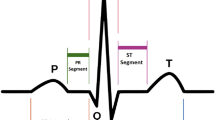
Abstract
Machine learning (ML) methods for the analysis of electrocardiography (ECG) data are gaining importance, substantially supported by the release of large public datasets. However, these current datasets miss important derived descriptors such as ECG features that have been devised in the past hundred years and still form the basis of most automatic ECG analysis algorithms and are critical for cardiologists’ decision processes. ECG features are available from sophisticated commercial software but are not accessible to the general public. To alleviate this issue, we add ECG features from two leading commercial algorithms and an open-source implementation supplemented by a set of automatic diagnostic statements from a commercial ECG analysis software in preprocessed format. This allows the comparison of ML models trained on clinically versus automatically generated label sets. We provide an extensive technical validation of features and diagnostic statements for ML applications. We believe this release crucially enhances the usability of the PTB-XL dataset as a reference dataset for ML methods in the context of ECG data.
Similar content being viewed by others
Background & Summary
Cardiovascular diseases continue to be one of the largest burdens for the population worldwide1. Due to its simplicity, non-invasive nature, widespread use and diagnostic value, the electrocardiogram (ECG) is one of the primary tools for the first assessment. However, it requires the analysis of a huge amount of time-series ECG-data. Therefore automatic analysis tools have become standard. The recent developments in machine learning/AI have demonstrated its potential in this direction2,3,4,5. Large freely available ECG databases6,7 are crucial for the development and benchmarking of AI algorithms for automatic classification. Consequently, they have been the basis of recent competitions and challenges8,9. Even though end-to-end trained deep learning models are on the rise, handcrafted features continue to play an important role in ECG analysis: They involve decades of engineering and encode valuable domain knowledge used for clinical diagnosis. Most of the ECG features are inherently interpretable for domain experts and represent a very efficient way to perform patient stratification. Furthermore, their availability allows investigating the extent to which deep models align with these features (concepts), or to directly compare to algorithms trained on manually extracted features, or potentially devise more robust algorithms relying on both. ECG features also represent a substantial reduction of the high-dimensional raw ECG time series and enable therefore comprehensive comparisons between different clinical ECG data bases. They may also be used for clinical validation of synthetic data sets stemming from simulations based on digital twins of individuals10,11,12 or virtual cohorts of realistic models13,14.
Electrocardiography is a unique domain with a long history of such handcrafted features and commercially available software packages that allow extracting them in a reliable way. However, as a practical obstacle, high-quality ECG features from commercial software are not accessible to the broader ECG research community. Furthermore, their comparative quality, also in comparison to available open-source toolkits, when applied to a comprehensive ECG dataset, is unknown. With this dataset, PTB-XL+, see Fig. 1 for a schematic overview, we aim to mitigate these shortcomings by releasing ECG features from two commercial and one open-source feature extractors for the entire PTB-XL6,15,16 dataset. Since its publication, the PTB-XL dataset quickly developed into one of the largest and most widely used publicly available 12-lead clinical ECG datasets covering a broad set of conditions with diverse signal quality and hence representative of real-world ECG data. By releasing accompanying ECG features, we hope to further strengthen the role of the PTB-XL dataset as a reference dataset for the development and evaluation of automatic ECG analysis algorithms. To increase the interoperability of the features from different ECG feature providers, we mapped features to a common naming scheme (including mapping to SNOMED CT17/LOINC18 ontologies) that allows using the corresponding feature sets as interchangeably as possible. Further metadata such as median beats or fiducial points further enhance the value of the dataset. Finally, the PTB-XL + dataset includes automatic diagnostic statements as provided by one of the most widely used commercial ECG algorithms, the Marquette 12SL (GE Healthcare, WI) algorithm. To also increase the interoperability in this respect, we provide mappings for these statements as well as for the original PTB-XL ECG statements to SNOMED CT statements as a common ontology and advocate this as a useful procedure to increase the interoperability of datasets that were labeled according to different ontologies. This has several important implications: First, mismatches between the 12SL statements and the original labels can be used to assess the label quality of the PTB-XL dataset itself. Second, it allows to directly compare the performance of models trained on the original PTB-XL labels provided by cardiologists to the predictions of the 12SL. The dataset was compiled with direct applicability for machine learning applications in mind and includes an extensive technical validation based on publicly available source code19, which can be used as a starting point for own analyses.
