Abstract
The crocodilians include true crocodiles, alligators, caimans, and gharial, and the trade of crocodilian products is regulated in accordance with the Convention of Wild Fauna and Flora (CITES). Hong Kong does not have her own wild crocodilians; thus, all crocodilians meat available is presumably imported with proper license. Here, we obtained a dataset of cytochrome oxidase I (COI) gene markers of 114 crocodilian meat samples (including frozen and dried crocodilian meat products) available in the contemporary market. We have also validated these barcodes in a phylogenetic approach with other data deposited on the GenBank, and detected 112 samples belonging to four crocodile species Crocodylus siamensis, C. porosus, C. niloticus and Alligator mississippiensis, and 2 samples belonging to snake Malayopython reticulatus. The dataset generated in this study will be useful for further studies including meat inspection, illegal trading, and enhancement of international and local legislations on illegal reptile importation.
Similar content being viewed by others
Background & Summary
The Crocodilia or Crocodylia is an order containing amphibious reptiles generally known as the crocodilians and can be found in the tropics, and certain areas in China and United States. The crocodilians include true crocodiles, alligators, caimans, and gharial, and the trade of crocodilian products is regulated in accordance with the CITES. For instance, commercial organisations such as alligator and crocodile farming need to demonstrate that they do not adversely impact the wild population. Nearly all parts of these animals are now traded, either as leather, meat, ornaments, and traditional Chinese medicine1,2,3,4,5,6.
The Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) aims to protect endangered species from over-exploitation by controlling international trade through a permit system that ensures the survival of species. CITES have generated three appendices, namely CITES Appendix I, II and III, which are lists of species at various levels or kinds of protection from over-exploitation. According to the International Union for Conservation of Nature (IUCN) Red List of Threatened Species, 11 out of 23 extant crocodile species are currently listed as either critically endangered or vulnerable (https://www.iucnredlist.org), and 17 species are listed in the Appendix I of CITES, i.e. commercial trade is prohibited. Captive-bred of these species from CITES registered operations and artificially propagated for commercial purposes are considered as Appendix II specimens, and their trade is permitted with a valid license. To date, the crocodilian meat products that are available in the global markets are largely manufactured from the captive farms in Madagascar, South Africa, Zambia, Zimbabwe, Thailand and Vietnam, with Crocodylus niloticus and C. siamensis being the most cultured species1. Nevertheless, hunting and illegal trading continues to harm the wild population and their associated habitats7.
Hong Kong does not have wild crocodiles nor crocodilian farms, and thus all the crocodilian meat selling in the market for various purposes are basically imported with proper permits from crocodile farms8,9,10,11,12,13,14,15. CITES in Hong Kong is implemented through the enforcement of the Animals and Plants (Protection of Endangered Species) Ordinance, Cap 187, where no person shall import, export, or possess any endangered species, or parts and derivatives of such species, except under and in accordance with a license issued by the Director of Agriculture, Fisheries and Conservation Department, Hong Kong SAR Government. According to the data retrieved from CITES, crocodilian products imported into Hong Kong from 2018 to 2021 included 4 species including Alligator mississipplensis (CITES Appendix II), C. niloticus (CITES Appendix I and II depending on the geographical locations, such as Botswana, Ethiopia, Kenya, Madagascar, Malawi, Mozambique, Namibia, South Africa, Uganda, the United Republic of Tanzania, Zambia and Zimbabwe), C. porosus (CITES Appendix I and II depending on the geographical locations, such as populations of Australia, Indonesia, Malaysia, Papua New Guinea); and C. siamensis (CITES Appendix I).
From time to time, there are news of illegal importing of crocodiles to Hong Kong. In 2013, customs officers seized 579 kg of endangered crocodile meat hidden in a truck container16. More than a decade later in 2019, there was also report of endangered Southeast Asia species of crocodile at the Bride’s Pool presumably via illegal pet trade17. Another issue goes to the authenticity of crocodile meat being sold in Hong Kong. In 2006, a survey revealed only 10% of dry crocodile meat available in Hong Kong are genuine based on 11 samples18, and the Consumer Council of Hong Kong SAR Government further revealed only 30% of 24 samples were found to be unadulterated dried crocodile meat19. Understanding the contemporary situation of crocodile meat being sold in Hong Kong, will provide necessary up-to-date baseline information to further enhance the current practice in executing CITES and law enforcement.
