).
US Antarctic Program R/V Laurence M. Gould cruise history. https://www.usap.gov/usapgov/vesselscienceandoperations/ (2020).
Iannuzzi, R. Conductivity temperature depth (CTD) sensor profile data binned by depth from PAL LTER annual cruises, 1991 - 2017 (ongoing). PAL-LTER https://doi.org/10.6073/pasta/12276fbc0d68568177702aed0d4b44bc (2018).
Oscar Schofield, B. P., Vernet, M. Photosynthetic pigments of water column samples and analyzed with high performance liquid chromatography (HPLC), collected aboard Palmer LTER annual cruises off the coast of the Western Antarctica Peninsula, 1991 - 2016. PAL-LTER https://doi.org/10.6073/pasta/4d583713667a0f52b9d2937a26d0d82e (2018).
Oscar Schofield, R. S., Vernet, M. Chlorophyll and phaeopigments from water column samples, collected at selected depths aboard Palmer LTER annual cruises off the coast of the Western Antarctic Peninsula, 1991 – 2019. PAL-LTER https://doi.org/10.6073/pasta/6bbce1e3264571463c0354874dba88cf (2020).
Hugh Ducklow, B. P. & Vernet, M. Particulate organic carbon and nitrogen measurements from water column sample bottles, collected aboard Palmer LTER annual cruises off the Western Antarctic Peninsula, 1991 - 2018. Cruise PD94-01 not included in time series for lack of samples. PAL-LTER https://doi.org/10.6073/pasta/d2de255ff3e7c6538c9d49c69c0ad377 (2017).
Walton, D. & Klepikov, A. CTD data from cruise RUB320161220. CCHDO https://cchdo.ucsd.edu/cruise/RUB320161220 (2019).
Werdell, P. J. et al. Unique data repository facilitates ocean color satellite validation. Eos, Transactions American Geophysical Union 84, 377–387, https://doi.org/10.1029/2003EO380001 (2003).
Werdell, P. J. & Bailey, S. The SeaWiFS bio-optical archive and storage system (SeaBASS): Current architecture and implementation. NASA 1–45 (2002).
Boss, E. & Talley, L. CLIVAR. SeaBASS https://doi.org/10.5067/SeaBASS/CLIVAR/DATA001 (2020).
Boss, E. & Talley, L. SOCCOM HPLC pigments. SeaBASS https://doi.org/10.5067/SeaBASS/SOCCOM/DATA001 (2020).
Arrigo, K. Measurements taken off the Antarctic coast in the ross sea between 1996 and 1998 under the research on ocean-atmosphere variability and ecosystem response in the ross sea (ROAVERRS). SeaBASS https://doi.org/10.5067/SeaBASS/ROAVERRS/DATA001 (2020).
Smith, W. 0. Bottle data including phosphate, nitrate, total nitrite and nitrate, ammonium, silicate, chlorophyll, particulate organic carbon and nitrogen (POC, PON), and biogenic silica from multiple cruises to the southern Ross Sea, 2001-2006 (IVARS project). BCO-DMO https://doi.org/10.26008/1912/bco-dmo.710112.1.14 (2017).
Felden, J. et al. PANGAEA – Data Publisher for Earth & Environmental Science. Sci. Data 10, 347 (2023).
Haëntjens, N., Boss, E. & Talley, L. D. Revisiting ocean color algorithms for chlorophyll a and particulate organic carbon in the southern ocean using biogeochemical floats. J. Geophys. Res. Oceans 122, 6583–6593 (2017).
Bianchi, T. S., Lambert, C. & Biggs, D. C. Distribution of chlorophyll a and phaeopigments in the northwestern Gulf of Mexico: A comparison between fluorometric and high-performance liquid chromatography measurements. Bull. Mar. Sci. 56, 25–32 (1995).
Dos Santos, A. C. A. et al. Comparison of three methods for chlorophyll determination: Spectrophotometry and fluorimetry in samples containing pigment mixtures and spectrophotometry in samples with separate pigments through high performance liquid chromatography. Acta Limnol. Bras 15, 7–18 (2003).
Kumari, B. Comparison of high performance liquid chromatography and fluorometric ocean colour pigments. J. Indian Soc. Rem. Sens. 33, 541–546 (2005).
Kratzer, S., Harvey, E. T. & Canuti, E. International intercomparison of in situ chlorophyll-a measurements for data quality assurance of the Swedish Monitoring Program. Front. Rem. Sens. 3, 866712 (2022).
Acknowledgements
This research was supported under Australian Research Council’s Special Research Initiative for Antarctic Gateway Partnership (Project ID SR140300001) and a 2019 Fellowship from the Scientific Committee of Antarctic Research. It was also partially supported by Geoscience Australia, in the form of paid study leave from employment. PS and PB were supported by the Australian Research Council Special Research Initiative, Australian Centre for Excellence in Antarctic Science (Project Number SR200100008). PS received support from the Australian Research Council Centre of Excellence for Climate Extremes, CE170100023. Data included in the data product were made available by the following data repositories; PANGAEA, AODN/IMOS, SeaBASS, CCHDO, AADC, GLODAP, PAL-LTER, CSIRO, MDGS and BCO-DMO. We acknowledge the use of the CSIRO Marine National Facility (https://ror.org/01mae9353) in undertaking this research. Data was sourced from Australia’s Integrated Marine Observing System (IMOS) – IMOS is enabled by the National Collaborative Research Infrastructure strategy (NCRIS). It is operated by a consortium of institutions as an unincorporated joint venture, with the University of Tasmania as Lead Agent. Records of data access dates, source addresses and digital object identifiers are recorded as metadata, alongside appropriate data citations. We acknowledge the enormous community effort undertaken in the collection, analysis and publication of this data and thank principal investigators for publishing their data in open access repositories.
Author information
Authors and Affiliations
Contributions
K.B. designed the data product, performed the data aggregation and wrote the manuscript. R.J. contributed to the data product design and manuscript. P.G.S. and P.W.B. contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baldry, K., Johnson, R., Strutton, P.G. et al. A biological ocean data reformatting effort. Sci Data 11, 215 (2024). https://doi.org/10.1038/s41597-024-03038-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03038-0