Methods
Considered algorithms
Before we describe the steps that were followed to create the PTB-XL + dataset, we give a brief overview of the different methodologies followed by the included ECG analysis algorithms.
University of Glasgow ECG analysis program (Uni-G) and Marquette 12SL (12SL)
The University of Glasgow ECG Analysis Program and Marquette 12SL (GE Healthcare, WI) are two commercial, state-of-the-art ECG analysis packages that are distributed in millions of ECG devices world-wide. Both follow a similar approach: In a first step, a median/template beat is calculated. In a second step, ECG features are extracted from this median beat (in addition to some features such as heart rate that are collected from the full ECG) and in a third step diagnostic statements are predicted from these features, see20 for details on the Uni-G approach and21 for details on the 12SL algorithm. Due to usage restrictions, the PTB-XL + dataset includes automatic diagnostic statements only from 12SL but the full feature sets from both algorithms. Both feature extraction algorithms are closed source and only accessible on special devices or after purchase. The decision rules followed by the 12SL algorithm are available from the Physician’s Manual21.
ECGDeli
ECGDeli is an open-source ECG delineation toolkit developed within the Institute of Biomedical Engineering at the Karlsruhe Institute of Technology, Germany. The feature extraction follows a different approach compared to the two approaches discussed before. It builds on the fiducial points obtained from the open-source ECGDeli22,23 software. ECG features are computed separately for each available beat. Even though the package is publicly available, its execution relies on MATLAB as proprietary software, which limits the range of potential users. In the dataset, we report only the median and the (0.25,0.75)-interquartile range across beats, which allows to assess the variability of features across different beats, as well as the total count of beats that were considered for each respective feature. In addition to amplitude and interval features, the dataset includes a number of morphological features.
Data processing
The records from the PTB-XL6,15,16 dataset were converted to appropriate input formats and processed by the Uni-G, the 12SL and the ECGDeli algorithms. For 12SL, all ECGs were imported into a custom-built MUSE Cardiology Information System (GE Healthcare, Wauwatosa, WI, USA) and upon import they were reanalyzed with the latest version of 12SL (v.243). Automatic diagnostic statements were directly exported from the GE software rather than re-implemented based on the reference manual. Uni-G features were exported from a custom-built version of the Glasgow software (R30.4.2). ECGDeli features were extracted from the publicly available version 1.1 of the software.
The output features were harmonized into a unified naming scheme and converted into compatible units (using mV for amplitudes and ms for intervals as base units). However, the output features still maintain their original form as produced by each respective algorithm. The ECG features for each of the three feature sets were converted into a tabular format with a single row per ECG record and a column for each ECG feature. Additional features that were provided by the different algorithms such as fiducial points or median beats were converted to appropriate output formats and are also distributed as part of this dataset. Finally, the automatic diagnostic statements provided by 12SL were converted to a format that makes them directly applicable for training ML algorithms. Additionally, we devised a mapping both from the original PTB-XL statements and of the 12SL automatic diagnostic statements to SNOMED CT17 and applied them to the original label sets. The details are described in the following section.
Data Records
Data released as part of this dataset
This section describes the components of the released data repository, which is hosted by PhysioNet16,24. For the three feature sets, Uni-G, 12SL and ECGDeli, we provide the following collection of features:
-
The Uni-G feature set includes ECG features and median beats from which most of the features were extracted.
-
The 12SL feature set includes ECG features and median beats from which most of the features were extracted. In addition, automatic diagnostic statements provided by the 12SL algorithm are included.
-
The ECGDeli feature set includes median feature values across beats, corresponding (0.25,0.75)-interquartile ranges across beats and counts across beats along with the fiducial points along the rhythm strip from which the features were extracted.
Generally, we refer to ECG features as a collection of amplitudes and intervals (global as well as lead-specific), onsets of ECG segments (global as well as lead-specific), areas and similar features. The precise composition of features only depends on the availability of features in the respective algorithms. The data is organized as follows:
-
ECG features (Uni-G, 12SL, ECGDeli): For each of the three feature providers, we provide feature tables as csv-files with the PTB-XL ECG identifier as key (unig_features.csv, 12sl_features.csv, ecgdeli_features.csv).
The columns follow a unified naming scheme (including mapping to SNOMED CT or LOINC where available), which allows using the three feature sets interchangeably provided the corresponding features are available in multiple datasets. A corresponding summary table (feature_description.csv) lists the available ECG features along with a short description and units of measurement. For all three feature sets, the ECG features include durations, amplitudes and on/off-sets of segments. Uni-G and 12SL include in addition area features and Uni-G also has vectorcardiographic measurements (calculated from I, aVF and V2 as quasi-orthogonal leads).