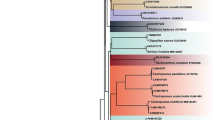
In this study, a total of 114 samples including frozen and dried meat products were collected and identified at species level using cytochrome oxidase I (COI) gene markers. The COI sequences were then subjected to phylogenetic analysis for the validation of species identity (Fig. 1a, Supplementary Figure S1). Among the 114 investigated samples, 112 of them were crocodilians including C. siamensis (75), C. porosus (30), C. niloticus (5), and A. mississipplensis (2) (Supplementary Table S1 & S2), which agrees with the CITES Trade records20 (Fig. 1b). In addition, we also found that 2 samples were Malayopython reticulatus, which is a python species under CITES Appendix II (Fig. 1b).
Methods
Sampling and storage of samples
A total of 114 samples including 15 frozen and 99 dried claimed crocodile meat were purchased in Hong Kong, including 46 of them purchased from e-commerce platform while 68 were obtained from dried food or seafood stores in different geographical areas. Each of the samples were labelled with a corresponding voucher number for this study upon receival. A small portion of samples were removed from the package for DNA analysis, while the remaining tissues were properly stored. The dried samples were stored in a sealed plastic box with desiccant packs under room temperature, while the frozen samples were sealed in zipper bags and stored at -20 °C freezer. The Voucher numbers were assigned for each sample, and the metadata could be found in Supplementary Table S1.
DNA extraction, PCR amplification of target markers and Sanger sequencing
All the equipment and consumables were autoclaved and cleaned with 75% ethanol, and disinfected under a flame prior to each sample experiment. Genomic DNA of the meat was isolated with a spin-column based extraction method using the PureLink™ Genomic DNA Mini Kit (Invitrogen, USA), following the manufacturer’s instructions. In brief, a small amount of tissue was excised from the package by a pair of disinfected scissors and placed in a 1.5 ml microcentrifuge tube. Tissue digestion buffer was then added into each tube and the tissue was homogenized thoroughly with autoclaved plastic homogenisers. The samples were then incubated in a dry bath with temperature set at 55 °C for 2–3 hours, or until the tissue completely disintegrated. The subsequent supernatant was obtained by centrifugation and subjected to ethanol precipitation, which was then passed through a spin-column to yield purified genomic DNA. A positive and negative (with no tissues) control tube was included in each extraction process.
The extracted DNA was subjected to quality and quantity control by 1% gel electrophoresis and One/OneC Microvolume UV-Vis Spectrophotometer (Thermo Scientific, NanoDrop, USA). The extracted DNA was kept in designated -20 °C freezer with cleared labels after sample preparation. The qualified genomic DNA was subjected to polymerase chain reaction (PCR) using a set of primer cocktails: (VF2_t1: 5′-TGTAAAACGACGGCCAGTCAACCAACCACAAAGACATTGGCAC-3′, FishF2_t1: 5′-TGTAAAACGACGGCCAGTCGACTAATCATAAAGATATCGGCAC-3′, FishR2_t1: 5′-CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA-3′ and FR1d_t1: CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCARAA-3′) for amplifying mitochondrial cytochrome c oxidase subunit I (COI) gene21. For sample #41 and #113, where COI gene could not be amplified with the above primers, universal primers LCO1490/HCO2198 (LCO1490: 5′-GGTCAACAAATCATAAAGATATTGG-3′, HCO2198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′)22 was used. PCR was carried out on a T100™ thermal cycler (Bio-Rad, USA) with the following parameters: an initial denaturation step at 95 °C for 3 minutes; followed by 36 amplification cycles of 30 seconds for denaturation at 95 °C, 30 seconds for primer annealing at 54 °C and 45 seconds for extension at 72 °C, and a final extension step at 72 °C for 5 minutes. The reaction mixture included PCR buffer, extracted genomic DNA sample, 2 mM dNTP, 1.5 mM MgCl2, 0.4 mM of each forward and reverse primers, and Taq DNA polymerase. A positive and negative (sample DNA is replaced by ddH2O) control tube was included in each PCR process. The amplified products were then validated by 1% agarose gel electrophoresis and sent to BGI Genomics Company Hong Kong for purification of PCR products by magnetic beads and Sanger sequencing on the platform ABI3730xl.