-
Fiducial points (ECGDeli): We provide fiducial points in PhysioNet’s wfdb annotation format25, both lead-specific and consensus annotations across all leads. The annotations are organized in subfolders following the structure of the PTB-XL dataset with filenames relating to the PTB-XL ECG identifier.
-
Median beats (Uni-G, 12SL): We provide median beats in PhysioNet’s wfdb signal format25 that can be processed analogously to the samples in the original PTB-XL dataset. As the fiducial points, the median beats are organized in subfolders following the structure of the PTB-XL dataset with filenames relating to the PTB-XL ECG identifier.
-
Automatic diagnostic statements (12SL): We provide the automatic diagnostic statements as a csv-file (12sl_statements.csv) indexed by PTB-XL ECG identifier, where we provide both the original ECG statements assigned by the 12SL-algorithm and the statements after mapping to SNOMED CT. For every statement, we also include all parent nodes and in this way propagate the label upwards in the SNOMED CT ontology until we reach the root node of the label tree. For the user’s convenience, we provide a similar file for the statements assigned in the PTB-XL dataset after application of a similar mapping (ptbxl_statements.csv). We also release the tables underlying the mappings to SNOMED CT codes (12slv23ToSNOMED.csv and ptbxlMapToSNOMED.csv). In addition, we provide the code to apply a potentially modified mapping at a later point in time (apply_snomed_mapping.py). Finally, we provide a human-readable description of the used SNOMED CT concept identifiers in SNOMED_description.csv. In this table, we also mark identifiers as informative if they neither perfectly correlate with another label nor are too unspecific such as “Finding of body region”. We propose to use only this reduced set for the training and evaluation of ML algorithms, see below. Finally, we stress that we provide for the first time a way to convert automatic 12SL’s diagnostic statements into a machine-readable format that can be directly used to train machine learning models. A complete description of the available label sets in ptbxl_statements.csv and 12sl_statements.csv is given in Table 1.
Table 1 Description of the provided label sets.
Descriptive statistics
With the exception of a small number of samples that could not be processed by particular algorithms, the feature sets cover the full PTB-XL dataset6,15,16, i.e., up to 21799 records from 18869 patients.
We summarize the available features in each of the three feature sets in terms of two figures: Fig. 2 shows the fraction of samples in the dataset for which a particular feature is present for lead-dependent features. Figure 3 shows the analogous plot for global, i.e., lead-independent, features. The features are labeled according to their abbreviations. The corresponding descriptions can be found in feature_description.csv. Here and in the following, we use X as a placeholder for the leads, i.e., X can take values from the set {I,II,III,aVR,aVL,aVF,V1,V2,V3,V4,V5,V6}. The figures visually demonstrate that there are 13 features (counting lead-specific features only once) that are present in all three feature sets and 39 features that are present in at least two feature sets, which allows for a large number of cross-comparisons for consistency checks, see Technical Validation.
Lead- and segment-specific features as provided in the different feature sets. Color-coding corresponds to the fraction of samples for which values are present whereas black corresponds to values present for all samples. We report average statistics across leads X. The used acronyms are described in feature_description.csv.
Global (sample-wise) ECG features as provided within the different feature sets. Color-coding as in Fig. 2.
In Fig. 4, we visualize the label distribution according to the automatic 12SL diagnostic statements (column statements_ext in 12sl_statements.csv). The acronyms used in Fig. 4 are described in 12slv23ToSNOMED.csv. The distribution of statements over the full PTB-XL dataset covers 117 statements and therefore provides a rich source of information - in particular in comparison to the original labels provided within the PTB-XL dataset. In the Technical Validation Section, we provide a first quantitative comparison between both label sets based on SNOMED CT terms as common vocabulary.
Technical Validation
The technical validation for the PTB-XL + dataset covers three different aspects. First, we assess the consistency of the different ECG features sets by comparing output distributions as well as comparisons on the level of individual samples. Second, we use the performance level of Random Forest classifiers trained on different feature sets on standard ECG prediction tasks26 as an indirect measure for the discriminative power of the different feature sets. Third, we investigate the correlation between the automatic 12SL ECG statements and the ECG statements provided within the PTB-XL dataset by cardiologists. Finally, we assess the performance of state-of-the-art deep learning models26 trained on the original PTB-XL labels and evaluated on 12SL-labels and vice-versa.