Species identification and Phylogenetic analyses
After obtaining the DNA sequences, the chromatogram of each sample was carefully examined. If the chromatogram shows the limit in resolution, no clear peaks, or multiple sequences/peaks, the sequence was considered as not suitable for analysis. The corresponding sample was then proceeded to re-extraction, re-amplification, and re-sequencing. If any unincorporated detection of peaks, dye blob, pull up, mobility errors, or background noise were observed in the chromatogram, the sequence was manually edited, and the edited content was recorded.
Each resultant DNA sequence was compared to the non-redundant (nr) database on National Center for Biotechnology Information (NCBI) for homology analysis. Only sequences showing a similarity match > 98% to the annotated sequence on the sequence database were assigned to that species.
For phylogenetic analysis, the DNA sequences were aligned with reference crocodilian sequences on NCBI GenBank database using software MEGA7.023. Neighbour-Joining (NJ) and Maximum-likelihood (ML) phylogenetic trees were constructed with 1,000 bootstraps in MEGA7.022 and IQ-TREE ver. 2.0.324 respectively. The trees were subsequently visualised and customized on iTOL tree25.
Data Records
The dataset has been deposited on Genbank under the BioProject accession number PRJNA105382726. An additional master file containing statistic information in this study is available on Figshare (https://doi.org/10.6084/m9.figshare.23283839)27, which consists of the following parts:
-
1.
A document of supplementary information includes Figure S1. Figure S1. Neighbor-joining phylogenetic tree of 114 crocodile samples purchased from Hong Kong.
-
2.
An excel of Supplementary Tables S1-S2. Supplementary Table S1. Voucher and product details of crocodilian meat samples Supplementary Table S2. Species identity of crocodilian meat samples, COI sequences and the approved NCBI accession numbers.
Technical Validation
Sequence validation
The chromatogram of each sequence obtained from BGI was examined base by base on the software SnapGene Viewer. Manual deletion of primer sequences at the 5’ and 3’ ends were performed for each barcoded sequence.
Species identification validation
Species identity of each sample was confirmed by phylogenetic analysis on COI marker gene. Only sequences having a similarity match > 98% to the annotated sequence on the database were assigned the identity.
Usage Notes
Not applicable
Code availability
There was no custom code used in this study.
References
Tosun, D. D. Crocodile farming and its present state in global aquaculture. J. fish. sci. 7, 43–57 (2013).
Koo, L. C. The use of food to treat and prevent disease in Chinese culture. Soc. sci. med. 18, 757–766 (1984).
Ng, T. P., Wong, M. L., Hong, C. Y., Koh, K. T. C. & Goh, L. G. The use of complementary and alternative medicine by asthma patients. Qjm 96, 747–754 (2003).
Chui, C. H. et al. Antiproliferative ability of a combination regimen of crocodile egg extract, wild radix ginseng and natural Ganoderma lucidum on acute myelogenous leukemia. Oncol. Rep. 16, 1313–1316 (2006).
Williams, V. L., Moshoeu, T. J. & Alexander, G. J. Reptiles sold as traditional medicine in Xipamanine and Xiquelene Markets (Maputo, Mozambique). S. Afr. J. Sci. 112, 1–9 (2016).
Ramdzan, S. N. et al. Perceptions of complementary/alternative medicine use and influence on evidence-based asthma medicine adherence in Malaysian children. NPJ Prim. Care Respir. Med. 29, 1–7 (2019).
Eaton, M. J. et al. Barcoding bushmeat: molecular identification of Central African and South American harvested vertebrates. Conserv. Genet. 11, 1389–1404 (2010).
Bolton, M. Crocodile farming in Papua New Guinea. Oryx 14, 365–369 (1978).
Magnusson, W. E. Economics, developing countries, and the captive propagation of crocodilians. Wildl. Soc. Bull. (1973-2006) 12, 194–197 (1984).
Mercado, V. P. Current status of the crocodile industry in the Republic of the Philippines. Nat. Mus. Pap. 14, 26–34 (2008).