ECG features: Consistency between different feature sets
In Fig. 5, we compare the different feature sets based on sample-wise Pearson correlation coefficients of those ECG features that are each contained in two of the feature sets under consideration, where we restrict ourselves for simplicity to continuous features. At this point, it is worth stressing again that this is to the best of our knowledge the first publicly available set which allows for a quantitative comparison between ECG features, in particular including those from two leading commercial providers. To simplify the presentation, we compute lead-specific correlation coefficients but only report average correlation coefficients across all 12 leads for lead-specific ECG features.
The left panel in Fig. 5 compares the two commercial algorithms 12SL and Uni-G and shows very good agreement among all common global features. Also most of the lead-specific standard amplitude and interval features show a good agreement with correlation coefficients above 0.9. The least agreement show features related to R’ and S’ (i.e., a second positive/negative wave after the R/S-wave), which are potentially more difficult to detect, and certainly are features for which some deviations might potentially also be due to different definitions. The center and the right panel of Fig. 5 show the comparison to the ECGDeli features. Again, one observes good agreement for the global features and many interval features, reasonable agreement for T and R amplitudes and least agreement for S, P and Q amplitudes.
ECG features: Assessing the discriminative power of different feature sets
Following the evaluation protocol established in26, we train Random Forest classifiers on the different feature sets to assess their discriminative power, when used as input features for comprehensive ECG classification tasks. As the three feature sets are composed differently, we also consider training on feature subsets that two feature extraction algorithms have in common, which in principle allows for a direct comparison of the discriminative power of features extracted by different algorithms. We assess the performance on the set of seven multi-label prediction tasks put forward in26 and report the macro-average (across labels) of the respective areas under the receiver operating curves, henceforth referred to as macro AUC, on the PTB-XL test set. For reference, we also report the published performance scores of the xresnet1d101, a convolutional neural network operating on the raw waveform data26.
First of all, the results compiled in Table 2 reveal that all three feature sets are highly predictive, reaching mean macro AUC values of 0.889, 0.871 and 0.879 for Uni-G, 12SL and ECGDeli, respectively. On their entire respective feature sets (denoted as “full”), the Uni-G features are most discriminative. Interestingly, while the feature-based approaches fail to reach the CNN performance on comprehensive classification tasks (such as “all”), ECGDeli outperforms the CNN baseline in the rhythm category. This is in slight tension to the results from27, where the authors found that feature-based and raw-signal-based approaches lead to comparable performance across several diagnostic categories. We also provide results for models trained on the set of features shared by two feature sets (line 5–10 Table 2), which allows for a more direct comparison between the two feature sets. The results reveal that Glasgow and 12SL features have comparable quality but both are superior to the ECGDeli features (leaving aside the rhythm category).
Automatic diagnostic statements: Agreement between 12SL and original PTB-XL labels
Descriptive analysis
We study the overlap between cardiologists’ annotations provided as part of the PTB-XL dataset and the automatic 12SL diagnostic statements. We use the provided mapping to SNOMED CT terms (12slv23ToSNOMED.csv and ptbxlMapToSNOMED.csv as described in Data Records) to obtain compatible label sets. We consider the set of SNOMED CT terms that are present in both label sets while only keeping informative terms, see the description in the Section Data Records. This leaves us with 94 SNOMED CT terms that can be directly compared across both label sets.
First, we visually compare the label distributions in Fig. 6, where we show the label occurrence for the common SNOMED identifiers in the 12SL vs. the original PTB-XL label set after mapping to SNOMED CT (ordered by occurrence in PTB-XL), which shows a rough overlap in terms of label distributions.
To investigate this in more detail on the per-sample level, we compute the Matthews Correlation Coefficient (MCC)28 between the binarized scores obtained from selecting the non-zero values of the continuous scores. The result of this analysis is listed in Tables 3, 4. The median of the correlation across all terms is 0.45. In particular, we find good agreement for atrial fibrillation, complete bundle branch blocks, sinus tachycardia (all with MCC above 0.8), which aligns with cardiologists’ knowledge as these conditions are rather clearly identifiable from a 12-lead ECG. On the other hand, there is also a range of statements, including myocardial infarctions with specific localization, with essentially no agreement. In any case, these findings provide valuable hints for future investigations of the label quality of the PTB-XL annotations and the 12SL statements.
Model training
To assess the quality of the 12SL labels, we conducted a series of model training experiments, the results of which are shown in Table 5. First, we used the original 12SL labels and trained an xresnet1d50 classification model, which is a modern convolutional neural network, which was found to perform well on PTB-XL across various prediction tasks26. We used the first eight stratified folds (training set) from PTB-XL for training, the ninth fold (validation set) for model selection via early stopping and report the macro AUC on the tenth fold (test set). Further, we discarded labels, that do not occur at least once in all of the before-mentioned splits, leaving us with 109 labels. The xresnet1d50 reaches a macro AUC of 0.956 demonstrating that the full input signals are very discriminative for the prediction of the 12SL labels.
To investigate the comparability of the 12SL labels with the original PTB-XL labels, we use the provided mapping to SNOMED CT labels (up-propagated in the label hierarchy) that was described above. After removing uninformative SNOMED CT labels close to the SNOMED CT root node (and SNOMED CT labels that show perfect correlation to other labels on both datasets) and discarding all those SNOMED CT labels that did not appear in each split, we reduced the label set to 168 SNOMED CT codes. Following the same procedure as described above, we report again the macro AUC on the test set in Table 5. In addition, we also report the results of cross-evaluation of models trained on the 12SL SNOMED CT labels and evaluated on the PTB-XL SNOMED CT labels and vice versa. Models trained and evaluated on labels stemming from the same original source show a high predictive performance (0.939 vs. 0.912 for 12SL vs. original PTB-XL labels). The cross-evaluation results are in both cases considerably weaker but very similar (0.867 in both cases). The precise understanding of this discrepancy is an interesting direction for future research.
Usage Notes
We structure the usage instructions according to the different components provided in the dataset:
-
ECG-features are provided as csv-files, which can be read by any standard software.
-
Median beats and fiducial points are provided in PhysioNet’s wfdb format25, which can be conveniently processed using toolkits in C, MATLAB and Python.
-
Automatic diagnostic statements are again provided as csv-files for easy accessibility.
For the user’s convenience, we release the classifier training code19 for the experiments presented in the Technical Validation Section. This should provide a good starting point for own explorations of the dataset. We believe that the availability of the additional features provided will significantly enhance the usability of the PTB-XL dataset due to the ability to train ML models on features and combinations of raw data and features, to look into the quality of features from different feature sets and into the strengths and weaknesses of diagnostic statements provided by state-of-the-art ECG analysis software.
Code availability
The ECG features directly correspond to the outputs of the respective algorithms up to minor harmonization. We provide code to apply the predefined SNOMED CT mappings to the labels in the dataset (apply_snomed_mapping.py released as part of the data repository24). Links to code samples facilitating the usage of the dataset are described under Usage Notes and are released in a dedicated code repository19.
References
Dagenais, G. R. et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. The Lancet (2019).
Hannun, A. Y. et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nature Medicine 25, 65–69 (2019).
Attia, Z. I. et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nature Medicine 25, 70–74 (2019).
Lima, E. M. et al. Deep neural network-estimated electrocardiographic age as a mortality predictor. Nature Communications 12 (2021).
Verbrugge, F. H. et al. Detection of left atrial myopathy using artificial intelligence–enabled electrocardiography. Circulation: Heart Failure 15 (2022).
Wagner, P. et al. PTB-XL, a large publicly available electrocardiography dataset. Scientific Data 7, 154 (2020).
Zheng, J. et al. A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients. Scientific Data 7, 1–8 (2020).
Alday, E. A. P. et al. Classification of 12-lead ECGs: the Physionet/Computing in Cardiology Challenge 2020. Physiological Measurement 41, 124003 (2020).
Reyna, M. A. et al. Will two do? varying dimensions in electrocardiography: the PhysioNet/Computing in Cardiology Challenge 2021. 2021 Computing in Cardiology (CinC) 48, 1–4 (2021).
Dossel, O., Luongo, G., Nagel, C. & Loewe, A. Computer modeling of the heart for ECG interpretation— a review. Hearts 2, 350–368 (2021).
Gillette, K. et al. A framework for the generation of digital twins of cardiac electrophysiology from clinical 12-leads ECGs. Medical Image Analysis 71, 102080 (2021).
Coorey, G. et al. The health digital twin to tackle cardiovascular disease—a review of an emerging interdisciplinary field. NPJ Digital Medicine 5, 1–12 (2022).