Hossain, M. S., Jaman, M. F., Ahmed, M., Rahman, M. M. & Rahman, M. S. High hatching success of saltwater crocodile (Crocodylus porosus) in a commercial Crocodile Farm of Bangladesh. Univ. j. zool., Rajshahi Univ. 31, 35–38 (2012).
Platt, S. G., Monyrath, V., Sovannara, H., Kheng, L. & Rainwater, T. R. Nesting phenology and clutch characteristics of captive Siamese crocodiles (Crocodylus siamensis) in Cambodia. Zoo Biol. 31, 534–545 (2012).
Getpech Y. Crocodile conservation and captive breeding in Thailand. In Crocodiles. Proceedings of the 1st Regional Species Meeting. Crocodile Specialist Group: Darwin: Australia; 27–28. (2011).
Manolis, S. C. & Webb, G. J. Best management practices for crocodilian farming. IUCN-SSC Crocodile Specialist Group http://www.iucncsg.org/pages/Best-Management-Practices-for-Crocodilian-Farming.html (2016).
Cawthorn, D. M. & Hoffman, L. C. Controversial cuisine: A global account of the demand, supply and acceptance of “unconventional” and “exotic” meats. Meat Sci. 120, 19–36 (2016).
France-Presse, A. HK$8m haul of smuggled seahorse and crocodile meat seized. https://www.scmp.com/news/hong-kong/article/1122179/hk8m-haul-smuggled-seahorse-and-crocodile-meat-seized?campaign=1122179&module=perpetual_scroll_0&pgtype=article (2013).
Mok, D. Small crocodile discovered in Hong Kong thought to have been released from illegal pet trade. https://www.scmp.com/news/hong-kong/society/article/2181780/small-crocodile-discovered-hong-kong-thought-have-been (2019).
Ng, T. Only 10% of crocodile meat genuine: Survey. https://www.chinadaily.com.cn/hkedition/2006-12/02/content_748786.htm (2006).
Customs and Excise Department Hong Kong Special Administrative Region. Customs and AFCD step up checks against illicit crocodile meat. https://www.info.gov.hk/gia/general/200711/14/P200711140140.htm (2007).
Convention on International Trade in Endangered Species of Wild Fauna and Flora. Version 2022.1. CITES Trade Database. https://trade.cites.org/ (2022).
Ivanova, N. V., Zemlak, T. S., Hanner, R. H. & Hebert, P. D. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 7, 544–548 (2007).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 3, 294–299 (1994).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, 293–296 (2021).
So, W. L. et al. Cytochrome oxidase I DNA barcode database of crocodilians meat selling in Hong Kong. Genbank https://identifiers.org/ncbi/bioproject:PRJNA1053827 (2023).
So, W. L. et al. Cytochrome oxidase I DNA barcode database of crocodilians meat selling in Hong Kong. figshare https://doi.org/10.6084/m9.figshare.23283839 (2023).
Acknowledgements
The authors would like to thank Beth Chan, Stacey Tsang and Candy Au for their assistance in sample collection. This work was supported by Innovation Technology Fund of Innovation Technology Commission: Funding Support to State Key Laboratory of Agrobiotechnology, Li Dak Sum Yip Yio Chin R & D Centre for Chinese Medicine and Direct Grant (4053547) of The Chinese University of Hong Kong.
Author information
Authors and Affiliations
Contributions
J.H.L.H conceived and supervised the study; PCS contributed the discussion of project at different stages; W.L.S., T.K.C., M.T.W.S., A.M.Y.L., S.T.C.L. and H.Y.Y. carried out sample collection; W.L.S. and I.H.T.L. optimized the P.C.R. protocol for COI amplification; W.L.S., T.K.C., I.H.T.L., M.T.W.S., A.M.Y.L., S.T.C.L. and W.C. performed COI amplification of the samples; W.L.S. and T.K.C. performed data curation on the COI sequences and phylogenetic analysis; J.H.L.H., W.L.S. and T.K.C. wrote the initial manuscript; all authors revised and contributed to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
So, W.L., Chong, T.K., Lee, I.H.T. et al. Cytochrome oxidase I DNA barcodes of crocodilians meat selling in Hong Kong. Sci Data 11, 46 (2024). https://doi.org/10.1038/s41597-023-02889-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02889-3