Niederer, S. et al. Creation and application of virtual patient cohorts of heart models. Philosophical Transactions of the Royal Society A 378, 20190558 (2020).
Gillette, K. et al. Medalcare-xl: 16,900 healthy and pathological 12 lead ecgs obtained through electrophysiological simulations. Preprint at https://arXiv.org/abs/2211.15997 (2022).
Wagner, P., Strodthoff, N., Bousseljot, R.-D., Samek, W. & Schaeffter, T. PTBXL, a large publicly available electrocardiography dataset. PhysioNet. https://doi.org/10.13026/qgmg-0d46 (2020).
Goldberger, A. L. et al. PhysioBank, PhysioToolkit, and PhysioNet. Circulation 101, e215–e220 (2000).
National Library of Medicine. Health Information Technology and Health Data Standards at NLM. National Library of Medicine. https://www.nlm.nih.gov/healthit/snomedct/index.html (2022).
Regenstrief Institute. Logic Observation Identifier Names and Codes. Regenstrief Institute. https://loinc.org/ (2022).
Mehari, T. & Strodthoff, N. Source code for: PTB-XL+, a comprehensive electrocardiographic feature dataset, version 1.0. Zenodo. https://doi.org/10.5281/zenodo.7817567 (2022).
Macfarlane, P., Devine, B. & Clark, E. The university of Glasgow (Uni-G) ECG analysis program. Computers in Cardiology 2005, 451–454 (2005).
GE Healthcare. Marquette 12SL ECG Analysis Program: Physician’s Guide, version 2056246-002c. General Electric Company (2019).
Pilia, N. et al. ECGdeli - an open source ECG delineation toolbox for MATLAB. SoftwareX 13, 100639 (2021).
Pilia, N. et al. Source code for: ECGdeli - ECG delineation algorithms, version 1.1. Zenodo. https://doi.org/10.5281/zenodo.7191379 (2020).
Strodthoff, N., Mehari, T. & Schaeffter, T. PTB-XL+, a comprehensive electrocardiographic feature dataset. PhysioNet. https://doi.org/10.13026/nqsf-pc74 (2023).
Moody, G., Pollard, T. & Moody, B. Wfdb software package. PhysioNet. https://physionet.org/content/wfdb/10.7.0/ (2022).
Strodthoff, N., Wagner, P., Schaeffter, T. & Samek, W. Deep learning for ECG analysis: Benchmarks and insights from PTB-XL. IEEE Journal of Biomedical and Health Informatics 25, 1519–1528 (2021).
Zvuloni, E., Read, J., Ribeiro, A. H., Ribeiro, A. L. P. & Behar, J. A. On merging feature engineering and deep learning for diagnosis, risk prediction and age estimation based on the 12-lead ecg. IEEE Transactions on Biomedical Engineering 1–10 (2023).
Matthews, B. Comparison of the predicted and observed secondary structure of t4 phage lysozyme. Biochimica et Biophysica Acta (BBA) - Protein Structure 405, 442–451 (1975).
ISO Central Secretary. Health informatics – Standard communication protocol – Part 91064: Computer-assisted electrocardiography. Standard ISO 11073-91064:2009, International Organization for Standardization, Geneva, CH (2009).
Acknowledgements
This work was supported by the EMPIR project 18HLT07 MedalCare. The EMPIR initiative is cofunded by the European Union’s Horizon 2020 research and innovation program and the EMPIR Participating States.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
N.S. conceived the creation of the dataset. C.N. provided ECGDeli features. A.S. and P.A. provided Uni-G features. J.K. and C.G. provided 12SL features. N.S. devised mappings to SNOMED CT. N.S. and T.M. converted/harmonized the data. T.M. performed the descriptive analysis and technical validation. N.S. drafted the first version of the manuscript. All authors discussed the results and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Strodthoff, N., Mehari, T., Nagel, C. et al. PTB-XL+, a comprehensive electrocardiographic feature dataset. Sci Data 10, 279 (2023). https://doi.org/10.1038/s41597-023-02153-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02153-8
This article is cited by
-
A hybrid ResNet-ViT approach to bridge the global and local features for myocardial infarction detection
Scientific Reports (2024)
-
Heptagonal Reinforcement Learning (HRL): a novel algorithm for early prevention of non-sinus cardiac arrhythmia
Journal of Ambient Intelligence and Humanized Computing (2024)
-
MedalCare-XL: 16,900 healthy and pathological synthetic 12 lead ECGs from electrophysiological simulations
Scientific Data (2023)